Computational Insights in DNA Methylation: Catalytic and Mechanistic Elucidations for Forming 3-Methyl Cytosine
Abstract
Methylation at C5 position of cytosine (5 mC) is the most abundantly occurring methylation process at CpG island, which has been well known as an epigenetic modification linked to many human diseases. Recently, another methylation approach has been discovered to show that DNA methyltransferases (DNMTs) promote the addition of the methyl group at position 3 to yield 3 mC. The existence of 3 mC can cause severe damages to the DNA strand, such as blocking its replication, repair, and transcription, affecting its stability, and initiating a double-strand DNA break. To gain a deeper insight into the formation of 3 mC, we have performed density functional theory (DFT) modeling studies at different levels of theory to clearly map out the mechanistic details for this new methylation approach. Our computed results are in harmony with pertinent experimental observations and shed light on a crucial off-target activity of DNMTs.
1. Introduction
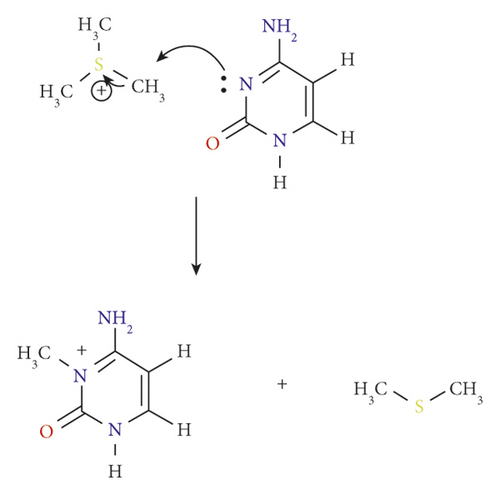
Although methylation of cytosine at position 5 is a hallmark of epigenetic modifications in organisms produced and conserved by DNA methyl transferases (DNMTs), other methylation pathways still possibly exist under the catalysis of the same enzyme, while the resulting methylated products can pose profoundly serious threats to the DNA helix. In 2018, a transmutative relationship between alkylation and DNMTs appeared to the surface, showing the methylation of cytosine can take place at position 3 as well to form 3-methyl cytosine (3 mC) as the product [1]. DNMT plays a catalytic role in this reaction as shown in Figure 1, and this methylation pathway has been found to result in a spontaneous DNA mutation in the cell.

Experimental analyses using ultrasensitive liquid chromatography/mass spectrometry (LC/MS) have unambiguously confirmed that DNMT can catalyze the methylation at the N3 position of cytosine with S-adenosyl-L-methionine (AdoMet). This finding reflects that the generation of 3 mC is a general reactivity enabled by cytosine methyltransferases [1].
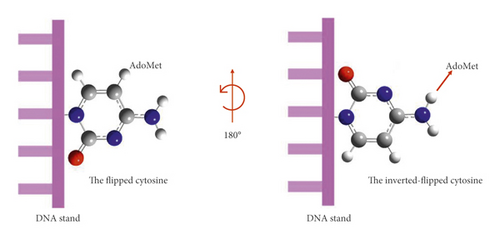
Mechanistic details of this newly observed methylation pathway, however, have not yet been disclosed. Duktaz and others. [2] recently conducted a mutational study of catalytically relevant amino acids in DNMT enzyme. Different amino acid residues were mutated in the active site of DNMT, and the catalytic roles that DNMT plays in methylation at C5 and N3 were assessed. Based on the experimental results, they proposed that the N3 methylation occurs through a mechanism of cytosine inversion step; that is, cytosine bound into the active site pocket of DNMT is flipped into an inverted orientation. The flipping allows the N3 site of cytosine to be positioned in a close proximity to the methyl group of AdoMet to facilitate methyl group transfer (see Figure 2vid infra). The role of amino acids present in the active site in the formation of 3 mC and 5 mC was varied; unlike the N3 position, methylation on the C5 position was found to be attenuated, suggesting these amino acids play vital roles in the catalytic mechanisms. Cysteine (C710) has no catalytic role in the mechanism of N3 methylation, while the mutation of Arg (R792) to Ala induces the formation of 3 mC with a lower rate of formation, which was explained by the loss of H-bond of R792 to the deoxyribose due to the inversion of the base by 180°. Glutamic acid (E756) has a neutral effect on producing both 5 mC and 3 mC. The generation of both products was catalyzed through the formation of a H-bond with the exocyclic-amino group (N4), which stabilizes the base [2]. Despite the repair system displayed by the cell, alkylation repair protein B (ALKB2/3) can erase alkylation adducts as 1-methyladenine and 3-methylcytosine to help the survival of the genome [3]. However, when ALKB2 loses its functionality, the level of 3 mC increases by around 15% [1]. It has also been reported that if the damage occurs during DNA replication and when the damage is beyond the limits of correction, ALKB is no longer capable of removing such a lesion [4].
