Association of Polymorphisms in Vitamin D-Metabolizing Enzymes DHCR7 and CYP2R1 with Cancer Susceptibility: A Systematic Review and Meta-Analysis
Abstract
The deficiency of vitamin D has been reported to be relevant to cancer risk. DHCR7 and CYP2R1 are crucial components of vitamin D-metabolizing enzymes. Thus, accumulating researchers are concerned with the correlation between polymorphisms of DHCR7 and CYP2R1 genes and cancer susceptibility. Nevertheless, the conclusions of literatures are inconsistent. We conducted an integrated review for the correlation of DHCR7 and CYP2R1 SNPs with cancer susceptibility. In the meanwhile, a meta-analysis was performed using accessible data to clarify the association between DHCR7 and CYP2R1 SNPs and overall cancer risk. Literatures which meet the rigid inclusion and exclusion criteria were involved. The association of each SNP with cancer risk was calculated by odds ratios (ORs). 12 case-control designed studies covering 23780 cases and 27307 controls were ultimately evolved in the present meta-analysis of five SNPs (DHCR7 rs12785878 and rs1790349 SNP; CYP2R1 rs10741657, rs12794714, and rs2060793 SNP). We found that DHCR7 rs12785878 SNP was significantly related to cancer risk in the whole population, Caucasian subgroup, and hospital-based (HB) subgroup. DHCR7 rs1790349 SNP was analyzed to increase cancer risk in Caucasians. Moreover, CYP2R1 rs12794714-A allele had correlation with a lower risk of colorectal cancer. Our findings indicated that rs12785878, rs1790349, and rs12794714 SNPs might potentially be biomarkers for cancer susceptibility.
1. Introduction
Vitamin D, also regarded as 1,25-dihydroxyvitamin D3, is a pivotal steroid prohormone which has a significant role to play in musculoskeletal health [1]. Additionally, compelling evidence reveals the roles of vitamin D on extraskeletal diseases, such as infectious disease [2], cardiovascular disease [3], autoimmune disease [4], neurodegeneration [5], and cancer [6]. Deficiency of vitamin D has been reported to be relevant to oral squamous cell carcinoma [7], breast cancer [8], colorectal cancer [9], prostate cancer [10], pancreas cancer [11], thyroid cancer [12], hepatocellular carcinoma [13], and ovarian cancer [14]. Furthermore, vitamin D supplementation may decrease the death of cancer by 16% [15].
There has an individual variability in serum vitamin D stores which cannot be explained alone by age, sunlight exposure, body mass index, or dietary intake [12]. Studies have demonstrated that vitamin D level is highly heritable [16]. Genetic and epigenetic factors can impact several crucial steps along the metabolic pathway of vitamin D. Genes who directly participate in the vitamin D pathway gene are DHCR7, CYP2R1, VDR, CYP24A1, CYP27B1, and so on, and the aberrant expressions of them have been demonstrated to be associated with vitamin D concentrations and cancer [17–21]. Genome-wide association studies (GWAS) have detected the correlations of 25-hydroxyvitamin D concentrations with single nucleotide polymorphisms (SNPs) on genes that participated in the vitamin D metabolic pathway [1, 16].
DHCR7, located on chromosome 11q13.4, encodes ultimate enzyme 7-dehydrocholesterol reductase which catalyzes the conversion of the vitamin D3 precursor (7-dehydrocholesterol) to cholesterol, instead of vitamin D3 [22]. Cytochrome P450 family 2 subfamily R member 1 (CYP2R1, on chromosome 11p15.2) encodes vitamin D 25-hydroxylase which catalyzes the initial hydroxylation reaction of vitamin D synthesis, converting vitamin D to 25-hydroxyvitamin D [9]. Increasing correlational studies were concerned with DHCR7 and CYP2R1 polymorphisms and susceptibility to cancer. Some studies confirmed the associations, whereas others remained skeptical or denied their correlations. The aim of the present study was to explore whether the DHCR7 or CYP2R1 SNPs are related to cancer risk.
We comprehensively reviewed the eligible studies and analyzed all available data. Our aim is to explore the association of DHCR7 and CYP2R1 SNPs with cancer risk, supplying clues to researchers for screening novel cancer biomarkers.
2. Materials and Methods
2.1. Retrieval Strategy
Two investigators (J.W. and J.L.), respectively, carried out a comprehensive literature retrieval in PubMed and Web of Science database up to February 2020, by using the following query terms: “CYP2R1/cytochrome P450 family 2 subfamily R member 1/DHCR7/7-dehydrocholesterol reductase”, “polymorphism/SNP/variant/variation”, and “cancer/carcinoma/neoplasm/tumor/”. All enrolled articles must satisfy inclusion standards: (1) case-control or nested case-control designed study; (2) in regard to the association of DHCR7 and CYP2R1 SNPs with predisposition to cancer. Meanwhile, publications meeting the following exclusion standards were removed: (1) letters or reviews; (2) repeated records; (3) irrelevant to DHCR7 and CYP2R1 SNPs or carcinoma; (4) without any available genotype distribution data.
2.2. Data Extraction
Data was collected by two investigators (J.W. and J.L.) independently and came to a consensus regarding all items. Essential characteristics extracted from each qualified publication comprised first author, year of publication, ethnicity, sample size, type of carcinoma, gene, SNPs, genotype distribution frequency of case and control groups, control group source (hospital-based (HB) or population-based (PB)), Hardy-Weinberg equilibrium (HWE), adjustment factors, and genotyping method. When multiple studies were conducted in one article, data were collected individually.
