[Retracted] Identification of Apple Disease Grades Based on the Attention Mechanism of Lesion Location and Improved Data Enhancement Method
Abstract
The output and quality of apples were greatly threatened by plant diseases. Identifying the types and grades of diseases in time was helpful to the management of diseases. When the disease occurs on a leaf, there was little change in the leaf except for the affected area. The traditional attention mechanism changed the weight of the network to all pixels in the image, which affected the ability of the network to extract the features of the lesion area. And the traditional method of data enhancement was easy to cause local similarity and local discontinuity of all sample features in the same disease grade. In this paper, the improved metric matrix for kernel regression (IMMKR) was used to reduce the influence of local similarity and local discontinuity of all sample features in the same class. Then, a new attention mechanism by fusing the lesion location based on visual features was proposed, and the attention of the model to the lesion area was strengthened. The experiments were carried out on three different diseases of apple named black rot, scab, and rust. The accuracy rate and recall rate of the new method on the combined dataset of PlantVillage and PlantDoc were 91.55% and 92.06%, respectively, which was superior to existing methods. This algorithm has important reference significance for the identification and promotion of crop diseases.
1. Introduction
Plant disease is one of the most common factors restricting the yield and quality of crops and has brought a great threat to agriculture [1]. The traditional disease diagnosis method is that experts observe the characteristics of diseases with naked eyes. For experienced pathologists and agronomists, it is difficult to identify diseases and their severity in time, because there is a large number of arable land crops [2]. Moreover, the diagnostic results of experts with different botanical attainments are also very different. In addition, pesticides remain the main means to protect crops from diseases. However, in most cases, regardless of the severity of the disease, crops suffering from the same disease are given the same amount of pesticides [3]. Therefore, identifying the types and severity of diseases in time is helpful to the management of diseases [4].
With the development of computer vision technology, pattern recognition has been widely used in plant disease diagnosis [5]. The traditional plant disease diagnosis methods include features extraction and analysis [6]. First, a single threshold or multi-threshold was used to obtain the lesion. Then, different methods were used to analyze the features of the lesion area. Principal component analysis, fuzzy logic, and other tools have been used to construct plant disease classification models [7]. And the fungal colonies were classified by genetic algorithm [8]. In 2004, Support Vector Machine, K-nearest neighbor algorithm, and decision tree were also widely used to solve the problem of disease classification [9]. In 2008, the chromaticity moment analysis was applied to plant disease identification [10]. In recent years, with the development of computer technology, deep learning methods have become popular because they can automatically extract features instead of manually extracting them [11]. The convolutional neural network was widely considered a reliable method for image classification, object detection, semantic segmentation, and other computer vision tasks. For the classification of plant disease images, some research has been put forward, and good results have been achieved [12]. And deep learning is widely used in disease classification, such as in cassava [13], tomato [14], and apple [15]. In addition, deep learning has gradually been applied to the detection of disease severity [16]. In 2018, Namita Sengar et al. took RLT as the disease grading standard of cherry powdery mildew [17]. Fang Tao et al. calculated the ratio of the number of pixels in the diseased area to the number of pixels in the diseased leaf, put it into Resnet-50 as the classification threshold of disease grade, and classified 10 diseases of 8 plants [18].
Deep learning gives equal weight to each pixel in the image, and the key pixels cannot be highlighted. To improve the performance of the network, attention mechanisms were introduced into deep learning. Gamaleldin F et al. proposed the saccader attention model, which narrows the performance gap with ImageNet and reduces the demanding standard of training data [19]. Xie et al. had comprehensive depth characteristics and local characteristics to build a composite insulator hydrophobicity grade discrimination model [20]. Ying et al. used the fusion of image and prior knowledge of clinical diagnosis to improve the classification performance of chronic obstructive pulmonary disease severity [21]. Wang et al. proposed a crop disease recognition model based on image and text dual-modal joint representation learning.
The traditional attention mechanism changes the weight of the network to all pixels in the image, which affects the ability of the network to extract the features of the lesion area. Zhang et al. said that 80% to 90% of crop diseases would appear on the leaves [22]. To apple scab, black rot, and rust [23–28], the image of apple leaves is simple. At the time of onset, except for the diseased area, the leaf area hardly changes. Moreover, traditional data enhancement methods are easy to cause local similarity and local discontinuity of all sample features in the same disease grade. Then, by fusing the lesion location based on visual features, the attention of the model to the lesion area was strengthened. Therefore, how to make use of the characteristics of diseases to improve the network’s attention to the lesion area is the focus of this paper. Based on a convolutional neural network, this paper proposes a new model of disease severity recognition which integrates visual features and neural networks.