On one side, it is well recognized that the occurrence of 5 mC is mutagenic and can cause base mispairing G-T by the deamination of 5 mC to thymine [5]. The deamination reactions of nucleic acid bases and related compounds have been extensively studied, the results of which are of great value in proposing the mechanisms investigated in this work [6–13]. On the other side, however, 3 mC lesion affects the stability of single-strand (ss) and double-strand (ds) DNA [3]. Rošić et al. reported that 3 mC results in double-strand DNA (dsDNA) breaking [1].
The damage incurred in the presence of 3 mC contradicts with replication, repair, and transcription of the DNA strand, in addition to spontaneous mutations of C to A and C to T [5]. In comparison with unmethylated cytosine, 3 mC can accelerate deamination to form 3-methyluracil (3mU) by a factor of 4 × 103 at pH 7.4 (physiological conditions) [14].
Alkylation of cytosine at position 3 with carcinogenic alkylating agents, such as methanediazonium and ethanediazonium ions, has been studied theoretically through the B3LYP density functional method. Carcinogenic alkylating agents are produced from the metabolic activation of methyl- and ethyl-substituted nitrosoureas, and they tend to add a methyl group to N3 and O2 of cytosine; N7, N3, and O6 of guanine; N1 of adenine; and O2 and O4 of thymine via the SN2 mechanism [5, 15].
Another exogenous strong alkylating agent is dimethyl sulfate (DMS). The results obtained from combustion analysis have been subjected to DFT theoretical investigations to understand the methylation of N3 of cytosine, N7 of guanine, and N1, N3, and N7 of adenine at room temperature using M06-2X/6-31<puncsp> </puncsp>+<puncsp> </puncsp>G(d) and B3LYP/6-311<puncsp> </puncsp>+<puncsp> </puncsp>G(2df,2p) levels of theory. The solvation effects were examined using the conductor-like polarizable continuum model (CPCM). The obtained results indicated that the most reactive site towards methylation in the gas phase is N7 of adenine with an activation energy of 70.42 kJ mol−1, while the activation energy of cytosine is 96.52 kJ mol−1 [16].
The methylation reaction at position 5 has been thoroughly studied computationally and experimentally, and the activation energy of the methylation chemical event at C5 was examined. For instance, Zang and others used QM/MM to illustrate the concerted uncatalyzed reaction of the nucleophilic attack of Cys-81-S¯ to C6 and AdoMet to C5 in an aqueous medium. The calculated activation energy of this reaction was found to be 34.7 kJ • mol-1 [17]. Another independent study by Yang et al. characterized the methylation reaction by QM/MM-MD calculations. The obtained free energy of activation of this step was 66.1 kJ • mol-1 [18]. Aranda and coauthors utilized QM/MM-MD to study this mechanism; the same reaction step was found to cost 79.9 kJ • mol-1 [19]. Most recently, Jerbi and coworkers have combined both MD and DFT approaches in order to study the methylation possibility of the methyl donor (SAM) and a series of analogues. The obtained free energy of activation calculated when SAM molecule is used was 56.7 kJ • mol-1 [20].
To the best of our knowledge, the catalytic activity of DNMTs towards the alkylation of cytosine at position 3 has not been investigated theoretically. Furthermore, the side reaction of 3 mC promoted by DNMT warrant theoretical analysis to assess the validity of the proposed “inverted base flipping” mechanism. In this study, we performed a computational study on the most plausible reaction pathway starting from the general mechanism shown in Scheme 1. Based on calculation made at high-level theories, mechanistic details concerning the reactivity of DNMTs towards N3 of cytosine were established, and the results were compared to the methylation reaction at conventional C5 position. The catalytic roles of amino acids present in the active site such as cystine (Cys), glutamic acid (Glu), arginine (Arg), and alanine (Ala) in the gaseous phase were determined as well.
In vivo abundant potential catalytic species, such as phosphate and carbonate ions, have been known to play a vital role in the biological process. Most recently, Kato and coworkers studied the deamination of glutamine residues that were catalyzed by hydrogen carbonate and dihydrogen phosphate catalysts (HCO3- and H2PO4-) [21]. In our study, the effects of such catalyst anions were also examined in order to understand their influence on the energy cost of methylation at various positions of cytosine ring, the structure of transition states, and potential energy barriers.