2.3. Methodology Quality Assessment
Two authors (J.W. and X.L.) scored the quality of each enrolled publication independently, based on a scoring scheme mentioned in prior literature [23, 24]. Six evaluation items were involved in the scoring scheme: representativeness of cases, control source, ascertainment of carcinomas, sample size, HWE in the control group, and quality assurance of genotyping methods. The quality assessment scores ranged from 0 to 10. Study with no less than 5 quality scores was recognized as an eligible study which could be enrolled in subsequent analysis.
2.4. False-Positive Report Probability
False-positive report probability (FPRP) was computed to estimate whether our study findings are “noteworthy.” Initially, we computed the statistic power of the test based on the sample size, ORs, and P values by using NCSS-PASS software (USA, version 11.0.7). Then, we drew the FPRP values from a calculation formula which had been reported in earlier researches, and FPRP < 0.5 was regarded as a noteworthy finding [25].
2.5. Statistical Analysis
The chi-square test (χ2 test) was conducted to compute the HWE for genotype frequency distribution of CYP2R1 and DHCR7 polymorphisms in controls. The correlation of each CYP2R1 and DHCR7 polymorphism with carcinoma risk was computed by odds ratio (OR) with its 95% confidence interval (95% CI). Cochran’s χ2-based Q test was adopted to estimate the heterogeneity of interstudy (significance set as P < 0.10, I2 > 50%). We pooled the results by means of a fixed-effects model when no interstudy heterogeneity arose; the random-effects model was adopted otherwise. Besides, the recessive and dominant genetic models were, respectively, considered as variant homozygote vs. heterozygote/wild homozygote, and heterozygote/variant homozygote vs. wild homozygote. Publication bias was estimated using the rank correlation test (Begg’s test) and linear regression methods (Egger’s test). Sensitivity analysis was calculated to show whether the merged findings were steady enough after removing those outlying studies. All the mentioned statistical analyses were calculated by STATA software (STATA Corp., College Station, TX, USA, version 11.0). All P values were for two-tailed tests, and less than 0.05 was regarded as statistically significant.
3. Results
3.1. Features of Eligible Studies and Analyzed SNPs
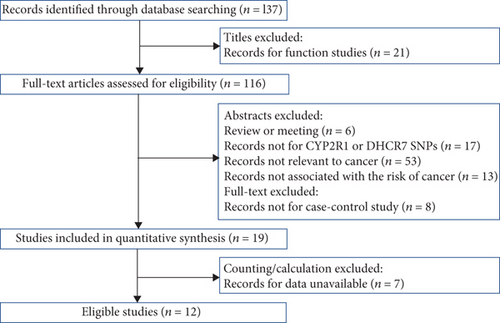
Totally 137 publications were gathered through database retrieval after removing duplicate hits. 125 articles were removed after browsing titles and abstracts: 21 were functional studies; 6 were review or meeting; 8 were not case-control studies; 17 were not related to DHCR7 or CYP2R1 SNPs; 53 were not concerned with carcinoma; and 13 were not correlated with carcinoma risk. Therefore, 19 studies are ought to be involved in the present analysis. Nevertheless, 7 publications lost original data, 5 of which were genome-wide association studies. And we were not able to contact with authors. Thus, 12 case-control designed studies were finally evolved in the present meta-analysis, covering 23780 cases and 27307 controls, which is shown in Figure 1. The features of these eligible studies which met the quality assessment criterion are listed in Table 1.
| No. | First author | Year | Ethnicity | Sample size | Source of control groups | Genotyping method | Adjusted factors | Citation | |
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | ||||||||
| 1 | Isabel S. Carvalho | 2019 | Caucasian (Portugal) | 500 | 500 | PB | PCR-RFLP | Age, sex | [12] |
| 2 | Prajjalendra Barooah | 2019 | Caucasian (Indian) | 60 | 102 | HB | PCR-RFLP | Age, sex | [13] |
| 3 | Jianzhou Yang | 2017 | Asian (China) | 565 | 557 | PB | GenomeLab SNPstream | Age, sex | [35] |
| 4 | Alison M. Mondul | 2015 | Caucasian (European) | 8618 | 9960 | HB | TaqMan or genome-wide scans | Age | [36] |
| 5 | Tess V. Clendenen | 2015 | Caucasian (Swedish) | 733 | 1432 | PB | Illumina GoldenGate assay | Age, menopausal status | [37] |
| 6 | Fabio Pibiri | 2014 | African (African-American) | 902 | 760 | PB | Sequenom MassARRAY | Age, sex, ancestry | [38] |
| 7 | Touraj Mahmoudi | 2014 | Caucasian (Iranian) | 290 | 354 | HB | PCR-RFLP | Age, BMI, sex | [9] |
| 8 | Wei Wang | 2014 | Caucasian (Hispanic) | 826 | 779 | PB | Illumina GoldenGate assay | Age, BMI | |
| Wei Wang | 2014 | Mixed (non-Hispanic) | 224 | 130 | PB | Illumina GoldenGate assay | Age, BMI | [39] | |
| 9 | Christian M. Lange | 2013 | Asian (Japanese) | 803 | 1253 | HB | Competitive allele-specific TaqMan PCR | Sex | [40] |
| 10 | Alison M. Mondul | 2013 | Caucasian | 9378 | 9986 | PB | TaqMan | Age, ethnicity | [41] |
| 11 | Laura N. Anderson | 2013 | Caucasian (Canada) | 628 | 1192 | PB | MassARRAY | Age, sex | [11] |
| 12 | Marissa Penna-Martinez | 2012 | Caucasian (Germany) | 253 | 302 | PB | TaqMan | NM | [42] |
- Note: HB: hospital based; PB: population based; PCR-RFLP: in reaction-restriction fragment length polymorphism; NM: not mentioned.