2. Methodology
The purpose of this study is to put forward a new disease grade identification model. The proposed method includes two main phases. In the first stage, an improved data enhancement method was used. In the second stage, a new attention mechanism was used. The details of each process are given below.
2.1. Improved Data Enhancement Method
When CNN is applied to the classification of plant diseases, to reduce the influence of too few images on classification results, several data enhancement methods, such as flipping images and rotating images, are introduced. Traditional data enhancement makes the number of pictures in different categories equal. When a leaf is diseased, the change of the spots will change with time. Under each category, if only the number of samples in each grade was considered, it was easy to cause local similarity and local discontinuity of all samples in each disease grade. Because the disease severity of samples may be too concentrated in the same growth stage, there may be data imbalance in the same category.
Metric learning for kernel regression is a supervised dimension reduction algorithm. In this paper, the metric learning of kernel regression is to fit the measurement matrix through the characteristics of the central samples and the adjoint labels. And the measurement matrix is used to make the feature vectors transformed from all sample features more distinguishable. In this paper, the center samples were selected manually. 100 samples are selected from each category, totaling 1,100. Based on the metric matrix, the distance between the center sample and other samples is calculated, and five intervals are divided according to the average distance. Then, traditional data enhancement methods are used to make the number of samples in each interval equal, to reduce the local similarity and local discontinuity of all sample features in the same disease grade.
C represents different disease grades. N is the number of center samples. L and k represent different parameters, respectively.
In this study, 32 parameters were selected to measure disease grades label more accurately, which are from color, texture, and shape of lesions. The color features include mean value, variance, and kurtosis of RGB value of the lesion. Texture characteristic parameters include contrast, entropy, angular second moment, and inverse differential moment of the gray level co-occurrence matrix in four directions of the lesion image. They reflect the depth of the texture of the image, including the randomness of the amount of information, the uniformity of the image gray distribution, the measurement of the texture thickness, and the clarity and regularity of the texture, respectively. Shape features include the seven invariant moments of Hu moment obtained by using the second-order and third-order central moments of the image, which reflects the invariance of the translation, expansion, and rotation of the shape parameters.
In Equation (5), m ∈ {1, 2, 3, 4, 5}. If the number of samples exceeds 300, the excess samples are randomly rounded off. If the number is less than 300, data enhancement is used to increase the number of samples to 300.
2.2. Improved Attention Mechanism
In the previous studies, many attention mechanisms were put forward. In the field of images classification, attention mechanism in the convolutional neural network includes Convolution Block Attention Module (CBAM) [29], SE block [30], and their mixture. There are also many attention mechanisms in target detection, such as Single Shot MultiBox Detector (SSD) [31] and Feature Pyramid Networks (FPN) [32]. The traditional attention mechanism changes the weight of the network to all pixels in the image, which affects the ability of the network to extract the features of the lesion area. In this paper, a new attention mechanism is proposed by integrating the acquired information of the lesion area with global information. This method can increase the attention of the network to the lesion area, while the influence of other areas on the attention of the network to the lesion area can be ignored. The details were given below. The proposed attention mechanism includes two main phases: (a) acquisition of lesion location; (b) fusion of lesion location features. The details of each process are given below.
2.2.1. Acquisition of Lesion Area
pi is the proportion of each P(x). When δ2(t) is the smallest, t is denoted as t0. The location of the lesion is the location of every pixel of the image belonging to [fI(x)min, t0].
2.2.2. Fusion of Lesion Area Features
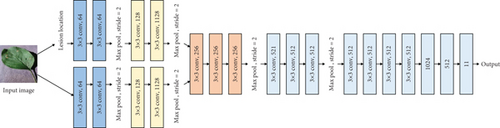
After obtaining the location of the lesion, to compare the influence of fusion depth on the classification effect, different fusion structures were explored. The purpose of this study is to put forward a new disease severity identification model by fusing the features of lesion area based on visual features and CNN. In recent years, some popular deep learning models, such as AlexNet, Inception, VGGNet, and ResNet, have been used to identify plant diseases. In these modes, VGG is a relatively simple network. Therefore, VGG 16 is selected to highlight the advantages of improvement.