2. Computational Methods
We conducted geometry optimizations using Gaussian 16 (G16) quantum chemistry package. The resulting geometries were confirmed to be energy minima or transition states (TSs) on the potential energy surface by calculating their vibrational frequencies, where the TSs were confirmed by showing one and only imaginary frequency. Furthermore, TSs were subjected to intrinsic reaction coordinate (IRC) calculations at the B3LYP/6-31G(d) level of theory in order to connect the TSs with the local minima of the reactants and products on the potential energy surface. Geometry optimization was performed using the B3LYP/6-31G(d), B3LYP/6-31G(d,p), M06-2X/6-31G(d), and APFD/6-31G(d) methods. We also performed APFD/6-31<puncsp> </puncsp>+<puncsp> </puncsp>G(d) and APFD/6-31G(2df,2p) calculations in order to detect the effect of the polarization and diffuse functions on the system. The proposed pathways have been studied in the aqueous medium using the solvation model based on density (SMD) at APFD/6-31G(d) level of theory. Thermodynamic parameters, ΔS, ΔH, and ΔG, as well as the kinetic parameters, activation energy (Ea), enthalpy of activation (ΔH‡), and the Gibbs energies of activation (ΔG‡), have been also calculated for each proposed reaction mechanism.
3. Results and Discussion
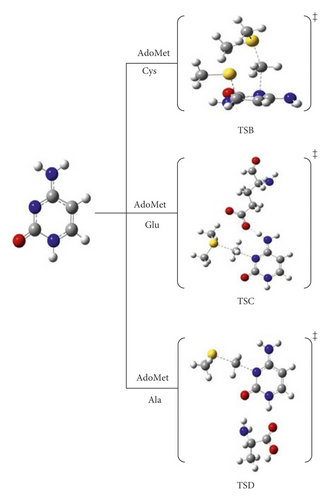
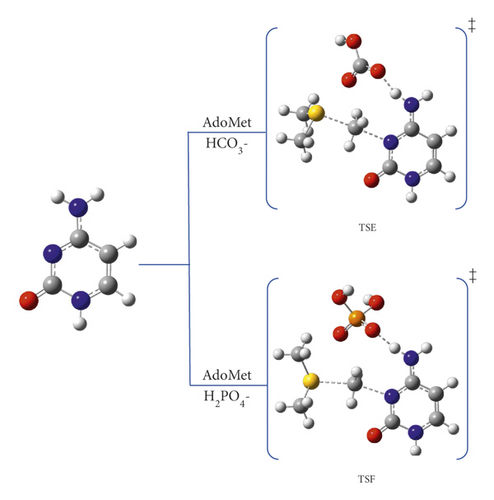
In this thorough study, comprehensive computational quantum chemistry calculations for 10 reaction pathway mechanisms were proposed for the methylation reaction of cytosine at position 3. We designated different pathways in order to study and observe how each component will behave and affect this off-target methylation reaction. Pathway A represents the cytosine and AdoMet system as a starting point to study the methylation reaction on N3 position, as shown in Figure 3. Pathways B⟶D include the participation of amino acids Cys, Glu, and Ala, respectively, as shown in Figure 4. Meanwhile, pathways E and F comprise the methylation reaction with our proposed catalytic anions such as hydrogen carbonate and dihydrogen phosphate, respectively, as depicted in Figure 5.
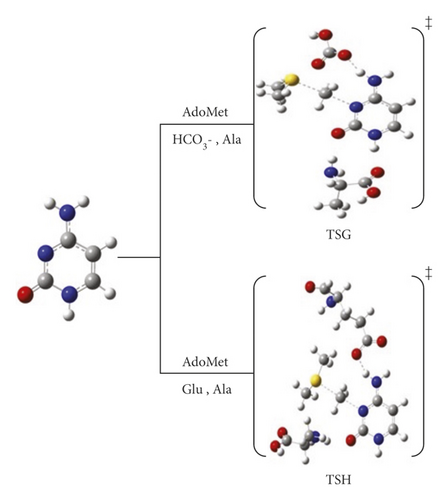
Furthermore, we studied the combined effect of amino acids and the proposed catalysis, in pathways G and H, as presented in Figure 6. It is worth observing that all pathways proceed through one-step mechanism, except pathway B. All investigated pathways are exothermic reactions.