Six polymorphisms were able to be involved in our systematic review, including rs10741657 G/A, rs12794714 G/A, rs2060793 G/A, rs3829251 G/A, rs12785878 T/G, and rs1790349 A/G. The frequency distribution of DHCR7 and CYP2R1 SNPs genotype is shown in Table 2. Six records, however, were removed from quantitative synthesis owing to the insufficient study number for some loci or being not conformed to HWE (PHWE < 0.05). Consequently, five SNPs were covered in the eventual meta-analysis. For DHCR7, the analyzed SNPs were rs12785878 T/G and rs1790349 A/G; for CYP2R1, the analyzed SNPs were rs10741657 G/A, rs12794714 G/A, and rs2060793 G/A.
| First author | Year | Gene | SNPsa | Type of cancer | Ethnicity | Sample size | Case | Control | Quality score | PHWE | Included in meta-analysis | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Homozygote wild | Heterozygote | Homozygote variant | Homozygote wild | Heterozygote | Homozygote variant | |||||||||
| Isabel S. Carvalho | 2019 | CYP2R1 | rs2060793 (G/A) | TC | Caucasian (Portugal) | 500 | 500 | 189 | 236 | 75 | 183 | 256 | 61 | 7 | 0.047 | Noc |
| 2019 | DHCR7 | rs12785878 (T/G) | TC | Caucasian (Portugal) | 500 | 500 | 150 | 251 | 99 | 197 | 234 | 69 | 7 | 0.971 | Yes | |
| Prajjalendra Barooah | 2019 | CYP2R1 | rs10741657 (G/A) | HCC | Caucasian (Indian) | 60 | 102 | 28 | 23 | 9 | 51 | 35 | 16 | 4.5 | 0.025 | Noc |
| Jianzhou Yang | 2017 | DHCR7 | rs3829251 (G/A) | ESCC | Asian (China) | 565 | 557 | 302 | 218 | 45 | 283 | 232 | 42 | 8 | 0.557 | Nob |
| Alison M. Mondul | 2015 | CYP2R1 | rs10741657 (G/A) | BC | Caucasian (European) | 8618 | 9960 | 3276 | 4041 | 1301 | 3708 | 4766 | 1486 | 6.5 | 0.475 | Yes |
| 2015 | DHCR7 | rs12785878 (T/G) | BC | Caucasian (European) | 9224 | 10560 | 4935 | 3620 | 669 | 5674 | 4052 | 834 | 5.5 | 0.003 | Noc | |
| Tess V. Clendenen | 2015 | CYP2R1 | rs10741657 (G/A) | BC | Caucasian (Swedish) | 733 | 1432 | 200 | 358 | 175 | 405 | 720 | 307 | 7.5 | 0.696 | Yes |
| 2015 | DHCR7 | rs12785878 (T/G) | BC | Caucasian (Swedish) | 733 | 1433 | 273 | 371 | 89 | 571 | 659 | 203 | 7.5 | 0.562 | Yes | |
| 2015 | DHCR7 | rs1790349 (A/G) | BC | Caucasian (Swedish) | 732 | 1431 | 348 | 326 | 58 | 752 | 571 | 108 | 7.5 | 0.978 | Yes | |
| Fabio Pibiri | 2014 | CYP2R1 | rs12794714 (G/A) | CRC | African (African-American) | 902 | 760 | 638 | 253 | 11 | 501 | 239 | 20 | 9.5 | 0.175 | Yes |
| Touraj Mahmoudi | 2014 | CYP2R1 | rs12794714 (G/A) | CRC | Caucasian (Iranian) | 290 | 354 | 93 | 135 | 62 | 110 | 167 | 77 | 7 | 0.364 | Yes |
| Wei Wang | 2014 | CYP2R1 | rs2060793 (G/A) | BC | Mixed (Hispanic) | 826 | 779 | 303 | 391 | 132 | 315 | 356 | 108 | 9.5 | 0.644 | Yes |
| 2014 | CYP2R1 | rs2060793 (G/A) | Caucasian (non-Hispanic) | 224 | 130 | 89 | 104 | 31 | 51 | 58 | 21 | 8.5 | 0.512 | Yes | ||
| 2014 | DHCR7 | rs12785878 (T/G) | BC | Mixed (Hispanic) | 826 | 779 | 189 | 410 | 227 | 185 | 353 | 241 | 7.5 | 0.013 | Noc | |
| 2014 | DHCR7 | rs12785878 (T/G) | BC | Caucasian (non-Hispanic) | 224 | 130 | 123 | 90 | 11 | 62 | 55 | 13 | 8.5 | 0.876 | Yes | |
| 2014 | DHCR7 | rs1790349 (A/G) | BC | Mixed (Hispanic) | 826 | 779 | 573 | 227 | 26 | 527 | 227 | 25 | 8.5 | 0.927 | Yes | |
| 2014 | DHCR7 | rs1790349 (A/G) | BC | Caucasian (non-Hispanic) | 224 | 130 | 168 | 52 | 4 | 95 | 30 | 5 | 8.5 | 0.195 | Yes | |