The original VGG16 is shown in Figure 1(a). Figures 1(b)–1(e) show four fusion structures that increase the information input of the sample lesion location on the original network, which are named C0, C1, C2, and C4. These fusion structures only replace the convolution layer corresponding to the original network and do not change the subsequent structure of the original network.
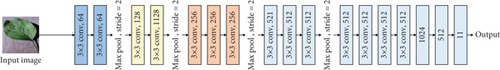
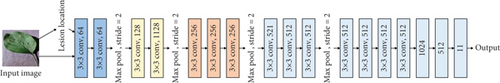
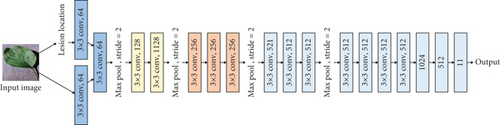
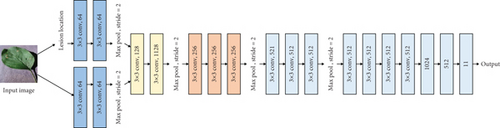
M is the number of categories. N is the number of samples. If the sample I belong to category c, yic = 1; otherwise, yic = 0, pic is the prediction probability that sample I belongs to category c.
3. Results
3.1. Experiment Setup
The experiment was carried out on a computer equipped with an NVIDIA GeForce RTX 2060Ti GPU. The CPU is Intel (R) Core (TM) i7-9700 CPU and runs under a 64-bit Windows 10 operating system. The programming language version is Python 3.6, the deep learning framework is TensorFlow 2.4.0, and the number of iterations is 100. The learning rate is 0. 001.
3.2. Data Acquisition
The dataset used in the experiment cones from PlantVillage and PlantDoc. The dataset contained three diseases of apple leaves named black rot, scab, and rust. The number of samples for each disease grade is 1500. Among them, the number of leaves belonging to black rot 4 in the original dataset is less. Moreover, data enhancement will cause the network to over-fit the samples under this grade, so the leaves belonging to black rot 4 will be discarded in the dataset. 90% of the dataset is the training set and 10% of the dataset is the test set. The performance of the proposed algorithm is evaluated on PlantVillage and the combination of PlantVillage and PlantDoc. Some sample lesions of this dataset are shown in Figure 2.

3.3. Performance Metrics
Average accuracy is the average value of the accuracy of four grades for each disease. The average recall is the average recall of four grades for each disease. As mentioned above, the leaves belonging to black rot 4 are removed.
3.4. Identification Results
In this paper, to reduce the influence of the random division of dataset on the experimental results, each group of experiments was repeated five times. And the average values of five groups of experimental results are shown in this paper.
3.4.1. Identification Results of the Attention Mechanism using Different Fusion Structures
In the Methodology section, a new attention mechanism was put forward and different fusion structures are put forward of the new attention mechanism. Table 1 shows the recognition results of attention mechanisms using different fusion structures. These experiments were compared with the experiment without fusion structure. Table 2 shows the accuracy of each category for each experiment. The dataset used in these experiments is the combined dataset of PlantVillage and PlantDoc, and the data enhancement method used in these experiments is traditional data enhancement method.
| Fusion structure name | Accuracy/(%) | Recall/(%) | Average accuracy/(%) | Average recall/(%) | ||||
|---|---|---|---|---|---|---|---|---|
| Black rot | Scab | Rust | Black rot | Scab | Rust | |||
| No fusion | 84.81 | 85.86 | 83.67 | 73.75 | 96.25 | 84.20 | 77.15 | 96.23 |
| C0 | 86.86 | 87.73 | 87.33 | 79.00 | 94.25 | 85.57 | 82.61 | 95.01 |
| C1 | 87.22 | 88.13 | 84.87 | 83.41 | 96.11 | 85.67 | 80.25 | 95.75 |
| C2 | 87.56 | 88.13 | 84.67 | 84.25 | 93.75 | 83.90 | 84.99 | 95.49 |
| C4 | 89.75 | 90.77 | 90.00 | 80.75 | 98.50 | 89.73 | 84.00 | 98.58 |
- Notes: All values are in percentage.