The activation energies (Ea), enthalpies of activation (ΔH‡), and Gibbs energies of activation (ΔG‡) were calculated at different levels of theory (Table 1 for all proposed pathways. Moreover, the potential energy diagrams (PEDs) were also constructed for studied pathways. The thermodynamic parameters for the proposed pathways are presented in Table 2. The potential energy diagram (PED) for pathways A⟶J calculated at different levels of theory in aqueous and gaseous phases (in kJ.mol−1 at 298.15 K) were included in the Supporting Information (SI) in Figures S1-S10.
| Transition state | B3LYP/6-31G(d) | B3LYP/6-311G(d,p) | M06-2x/6-31G(d) | APFD/6-31G(d) | SMD∗ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ea | ΔG‡ | Ea | ΔG‡ | Ea | ΔG‡ | Ea | ΔG‡ | Ea | ΔG‡ | |
| TSA | 103.5 | 105.2 | 104.2 | 105.5 | 118.0 | 117.2 | 110.8 | 114.9 | 92.2 | 98.8 |
| TSB | 140.6 | 150.1 | 144.4 | 153.9 | 125.3 | 136.6 | 123.9 | 134.3 | 132.2 | 140.9 |
| TSC | 86.1 | 89.7 | 83.6 | 87.6 | 107.3 | 108.9 | 101.8 | 100.1 | 97.6 | 99.0 |
| TSD | 79.1 | 81.1 | 97.4 | 94.1 | 104.6 | 93.1 | 95.6 | 86.4 | 86.4 | 95.3 |
| TSE | 64.0 | 60.3 | 61.7 | 58.3 | 62.3 | 69.0 | 56.5 | 62.5 | 77.2 | 88.1 |
| TSF | 66.5 | 79.9 | 64.7 | 72.6 | 78.8 | 84.9 | 72.2 | 74.1 | 97.6 | 98.5 |
| TSG | 59.5 | 54.3 | 57.4 | 52.0 | 64.4 | 70.7 | 96.6 | 91.2 | 82.3 | 96.3 |
| TSH | 76.8 | 70.9 | 74.9 | 66.9 | 75.5 | 76.4 | 97.3 | 82.3 | 91.2 | 84.4 |
- ∗Calculated at APFD/6-31G(d).
| Pathway | B3LYP/6-31G(d) | B3LYP/6-31G(d,p) | M06-2x/6-31G(d) | APFD/6-31G(d) | SMD | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| ΔH | ΔG | ΔH | ΔG | ΔH | ΔG | ΔH | ΔG | ΔH | ΔG | |
| A | −32 | −35 | −31 | −34 | −33 | −35 | −30 | −30 | −47 | −48 |
| B | −97 | −99 | −91 | −93 | −121 | −117 | −107 | −106 | −65 | −60 |
| C | −79 | −97 | −82 | −98 | −77 | −83 | −84 | −77 | −37 | −43 |
| D | −50 | −61 | −31 | −44 | −41 | −52 | −31 | -49 | −47 | −50 |
| E | −114 | −133 | −117 | −134 | −124 | −127 | −114 | −120 | −49 | −38 |
| F | −83 | −86 | −89 | −96 | −82 | −91 | −71 | −82 | −9 | −18 |
| G | −124 | −131 | −128 | −134 | −125 | −133 | −77 | −86 | −44 | −43 |
| H | −89 | −97 | −91 | −101 | −126 | −121 | −84 | −89 | −51 | −54 |
3.1. The Original Reaction: Pathway A
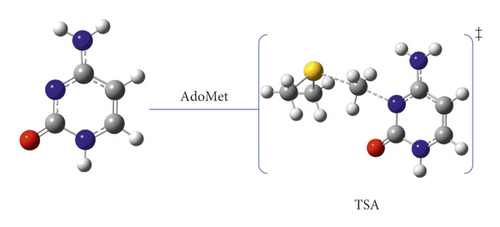
As mentioned above, DNMT can catalyze the substitution of a methyl group at the N3 position of the cytosine ring through a straightforward electrophilic addition mechanism [15]. The details of this one-step reaction are outlined as pathway A (Figure 2, and Scheme 2.