| Christian M. Lange | 2013 | CYP2R1 | rs10741657 (G/A) | HCC | Asian (Japanese) | 803 | 1253 | 320 | 377 | 106 | 482 | 597 | 174 | 8.5 | 0.615 | Yes |
| 2013 | DHCR7 | rs12785878 (T/G) | HCC | Asian (Japanese) | 803 | 1253 | 84 | 336 | 383 | 153 | 543 | 557 | 8.5 | 0.247 | Yes | |
| 2013 | DHCR7 | rs12785878 (T/G) | HCC | Caucasian (German) | 116 | 208 | 63 | 44 | 9 | 113 | 77 | 18 | 8.5 | 0.353 | Yes | |
| Alison M. Mondul | 2013 | CYP2R1 | rs10741657 (G/A) | PC1 | Caucasian | 9378 | 9986 | 3481 | 4392 | 1505 | 3789 | 4667 | 1530 | 9 | 0.137 | Yes |
| 2013 | DHCR7 | rs12785878 (T/G) | PC1 | Caucasian | 9620 | 10225 | 4979 | 3816 | 825 | 5221 | 4047 | 957 | 8 | 2.401E − 05 | Noc | |
| Laura N. Anderson | 2013 | CYP2R1 | rs10741657 (G/A) | PC2 | Caucasian (Canada) | 625 | 1191 | 262 | 286 | 77 | 451 | 550 | 190 | 8 | 0.304 | Yes |
| 2013 | CYP2R1 | rs12794714 (G/A) | PC2 | Caucasian (Canada) | 628 | 1192 | 180 | 307 | 141 | 399 | 559 | 234 | 8 | 0.131 | Yes | |
| Marissa Penna-Martinez | 2012 | CYP2R1 | rs10741657 (G/A) | TC | Caucasian (German) | 253 | 302 | 96 | 110 | 47 | 119 | 139 | 44 | 7.5 | 0.742 | Yes |
| 2012 | CYP2R1 | rs12794714 (G/A) | TC | Caucasian (German) | 253 | 302 | 78 | 130 | 45 | 94 | 144 | 64 | 7.5 | 0.522 | Yes | |
- Note: PHWE: the P value for Hardy-Weinberg equilibrium in control groups; amajor/minor; bexcluded due to the limited number for this locus; cexcluded due to the SNP not being in accordance with HWE. The results are in bold if P < 0.05; TC = thyroid cancer; HCC = hepatocellular carcinoma; ESCC = esophageal squamous cell carcinoma; BC = breast cancer; PC1 = prostate cancer; PC2 = pancreas cancer.
3.2. Quantitative Data Synthesis of Five SNPs in DHCR7 and CYP2R1 Genes
3.2.1. Two Polymorphisms in DHCR7 Gene
Five eligible studies were collected to evaluate the relationships between DHCR7 SNPs and risk of carcinoma, on the basis of entire population. The rs12785878 T/G SNP was illustrated to be associated with incremental cancer risk. The correlation of rs12785878 T/G SNP was discovered under the heterozygote genotype model (TG vs. TT: OR (95%CI) = 1.168 (1.027-1.328), P = 0.018, Table 3). The relationship between rs1790349 A/G SNP and carcinoma risk was not found in the initial analysis.
| Stratification | N | Heterozygote vs. wild-type | Mutation homozygote vs. wild-type | Dominant model | Recessive model | Allelic model | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | I2 (%) | OR (95% CI) | P | I2 (%) | OR (95% CI) | P | I2 (%) | OR (95% CI) | P | I2(%) | OR (95% CI) | P | I2 (%) | ||
| CYP2R1 | ||||||||||||||||
| rs10741657 (G/A) | 6 | 0.987 (0.947-1.028) | 0.522 | 0 | 1.006 (0.906-1.117) | 0.905 | 50.9 | 0.995 (0.957-1.034) | 0.799 | 18.4 | 1.030 (0.978-1.084) | 0.264 | 41.2 | 0.998 (0.949-1.050) | 0.943 | 51.2 |
| Ethnicity | ||||||||||||||||
| Caucasian | 5 | 0.989 (0.948-1.031) | 0.587 | 0 | 1.016 (0.905-1.142) | 0.785 | 58.2 | 0.997 (0.959-1.038) | 0.901 | 30.8 | 1.030 (0.936-1.133) | 0.543 | 50.2 | 1.003 (0.948-1.061) | 0.917 | 58.5 |
| Asian | 1 | 0.951 (0.786-1.152) | 0.608 | NA | 0.918 (0.694-1.214) | 0.547 | NA | 0.944 (0.787-1.131) | 0.531 | NA | 0.943 (0.727-1.223) | 0.658 | NA | 0.957 (0.840-1.089) | 0.503 | NA |
| Type of cancer | ||||||||||||||||