| No fusion | C0 | C1 | C2 | C4 | |
|---|---|---|---|---|---|
| Black rot 1 | 87.33 | 84.00 | 76.00 | 79.33 | 83.33 |
| Black rot 2 | 66.00 | 80.00 | 84.00 | 78.00 | 90.00 |
| Black rot 3 | 98.00 | 98.00 | 97.33 | 97.33 | 97.33 |
| Scab 1 | 68.00 | 74.67 | 75.33 | 74.00 | 74.00 |
| Scab 2 | 64.00 | 68.00 | 74.00 | 79.33 | 78.00 |
| Scab 3 | 69.33 | 71.33 | 78.00 | 90.00 | 81.33 |
| Scab 4 | 94.00 | 94.00 | 94.00 | 94.00 | 94.00 |
| Rust 1 | 93.33 | 93.33 | 98.00 | 97.33 | 98.67 |
| Rust 2 | 92.00 | 86.00 | 83.33 | 92.00 | 94.00 |
| Rust 3 | 98.67 | 98.00 | 99.33 | 97.33 | 98.00 |
| Rust 4 | 98.00 | 96.67 | 98.00 | 98.00 | 98.00 |
- Notes: All values are in percentage.
3.4.2. Identification Results Using Different Data Enhancement Methods
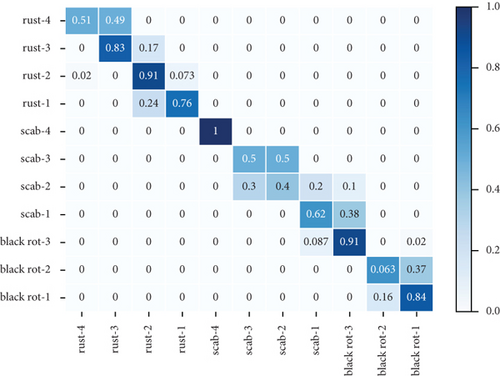
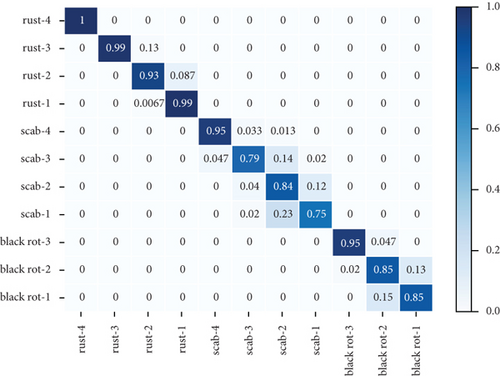
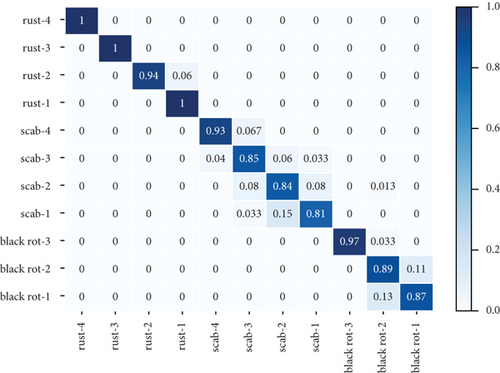
Table 3 shows the accuracy rate and recall rate of three experiments using different data enhancement methods with the new attention mechanism proposed in this paper. The first experiment did not use the method of data enhancement. The second experiment used the traditional method of data enhancement, and the third experiment used the method of data enhancement proposed in this paper. Figure 3 shows the confusion matrix of three groups of experiments. The dataset used in these experiments is the combined dataset of PlantVillage and PlantDoc.
| Accuracy/(%) | Recall/(%) | Average accuracy/(%) | Average recall/(%) | |||||
|---|---|---|---|---|---|---|---|---|
| Black rot | Scab | Rust | Black rot | Black rot | Scab | |||
| No data enhancement | 72.14 | 73.05 | 78.67 | 63.00 | 74.75 | 70.48 | 68.96 | 79.71 |
| Original data enhancement | 89.75 | 90.77 | 90.00 | 80.75 | 98.50 | 89.73 | 84.00 | 98.58 |
| New methods proposed in this paper | 91.55 | 92.06 | 90.33 | 85.75 | 98.67 | 89.67 | 87.32 | 98.58 |
- Notes: All values are in percentage.