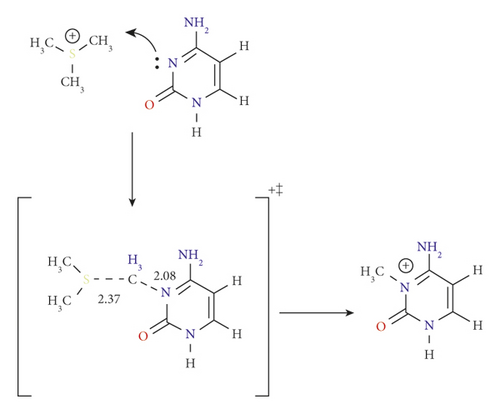
In this pathway, the reactants are placed to form electrostatic interaction between the positively charged AdoMet and the partially negative carbonyl oxygen of cytosine. For the TSA, the methyl group approaches the N3 atom at a semilinear angle of 174.9° in a side-way manner to the ring. The methyl carbon shows a distance of 2.08 Å to N3, which is indicative of a loosely bound SN2 transition state, as shown in Scheme 2 of pathway A reaction mechanism.
The activation energy was calculated to be 103.5 kJ mol−1 at the B3LYP/6-31G(d) level of theory, which was highly correlated with the value obtained at B3LYP/6-31G(d,p) with 104.2 kJ mol−1 (see Table 1). We attributed this high activation energy to the presence of a positive charge on the cytosine ring. The effect of solvent (water) was studied using the SMD solvation model. The activation energy of TSA was noticeably decreased in the aqueous environment with 92.2 kJ mol−1.
It is worth noting that the free energy of activation for the 5 mC generation by SAM molecule calculated using MD and DFT approaches was 56.7 kJ•mol−1 [20].
3.2. The Effect of Amino Acids Cys, Glu, Arg, and Ala : Pathways B, C, and D
Herein, we performed a study that investigates the effect of adding amino acids (Cys, Glu, and Ala) on the activation energy. We started by mutating all amino acids and activated Cys (C710) as in pathway B. Then, we activated Glu (E756) in pathway C and then Ala (A791) in pathway D. We intended to compare the calculated activation energy of each pathway with the main reaction activation energies (pathway A).
Previous studies showed that cysteine (C710) residue significantly influences the cytosine ring activation for conventional 5 mC generation, in which Cys initiates a nucleophilic attack at C6 to activate the C5 for methyl group transfer [17, 22–24]. In the case of 3 mC generation, however, its catalytic effects are insignificant. This was observed to noticeably increase the activation barrier for methyl addition, which was consistent with the results obtained experimentally by Dukatz and coworkers. Methylation of N3 in the presence of C710 denoted as pathway B was initiated by nucleophilic addition of Cys-S- residue at C6 of the ring in concert with the methyl addition to the target N3 followed by the departure of Cys-S-.
Unlike the mechanism in pathway A, pathway B proceeds through a nearly perpendicular attack at 113.1° and syn-addition of AdoMet and C710, concerning the ring. In TSB, the distance between Cys-S- and C6 was 2.00 Å, and the methyl group of AdoMet was 2.30 Å from N3 (see Scheme 3).
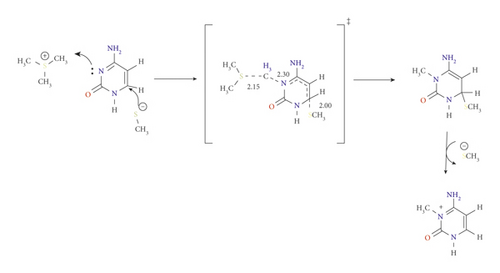
The activation energy of TSB was calculated as 140.6 kJ mol−1 using the B3LYP/6-31G(d) and was 144.4 kJ mol−1 at B3LYP/6-311G(d,p) level of theory. It has also been found that the M06-2X/6-31G(d) level of theory is much more sensitive towards this step and produces good results, with the activation energy being 125.3 kJ mol−1 comparable results were obtained using APFD/6-31G(d) level of theory (see Figure S2 and Table 1). The solvation model (SMD) showed an increase in the activation energy of TSB to 130.5 kJ mol−1.
The increase in energy was attributed to the loss of π-conjugation over the ring. In comparison with pathway A, pathway B is unfavorable, and C710 has no catalytic effect. Considering pathways with lower values are taken as the most plausible mechanism.
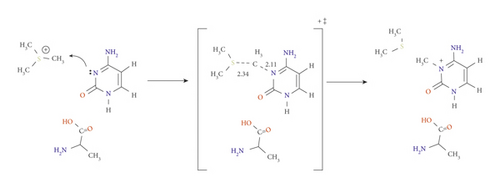
The catalytic role of glutamic acid residue (E756) was observed to be unique, although its role in conventional C5 methylation is still debatable [19, 25]. Herein, it enhances the reaction through a new approach, in which a proton (H4) abstraction occurs on the exocyclic amine group in concert with the methyl group addition. Removing H4 enhances the nucleophilicity of the ring to easily undergo the methylation reaction.