| Breast cancer | 2 | 0.963 (0.907-1.023) | 0.227 | 0 | 1.008 (0.927-1.095) | 0.858 | 20.7 | 0.974 (0.920-1.031) | 0.361 | 0 | 1.030 (0.955-1.111) | 0.442 | 14.9 | 0.995 (0.957-1.036) | 0.817 | 31.1 |
| Pancreas cancer 2 | 1 | 0.895 (0.726-1.103) | 0.289 | NA | 0.698 (0.514-0.947) | 0.021 | NA | 0.844 (0.693-1.029) | 0.093 | NA | 0.740 (0.557-0.984) | 0.038 | NA | 0.848 (0.736-0.978) | 0.023 | NA |
| Prostate cancer | 1 | 1.024 (0.963-1.090) | 0.445 | NA | 1.071 (0.984-1.165) | 0.114 | NA | 1.036 (0.977-1.098) | 0.236 | NA | 1.057 (0.978-1.142) | 0.165 | NA | 1.033 (0.992-1.076) | 0.118 | NA |
| Hepatocellular carcinoma | 1 | 0.951 (0.786-1.152) | 0.608 | NA | 0.918 (0.694-1.214) | 0.547 | NA | 0.944 (0.787-1.131) | 0.531 | NA | 0.943 (0.727-1.223) | 0.658 | NA | 0.957 (0.840-1.089) | 0.503 | NA |
| Thyroid caner | 1 | 0.981 (0.679-1.416) | 0.918 | NA | 1.324 (0.810-2.164) | 0.263 | NA | 1.063 (0.755-1.499) | 0.725 | NA | 1.338 (0.853-2.098) | 0.205 | NA | 1.122 (0.881-1.429) | 0.352 | NA |
| Source of controls | ||||||||||||||||
| HB | 2 | 0.959 (0.903-1.018) | 0.169 | 0 | 0.984 (0.905-1.070) | 0.709 | 0 | 0.965 (0.912-1.021) | 0.214 | 0 | 1.007 (0.933-1.088) | 0.85 | 0 | 0.984 (0.946-1.024) | 0.437 | 0 |
| PB | 4 | 1.012 (0.957-1.071) | 0.677 | 0 | 1.020 (0.829-1.256) | 0.849 | 64.6 | 1.022 (0.969-1.078) | 0.415 | 23.5 | 1.032 (0.867-1.229) | 0.723 | 60.7 | 1.007 (0.913-1.110) | 0.89 | 62.3 |
| rs12794714 (G/A) | 4 | 1.007 (0.825-1.231) | 0.942 | 51 | 0.908 (0.609-1.352) | 0.634 | 68 | 0.993 (0.789-1.249) | 0.953 | 66.2 | 0.907 (0.664-1.239) | 0.539 | 58.9 | 0.968 (0.807-1.160) | 0.723 | 72.5 |
| Ethnicity | ||||||||||||||||
| Caucasian | 3 | 1.127 (0.951-1.336) | 0.167 | 0 | 1.134 (0.921-1.397) | 0.236 | 40.9 | 1.130 (0.964-1.326) | 0.132 | 10.5 | 1.056 (0.881-1.265) | 0.558 | 24.8 | 1.074 (0.967-1.192) | 0.183 | 41.2 |
| African | 1 | 0.831 (0.672-1.028) | 0.088 | NA | 0.432 (0.205-0.910) | 0.027 | NA | 0.800 (0.650-0.985) | 0.036 | NA | 0.457 (0.217-0.959) | 0.039 | NA | 0.800 (0.666-0.960) | 0.017 | NA |
| Type of cancer | ||||||||||||||||
| Colorectal cancer | 2 | 0.862 (0.718-1.035) | 0.111 | 0 | 0.681 (0.317-1.466) | 0.326 | 69.1 | 0.841 (0.705-1.003) | 0.054 | 0 | 0.717 (0.344-1.493) | 0.374 | 68.9 | 0.866 (0.753-0.997) | 0.046 | 44.1 |
| Pancreas cancer | 1 | 1.217 (0.973-1.524) | 0.086 | NA | 1.336 (1.016-1.775) | 0.038 | NA | 1.252 (1.014-1.546) | 0.036 | NA | 1.185 (0.936-1.500) | 0.157 | NA | 1.167 (1.017-1.339) | 0.028 | NA |
| Thyroid caner | 1 | 1.088 (0.742-1.595) | 0.666 | NA | 0.847 (0.522-1.377) | 0.504 | NA | 1.014 (0.706-1.455) | 0.94 | NA | 0.805 (0.526-1.230) | 0.315 | NA | 0.939 (0.740-1.191) | 0.604 | NA |
| Source of controls | ||||||||||||||||
| HB | 1 | 0.956 (0.669-1.367) | 0.806 | NA | 0.952 (0.617-1.469) | 0.825 | NA | 0.955 (0.684-1.333) | 0.787 | NA | 0.978 (0.671-1.427) | 0.909 | NA | 0.973 (0.780-1.213) | 0.806 | NA |
| PB | 3 | 1.023 (0.787-1.331) | 0.864 | 66.9 | 0.856 (0.476-1.538) | 0.602 | 78 | 1.004 (0.742-1.360) | 0.978 | 77.3 | 0.839 (0.522-1.348) | 0.468 | 72.5 | 0.963 (0.754-1.231) | 0.765 | 81.6 |
| rs2060793 (G/A) | 2 | 1.121 (0.923-1.362) | 0.247 | 0 | 1.184 (0.902-1.554) | 0.223 | 19 | 1.136 (0.946-1.364) | 0.172 | 0 | 1.113 (0.866-1.430) | 0.402 | 6.3 | 1.098 (0.964-1.250) | 0.16 | 8.4 |
| DHCR7 | ||||||||||||||||