3.4.3. Identification Results Using Different Attention Mechanisms
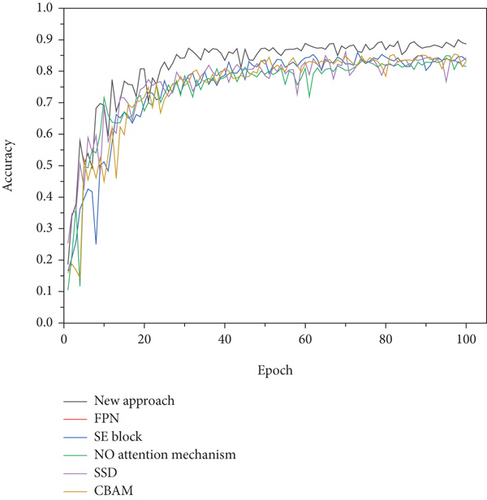
Figure 4 shows the convergence curves of the attention mechanism proposed in this paper and recent classic attention mechanisms. Among them, the attention mechanism proposed in this paper uses the fusion structure C4. Tables 4 and 5 show how identification results that different attention mechanisms were tested on the dataset of PlantVillage and the combination of PlantVillage and PlantDoc respectively.
| NO attention mechanism | CBAM | SE block | SSD | FPN | New approach | |
|---|---|---|---|---|---|---|
| Identification accuracy | 84.81 | 85.36 | 85.62 | 85.72 | 84.72 | 91.55 |
- Notes: All values are in percentage.
| NO attention mechanism | CBAM | SE block | SSD | FPN | New approach | |
|---|---|---|---|---|---|---|
| Identification accuracy | 85.26 | 86.37 | 86.33 | 87.01 | 85.23 | 92.38 |
- Notes: All values are in percentage.
4. Discussion
From Table 1, it can be seen that the experiment with the fusion structure C4 has the best performance. The accuracy of the experiment with the fusion structure C4 is 89.75% and the recall rate is 90.77%. Compared with the experiment without attention mechanism, the accuracy rate and recall rate were improved by 4.94% and 4.91%, respectively. The lowest average accuracy was 80.75% and the highest average accuracy was 98.50%. The lowest average recall rate is 84.00%, and the highest average recall rate is 98.58%. And all average accuracy and average recall of the three diseases are higher than those of the experiment without attention mechanism. Therefore, the location of the fused lesion strengthens the difference between lesion area and other area, which makes the network pay more attention to lesion area.
Compared with fusion structure C0, the accuracy and recall rate of the fusion structure C1 increased by 3.12% and 2.46%, respectively. Compared with fusion structure C1, the accuracy and recall rate of the fusion structure C2 increased by 0.52% and 0.09%, respectively. Compared with fusion structure C2, the accuracy and recall rate of fusion structure C4 are improved by 0.48% and 0.97%, respectively. It can be seen from Figures 1(b)–1(e) that the fusion structures CO, C1, C2, and C4 are different in the location of regional information fusion. With the deepening of the fusion localization, the classification effect of this model is getting better and better. The reason is that the deeper feature vector is derived from the intermediate feature vector by convolution operation and the deeper feature vector is a better expression than the intermediate feature vector.
As can be seen from Table 2, the classification accuracy of the experiment without attention mechanism on the samples of black rot 1, black rot 2, scab 1, scab 2, and scab 3 is low, which are no higher than 70%. Most of the mistaken identification comes from slight disease grade to mild disease grade. In the early stage of disease growth, the change of disease characteristics is vaguer than that in the late stage of disease, and it is more likely to lead to the wrong judgment of disease grade. The accuracy of these experiments with attention mechanism using different fusion structures was higher than the experiment without attention mechanism. After using the attention mechanism proposed in this paper, CNN pays more attention to the lesion area, which improves the feature extraction ability of the network.
It can be seen from Table 3 that the accuracy and recall are 72.14% and 73.05%, respectively, before using data enhancements to the dataset. In the experiment that the traditional data enhancement method was used, the accuracy rate and recall rate are 84.81% and 85.86%, respectively. And the accuracy rate and recall rate were improved by 12.67% and 12.81%, respectively, compared to the experiments without data enhancement method. In the experiment that the new data enhancement method was used, the accuracy and recall are 91.55% and 92.06%, respectively, which are increased by 1.68% and 0.72% compared with the experiment with the traditional data enhancement method.