In TSC, H4 approaches the Glu acidic oxygen at 1.60 Å and C4-N4 become 1.33 Å for the double bond to be formed. Meanwhile, the methyl group is transferred at a distance of 2.11 Å from N3, as displayed in Scheme 4. The activation energy of TSC is lower than those of the previous pathways, with a value of 86.1 and 83.6 kJ mol−1 at the B3LYP/6-31G(d) and B3LYP/6-31G(d,p) level of theory, respectively (Figure S3 and Table 1). The energy of the resulting imino-tautomeric product PC was calculated as −77.3 kJ mol−1 at the M06-2X/6-31G(d) level, which is much more stable than PA (−32.7 kJ mol−1 calculated at the same level of theory).
The PC is neutral product, which provides extra stability, unlike the positively charged product of pathway A. This result is comparable with the data obtained from double proton transfer between DNA base pair (guanine-cytosine), which led to a stable but rare imino-tautomer. Furthermore, this has been suggested to cause a spontaneous mutation [26–28].
The presence of Arg (R792) hinders the methylation of N3 and offers no assistance in 3 mC production, despite the catalytic role and the stabilization effect that it can provide for the 5 mC generation [17]. On the other hand, the mutation of Arg to Ala was seen to boost the reaction dramatically, particularly, pathway D. This pathway proceeded as pathway A in one step. In TSD (Scheme 5), Ala provided extra stabilization for the cytosine to react through forming H-bonding with H1 of cytosine at 1.80 Å and at 2.24 Å from O2. The activation barrier is lowered to 79.1 kJ mol−1 and 97.4 kJ mol−1 at B3LYP/6-31G(d) and B3LYP/6-31G(d,p), respectively (Figure S4 and Table 1). These results were consistent with the experimental findings observed by Dukatz et al. after the substantial increase in 3 mC/5 mC ratio upon mutating Arg to Ala. Fortunately, this can support the proposed “inverted flipped base” approach, as the role of Arg is lost in stabilizing the ring and catalyzing the reaction. In light of this, the pronounced stability of the ring is attributed to Ala amino acid. The activation energy of TSD is lower than those calculated for TSA and TSB, and then, the existence of Ala is favorable for this reaction much more than the Glu (and certainly Cys).
3.3. The Catalytic Effect of Hydrogen Carbonate and Dihydrogen Phosphate Anions: Pathways E and F
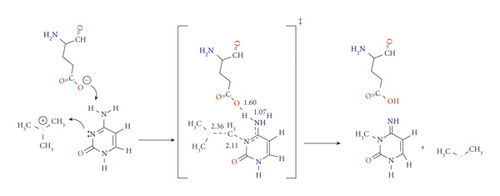
The role of potential catalyst molecules in the methylation of N3 was investigated; meanwhile, other amino acids are mutated, which are abundantly present in vivo. Hydrogen carbonate and dihydrogen phosphate ions can act as either acids or bases depending on the pH. At biological pH, phosphate can exist as dihydrogen phosphate ion (H2PO4-, pKa 2.15), which is known to speed up the aging process, while carbonates exist as HCO3- (pKa 10.25) [21]. These ions tend to follow the same proton transfer approach that has been described in pathway C (with E756) by forming the stable imino-tautomer concertedly with methylation at N3 (pathways E and F for carbonate and phosphate, respectively). Scheme 6 demonstrates the mechanism of pathways E. Structurally, both TSE and TSF were remarkably similar. The reaction mechanism of pathway F has been presented in the supplementary information as Scheme S1.
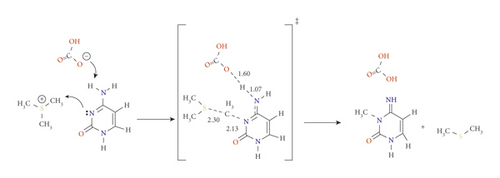
Although pathways E and F proceeded as pathway C mechanism, the ability of these proposed catalysts in lowering the activation barrier of the reaction is more significant and even much better than E756. Hydrogen carbonate is the most basic (pKa = 10.25); therefore, its ability to abstract H4 is higher than E756 (pKa around 5) and H2PO4- (pKa = 2.15). On the other hand, the polarizability of the phosphorus of H2PO4- enhances its ability to abstract the proton than E756.