| rs12785878 (T/G) | 5 | 1.168 (1.027-1.328) | 0.018 | 8.3 | 1.074 (0.736-1.569) | 0.71 | 73.1 | 1.136 (0.935-1.381) | 0.2 | 53.2 | 1.017 (0.762-1.357) | 0.91 | 67.5 | 1.064 (0.906-1.250) | 0.448 | 70.1 |
| Ethnicity | ||||||||||||||||
| Caucasian | 4 | 1.178 (1.021-1.358) | 0.024 | 30.1 | 0.980 (0.562-1.710) | 0.944 | 79.2 | 1.108 (0.854-1.436) | 0.441 | 64.8 | 0.929 (0.584-1.477) | 0.756 | 73.7 | 1.031 (0.814-1.305) | 0.802 | 77.2 |
| Asian | 1 | 1.127 (0.836-1.520) | 0.433 | NA | 1.252 (0.931-1.684) | 0.136 | NA | 1.191 (0.898-1.579) | 0.226 | NA | 1.139 (0.954-1.361) | 0.15 | NA | 1.120 (0.980-1.281) | 0.097 | NA |
| Type of cancer | ||||||||||||||||
| Breast cancer | 2 | 1.048 (0.756-1.454) | 0.778 | 50.1 | 0.699 (0.341-1.433) | 0.328 | 63.5 | 0.961 (0.658-1.404) | 0.838 | 63.9 | 0.795 (0.616-1.025) | 0.077 | 42.4 | 0.899 (0.665-1.215) | 0.488 | 66.1 |
| Hepatocellular carcinoma | 2 | 1.098 (0.851-1.415) | 0.472 | 0 | 1.209 (0.913-1.599) | 0.185 | 0 | 1.135 (0.893-1.442) | 0.302 | 0 | 1.127 (0.947-1.341) | 0.177 | 0 | 1.102 (0.972-1.250) | 0.129 | 0 |
| Thyroid caner | 1 | 1.409 (1.068-1.859) | 0.015 | NA | 1.884 (1.297-2.738) | 0.001 | NA | 1.517 (1.167-1.972) | 0.002 | NA | 1.542 (1.102-2.158) | 0.012 | NA | 1.376 (1.150-1.645) | <0.001 | NA |
| Source of controls | ||||||||||||||||
| HB | 2 | 1.098 (0.852-1.415) | 0.471 | 0 | 1.209 (0.913-1.599) | 0.185 | 0 | 1.135 (0.893-1.442) | 0.302 | 0 | 1.127 (0.947-1.341) | 0.177 | 0 | 1.102 (0.972-1.250) | 0.129 | 0 |
| PB | 3 | 1.193 (1.028-1.385) | 0.02 | 49.3 | 0.988 (0.501-1.945) | 0.971 | 85.9 | 1.125 (0.815-1.552) | 0.474 | 75.3 | 0.925 (0.528-1.623) | 0.787 | 82.3 | 1.038 (0.777-1.388) | 0.8 | 84.5 |
| rs1790349 (A/G) | 3 | 1.060 (0.850-1.323) | 0.605 | 52 | 1.056 (0.793-1.407) | 0.71 | 0 | 1.043 (0.837-1.300) | 0.705 | 55.1 | 0.998 (0.754-1.319) | 0.986 | 0 | 1.048 (0.942-1.167) | 0.391 | 45.7 |
| Ethnicity | ||||||||||||||||
| Caucasian | 2 | 1.201 (1.008-1.431) | 0.04 | 0 | 1.094 (0.784-1.526) | 0.598 | 44 | 1.180 (0.998-1.396) | 0.053 | 21.2 | 1.003 (0.727-1.386) | 0.983 | 30.6 | 1.110 (0.972-1.266) | 0.122 | 37.4 |
| Mixed | 1 | 0.920 (0.739-1.145) | 0.453 | NA | 0.957 (0.545-1.677) | 0.877 | NA | 0.923 (0.748-1.140) | 0.458 | NA | 0.980 (0.561-1.712) | 0.944 | NA | 0.940 (0.783-1.128) | 0.505 | NA |
- Note: OR: odds ratio; CI: confidence interval. The results are in bold if P < 0.05.
In stratified analyses, rs12785878 T/G SNP was quantitatively analyzed in “ethnicity,” “type of carcinoma,” and “source of control group” subgroups, and the rs1790349 A/G SNP was analyzed in the “ethnicity” subgroup. For rs12785878 T/G SNP, correlations calculated under the heterozygote genotype model (TG vs. TT) were observed in “Caucasian population” and “PB” subgroups (Caucasian: OR (95%CI) = 1.178 (1.021-1.358), P = 0.024; PB: OR (95%CI) = 1.193 (1.028-1.385), P = 0.020, Table 3). For rs1790349 A/G SNP, association was only manifested in the “Caucasian population” subgroup (AG vs. AA: OR (95%CI) = 1.201 (1.008-1.431), P = 0.040, Table 3).
3.2.2. Three Polymorphisms in CYP2R1 Gene
Nine eligible publications were involved to estimate the association intensity of CYP2R1 polymorphisms and overall carcinoma risk. Nevertheless, none of these SNPs manifest significant correlations with risk of carcinoma in any genetic models.