Figures 3(a)–3(c) show the confusion matrix of three groups of experiments, which introduces in detail the accuracy predicted image scores of three apple leaf diseases in each grade. Compared with the experiments without data enhancement, the average accuracy of black rot, scab, and rust of the experiment with traditional data enhancement methods increased by 11.33%, 16.25%, and 23.75%, respectively. Compared with the experiments without data enhancement, the average accuracy of black rot, scab, and rust of the experiment with traditional data enhancement methods increased by 11.67%, 21.25%, and 24.08%, respectively. The analysis of the above three diseases shows that, compared with the traditional data enhancement method, the data enhancement method proposed in this paper can better improve the classification effect of scab disease grades. The reasons can be divided into two parts: (1) Traditional data enhancement can reduce the influence of too few images on classification results, by making the number of pictures in different categories equal. (2) The local similarity and local discontinuity of all sample features in the same disease grade were reduced.
Figures 3(a)–3(c) show the identification accuracy of different categories in the method proposed in this paper. Most of the mistaken identification comes from disease grade 1 to disease grade 2. In the early stage of disease growth, the change of disease characteristics is vaguer than that in the late stage of disease, and it is more likely to lead to the wrong judgment of disease grade.
The attention mechanism proposed in this paper was compared with the classical attention mechanism on the PlantVillage dataset and the combined dataset of PlantVillage and PlantDoc. These classical attention mechanisms include CBAM, SE block, SSD, and FPN. Figure 4 shows the convergence curve of the accuracy of these experiments. As shown in Figure 4, the network without attention mechanism, four classic attention mechanism networks, and the proposed attention mechanism began to converge after a certain period of time, and finally achieved the best recognition performance. Generally speaking, the new attention mechanism is basically stable after 30 stages, while other models have satisfactory convergence after 40 stages.
Tables 4 and 5 show identification results that different attention mechanisms were tested on the dataset of PlantVillage and the combination of PlantVillage and PlantDoc, respectively. The recognition accuracy of 11 categories without attention mechanism is 84.81% on the combined dataset of PlantVillage and PlantDoc. The recognition accuracy with CBAM, SE block, SSD, and FPN is 85.36%, 85.62%, 85.72%, and 84.72%, respectively, on the combined dataset of PlantVillage and PlantDoc. And the recognition accuracy of 11 categories with attention mechanisms is 91.55%, which is superior to existing methods.
The traditional attention mechanism changes the weight of the network to all pixels in the image, which affects the ability of the network to extract the features of the lesion area. The new attention mechanism is proposed by integrating the acquired information of the lesion area with global information. This method can increase the attention of the network to the lesion area, while the influence of other areas on the attention of the network to the lesion area can be ignored. Compared with the classic attention mechanism, the new attention mechanism improves the convergence speed of the network model, and provides higher recognition accuracy for apple leaf disease grades. Those methods were also verified on the dataset of PlantVillage.
5. Conclusions
Images of apple leaf diseases collected under natural conditions have similar characteristics, but with different degrees. It is difficult to identify apple leaf diseases and their severity quickly and accurately by the existing methods. The traditional attention mechanism changes the weight of the network to all pixels in the image, which affects the ability of the network to extract the features of the lesion area. And the traditional method of data enhancement is easy to cause local similarity and local discontinuity of all sample features in the same disease grade. In this paper, the improved metric matrix for kernel regression (IMMKR) proposed was firstly used to reduce the influence of local similarity and local discontinuity of all sample features in the same class. Then, a new attention mechanism by fusing the lesion location based on visual features was proposed, and the attention of the model to the lesion area was strengthened. The experimental results show that the algorithm can improve the recognition accuracy of apple leaf disease grades, and meet the intelligent recognition requirements of different apple leaf diseases. This algorithm has good reference significance for the identification and promotion of crop diseases. In the future, the algorithm proposed in this study will be introduced into software or hardware products, such as apple leaf disease identification software, fine-grained visual classification, and temporal prediction [33–41].
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Open Research
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.