At the APFD/6-31G(d) level of theory, the activation energy of TSE was calculated as 56.5 kJ mol−1 and reduced to 61.7 kJ mol−1 at B3LYP/6-31G(d,p) (see Table 1). In contrast, the activation energies of TSF are 66.5 and 64.7 kJ mol−1 at B3LYP/6-31G(d) and B3LYP/6-31G(d,p), respectively (see Figure S5 and S6). It is worth noting that the products PE and PF are very stable with relative energies of -113.7 and -71.4 kJ mol−1, respectively, at APFD/6-31G(d) (which are the methylated rare tautomer).
It is important to address that the proton transfer mechanism is affected by the environment [24]. Despite many tries, using the SMD model, TSC, TSE, and TSF did not show proton transfer. This may be reflected in the higher activation energy of these transition states. Taking the effect of solvation into account, the activation energies of TSE and TSF using SMD model were 77.2 and 97.6 kJ mol−1, respectively.
3.4. The Collaborative Role of Amino Acids and Catalysts: Pathways G⟶J
So far, we have studied the impact of subunits available in the active site and mutate others to the methylation reaction to obtain the complete picture of the N3 methylation. According to the results obtained from the previously discussed pathways (A⟶F), we have considered the calculations including these units as an entire system to elucidate the properties of the active sites. Therefore, pathways G and H were assigned to mutate Arg to Ala simultaneously with activating HCO3- and Glu, respectively. While pathways I and J were designated by activating Arg with HCO3- and Glu. The reaction mechanism of pathways G and H are shown in Schemes 7A and 7B, respectively. The PED for pathways G and H are reported in Figures S7 and S8. The data for pathways I and J are included in the Supplementary Information in Table S1. Consistent with our previous results, the presence of Ala stabilizes the cytosine ring and catalyzes the reaction to significantly drop down the activation energy barrier compared to that with R792. In TSG, Ala formed H-bonding with H1 at 1.87 Å and the carbonyl carbon O2 of cytosine at 2.04 Å, offering further stabilization to the transition state.
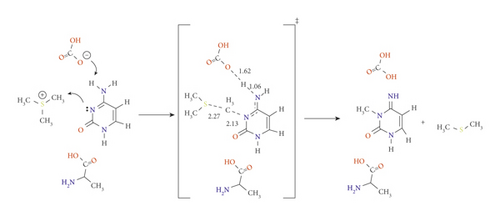
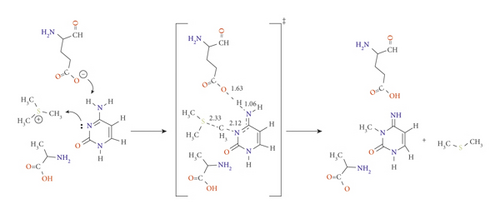
In TSH, the amine hydrogen from Ala side approaches the O2 of the cytosine ring with 2.01 Å as shown in Scheme 7B
It was also found that HCO3- can reduce the energy values more than E756, as E756 is less basic than HCO3- and adds extra bulkiness around the ring.
The lowest activation energies obtained for TSG and TSH are 59.5 and 76.8 kJ mol−1 at the B3LYP/6-31G(d) level of theory, and 64.4 and 75.5 kJ mol−1 at M06-2X/6-31G(d) level of theory, respectively. The results calculated using B3LYP/6-31G(d) and M06-2X/6-31G(d) are highly correlated for these pathways. However, utilizing APFD/6-31G(d) shows an increase in the activation energy with no reducing effect when a diffuse or polarization functions are considered (see Table 1, Figures S6 and S7). Taking the effect of solvation using the SMD model, the barrier for TSG was dramatically lowered to -63.3 kJ mol−1 while found at 91.2 kJ mol−1 for TSH. Consequently, Ala and HCO3- are the most suitable candidates for catalyzing the reaction and lowering the activation energy.
Despite the occurrence of the proton transfer during the methyl transfer in pathways I and J, the effect of Arg was much more pronounced in increasing the activation energy of TSI and TSJ and diminishing the catalytic role of HCO3- and Glu. By the withdrawal effect of positively charged Arg making the ring more electrophilic. The activation energy of TSI and TSJ were studied using B3LYP/6-31G(d) and M06-2X/6-31G(d) as displayed in Table S1 and Figures S8 and S9 (supplementary).