Then, stratified analyses of rs10741657 G/A and rs12794714 G/A SNPs were conducted based on “ethnicity,” “type of carcinoma,” and “source of control group,” on account of the presence of between-study heterogeneity. For rs12794714 G/A SNP, its allelic models had correlation with a decreased genetic predisposition to colorectal cancer (A vs. G: OR (95%CI) = 0.866 (0.753-0.997), P = 0.046, Table 3). Correlations could not be elucidated among any of the stratified analyses of rs10741657 G/A SNP.
3.3. Sensitivity Analysis
Sensitivity analysis was adopted to assess the impact of each study on summarized findings, by means of calculating the OR (95% CI) before and after deleting each article from the pooled analysis. For rs12785878 T/G SNP, it made no sense after the removal of two articles (Isabel S. Carvalho 2019, Tess V. Clendenen 2015) individually (Supplementary Table S1).
3.4. Publication Bias
Potential publication bias was evaluated for all covered publications by means of two test methods mentioned above. The publication bias was found in rs12794714 G/A SNP under the recessive model, for P < 0.1 in both tests, which might be because of the deficient publications with negative results or the defective methodological design for small-scale studies (Table 4).
| Comparison type | Begg’s test | Egger’s test | ||
|---|---|---|---|---|
| Z value | P value | t value | P value | |
| CYP2R1 rs10741657 (G/A) | ||||
| Heterozygote vs. homozygote wild | 0 | 1 | -0.7 | 0.521 |
| Homozygote variant vs. homozygote wild | 0.38 | 0.707 | -0.73 | 0.503 |
| Dominant model | 0 | 1 | -0.53 | 0.627 |
| Recessive model | 0 | 1 | -0.29 | 0.787 |
| Allelic model | 0.75 | 0.452 | -0.38 | 0.722 |
| CYP2R1 rs12794714 (G/A) | ||||
| Heterozygote vs. homozygote wild | 0.34 | 0.734 | 0.21 | 0.851 |
| Homozygote variant vs. homozygote wild | 1.02 | 0.308 | -2.84 | 0.105 |
| Dominant model | 0.34 | 0.734 | -0.01 | 0.994 |
| Recessive model | 1.7 | 0.089 | -9.45 | 0.011 |
| Allelic model | 0.34 | 0.734 | -1.12 | 0.38 |
| CYP2R1 rs2060793 (G/A) | ||||
| Heterozygote vs. homozygote wild | 0 | 1 | NA | NA |
| Homozygote variant vs. homozygote wild | 0 | 1 | NA | NA |
| Dominant model | 0 | 1 | NA | NA |
| Recessive model | 0 | 1 | NA | NA |
| Allelic model | 0 | 1 | NA | NA |
| DHCR7 rs12785878 (T/G) | ||||
| Heterozygote vs. homozygote wild | 0.24 | 0.806 | -1.64 | 0.2 |
| Homozygote variant vs. homozygote wild | -0.24 | 1 | -1.76 | 0.177 |
| Dominant model | -0.24 | 1 | -1.74 | 0.18 |
| Recessive model | 0.24 | 0.806 | -1.15 | 0.332 |
| Allelic model | 0.24 | 0.806 | -1.56 | 0.217 |
| DHCR7 rs1790349 (A/G) | ||||
| Heterozygote vs. homozygote wild | 0 | 1 | 0.18 | 0.884 |
| Homozygote variant vs. homozygote wild | 0 | 1 | 0.13 | 0.92 |
| Dominant model | 0 | 1 | -0.44 | 0.737 |
| Recessive model | 0 | 1 | -2.34 | 0.257 |
| Allelic model | 1.04 | 0.296 | -0.9 | 0.532 |
- Note: the results are in bold if P < 0.1.
3.5. FPRP Analyses
Eventually, we assessed the FPRP for our significant findings. For studies of uncommon neoplasm or common tumors with small sample size, the FRPR value less than 0.5 would make a massive improvement over previous practice, based on the professional guide of FPRP calculation. Since the present study is the first meta-analysis to estimate the association between DHCR7 and CYP2R1 SNPs and cancer risk, we consider 0.5 as the FPRP threshold. The FPRP values of rs12785878 SNP (prior probability 0.25/0.1) were less than 0.5, and FPRP values of rs1790349 and rs12794714 SNPs were also less than 0.5 (prior probability 0.25), suggesting these significant associations are deserving of attention (Table 5).
| Genotype | OR (95% CI) | P value | Statistical powera | Prior probabilityb | ||||
|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| rs12785878 (T/G) | ||||||||
| GT vs. TT (overall) | 1.168 (1.027-1.328) | 0.018 | 0.312 | 0.235 | 0.390 | 0.853 | 0.983 | 0.998 |
| GT vs. TT (Caucasian) | 1.178 (1.021-1.358) | 0.024 | 0.271 | 0.321 | 0.496 | 0.899 | 0.989 | 0.999 |
| GT vs. TT (PB) | 1.193 (1.028-1.385) | 0.02 | 0.264 | 0.288 | 0.457 | 0.885 | 0.986 | 0.999 |
| rs1790349 (A/G) | ||||||||
| GA vs. AA (Caucasian) | 1.201 (1.008-1.431) | 0.04 | 0.290 | 0.424 | 0.605 | 0.933 | 0.993 | 0.999 |
| rs12794714 (G/A) | ||||||||
| AA vs. GG (CRC) | 0.866 (0.753-0.997) | 0.046 | 0.367 | 0.401 | 0.582 | 0.927 | 0.992 | 0.999 |
- Note: CI: confidence interval; OR: odds ratio; astatistical power was computed using the sample size of case and control, OR, and P values; bthe false-positive report probability is in italics if the value < 0.5.