Using B3LYP/6-31G(d), the activation energies were 183.2 and 175.2 kJ mol−1 of TSI and TSJ, respectively. In comparison, the calculated values at M06-2X/6-31G(d) were 162.9 and 187.6 kJ mol−1. The obtained results were highly correlated and significantly higher than those observed while mutating Arg to Ala. Therefore, we have decided not to investigate these pathways further as they have higher activation barriers.
According to the experimental results obtained for 3 mC generation, it was observed that the rate of 3 mC generation was decreased when the impact of Arg is considered and further explained by the loss of the stabilizing H-bond between this amino acid and the ring [2].
Regarding the effect of polarization and diffuse functions, we have performed geometry optimization using APFD/6-31<puncsp> </puncsp>+<puncsp> </puncsp>G(d) and APFD/6-31G(2df,2p) levels of theory. According to our results, these functions did not decrease the activation barriers for the studied pathways (see Table S2 in the Supplementary Information (SI)).
3.5. Thermodynamic Parameters for Methylation of Cytosine at N3
The thermodynamic parameters (DH and DG) for the methylation of cytosine at position 3 along with its proposed pathways are given in Table 2, at all studied levels of theory.
The methylation reaction of cytosine at position 3 (pathway A) and in the presence of amino acids (Glu and Ala) as well as when catalytic anions (hydrogen carbonate and dihydrogen phosphate) are utilized in pathways (A ⟶ H) is found to be exothermic and exergonic at all levels of theory.
However, the cooperation of Arg in pathway I yielded endothermic and endergonic results at B3LYP/6-31G(d) level of theory. While pathway J, it results in exothermic and exergonic reactions at studied levels of theory but with higher energy than other pathways. As a result, both pathways are not favored for the 3 mC production direction.
According to the results obtained in Table 2, pathways E and G had the lowest thermodynamic parameter values; therefore, they are more spontaneous and plausible reactions to occur.
The solvation process, modeled by SMD, did not decrease but increase the thermodynamical parameters of all pathways.
4. Conclusions
In this work, we have performed an extensive study to investigate and provide mechanistic details concerning the methylation reaction of cytosine on position 3 utilizing accurate quantum chemical DFT calculations. The role of conserved residues and abundant anionic particles (hydrogen carbonate and dihydrogen phosphate) was investigated through designing pathways A ⟶ J for 3 mC generation. The potential energy diagram (PED) for each pathway was constructed employing the B3LYP/6-31G(d), M06-2X, and APFD methods in the gaseous phase besides considering the aqueous environment SMD model. The thermodynamic functions (ΔH and ΔG) and kinetic parameters (Ea, ΔHǂ, and ΔGǂ) were calculated, using these DFT methods, for each proposed pathway. The connections of the TS’s with the I’s, R’s, and P’s of every pathway have been confirmed using the intrinsic reaction coordinate (IRC) calculations.
In this work, we have conducted a mechanistic exploration of the methylation of N3 position at the cytosine ring. Our results suggest that the factors that influence the N3 methylation play different roles in C5 methylation. For instance, for 3 mC formation Ala and Glu showed catalytic activity towards the N3 reaction while Cys and Arg did not show any catalytic activity in the formation of 3 mC. On the other hand, Cys and Arg play a key role in 5 mC formation reaction. Methylation of N3 occurs through electrophilic addition mechanism concerted with proton H4 transfer to the surrounding employed residues, which were Glu, HCO3-, and H2PO4-. The calculated results showed that HCO3- and Ala residue proved a powerful catalytic activity in lowering the activation energy to 59.5 kJ mol−1 employing B3LYP/6-31G(d) level of theory. The integrated proton transfer mechanism between cytosine and the surrounding molecules and methylation of N3 can double the trouble through introducing a serious mutagenic lesion.
The agreement of our results with the experimental findings obtained by Dukatz and coworkers gives credence to our framework, which will pave the road to control these reactions and help drug design processes. Disclosure of the mechanistic details of N3 methylation of cytosine opens the door to clearly understand the recovery process by ALKB2 enzyme.
Disclosure
This project was published as preprints by ChemRxiv to share our early results and findings with colleagues [29].
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Acknowledgments
Mansour H. Almatarneh is grateful to the Deanship of Academic Research at the University of Jordan for the grant. The authors also gratefully acknowledge Compute Canada for the computer time.
Open Research
Data Availability
The optimized structures of all pathways for DNA methylation are available at https://github.com/matarneh/DNA-Methylation to facilitate reproducibility of the results.