4. Discussion
In the present article, a comprehensive review was performed for the correlation of SNPs in DHCR7 and CYP2R1 genes with overall cancer risk. And a meta-analysis was conducted for five prevalent SNPs (DHCR7: rs12785878 T/G and rs1790349 A/G; CYP2R1: rs10741657 G/A, rs12794714 G/A, and rs2060793 G/A) for the first time. Our findings showed that rs12785878, rs1790349, and rs12794714 SNPs were related to cancer susceptibility in the whole population or in some subgroups, which means they might participate in cancerogenesis. No associations were discovered in other polymorphisms.
4.1. Polymorphisms in DHCR7
DHCR7 encodes an enzyme 7-dehydrocholesterol reductase which converts 7-dehydrocholesterol into cholesterol. This enzyme is a critical regulatory switch between vitamin D3 and cholesterol, for both biosynthesis processes require 7-dehydrocholesterol as substrate [26]. Moreover, DHCR7 has been assumed to be a correlated gene for vitamin D concentration and carcinoma risk [1].
Regarding rs12785878 T/G, it has been illustrated to be a 25(OH) D concentration-related SNP [1]. We found significant correlations between rs12785878 SNP and cancer susceptibility in the whole population, Caucasian subgroup, and population-based subgroup. rs12785878 SNP is located 8000 bases upstream from 5 prime UTR region of DHCR7, and it is still unclear whether it has an impact on gene expression or has a linkage disequilibrium with some other functional SNPs. The present meta-analysis of rs12785878 SNP encompasses 5 case-control studies. Only one of the five studies, however, was in accordance with our consequence. For the rs1790349 A/G SNP, it was computed to be associated with cancer risk in the Caucasian subgroup under heterozygote genotype. The rs1790349 SNP is located in the intergenic region near DHCR7 and has also been identified to be a 25(OH) D concentration-associated SNP in genome-wide association study [16, 27, 28]. Our analysis of rs1790349 SNP involves only 2 case-control studies, so further expansion of sample volume is needed.
4.2. Polymorphisms in CYP2R1
CYP2R1, as a vital important 25-hydroxylase, metabolizes vitamin D to 25(OH) D in the liver [29]. The genetic variations in CYP2R1 were correlated with the impaired activity of 25-hydroxylases, which influence the serum 25(OH) D level [30]. Association of serum 25(OH) D level with cancer susceptibility has been revealed in breast cancer [20], gastric cancer [31], thyroid cancer [32], prostate cancer [33], colorectal cancer [34], and so on. Thus, accumulating researchers were concerned with the correlation between CYP2R1 SNPs and cancer susceptibility.
For rs12794714 (G/A) SNP, we analyzed a significant relationship between A allele-rs12794714 SNP and decreased risk of colorectal cancer (CRC). Located in exon 1 region of CYP2R1, rs12794714 G/A SNP may function as an exon splicing enhancer (ESE)/exon splicing silencer (ESS) to impact gene expression, whereas it is a synonymous variant (https://snpinfo.niehs.nih.gov/). The A allele-rs12794714 SNP has been illustrated to be associated with higher serum 25-hydroxyviatamin D concentrations [16]; thus, it may reduce the cancer risk. Thus far, the protective effect of rs12794714 has only been demonstrated in CRC. Further studies remain desired concerning rs12794714 and cancer.
4.3. Limitations and Conclusions
It ought to be mentioned that the present study has several limitations. First and foremost, association studies of DHCR7 and CYP2R1 polymorphisms with cancer predisposition remain limited. Further researches are demanded for updated meta-analyses. Moreover, several items without accessible original records were removed from ultimate analysis, which might cause publication bias.
Overall, we comprehensively assessed the correlation of DHCR7 and CYP2R1 SNPs with carcinoma risk. Additionally, a meta-analysis was conducted based on all accessible data for five polymorphisms. The consequence demonstrated that 3 (re12794714, rs12785878, and rs1790349) of the 5 SNPs were associated with cancer risk in whole population or in some subgroups, indicating that they might be feasible biomarkers for cancer susceptibility.
Abbreviations
-
- SNP:
-
- Single nucleotide polymorphism
-
- GWAS:
-
- Genome-wide association studies
-
- ORs:
-
- Odds ratios
-
- CI:
-
- Confidence intervals
-
- HWE:
-
- Hardy-Weinberg equilibrium
-
- FPRP:
-
- False-positive report probability
-
- ESE:
-
- Exon splicing enhancer
-
- ESS:
-
- Exon splicing silencer.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Authors’ Contributions
Aiping Wang conceived and designed the study. Jing Wen and Lia Li were responsible for the data extraction. Jing Wen and Xinyuan Liang were responsible for the quality assessment. Jing Wen and Aiping Wang wrote the manuscript, and Aiping Wang revised the manuscript.
Open Research
Data Availability
The authors declare that all relevant data are presented within the paper.