Identification of Chemical Components of Qi-Fu-Yin and Its Prototype Components and Metabolites in Rat Plasma and Cerebrospinal Fluid via UPLC-Q-TOF-MS
Abstract
Qi-Fu-Yin, a traditional Chinese medicine formula, has been used to treat Alzheimer’s disease (AD, a neurodegenerative disorder) in clinical setting. In this study, the chemical components of Qi-Fu-Yin and its prototype components and metabolites in rat plasma and cerebrospinal fluid, after oral administration, were preliminarily characterized via ultrahigh-performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry (UPLC-Q-TOF-MS). A total of 180 compounds, including saponins, flavonoids, organic acids, sucrose esters, oligosaccharide esters, phthalides, phenylethanoid glycosides, alkaloids, xanthones, terpene lactones, ionones, and iridoid glycoside, were tentatively characterized. For the first time, 51 prototypical components and 26 metabolites, including saponins, phthalides, flavonoids, sucrose esters, organic acids, alkaloids, ionones, terpene lactones, iridoid glycoside, and their derivatives, have been tentatively identified in the plasma. Furthermore, 10 prototypical components (including butylidenephthalide, butylphthalide, 20(S)-ginsenoside Rh1, 20(R)-ginsenoside Rh1, and zingibroside R1) and 6 metabolites were preliminarily characterized in cerebrospinal fluid. These results were beneficial to the discovery of the active components of Qi-Fu-Yin anti-AD.
1. Introduction
Traditional Chinese medicine (TCM) plays a vital role in the treatment of various complex chronic diseases owing to the synergistic effects of the formulations and has, accordingly, garnered increasing attention worldwide [1, 2]. Qi-Fu-Yin, a TCM prescription, was first recorded in the book Jingyue Encyclopedia written by Jingyue Zhang during the Ming Dynasty. It is composed of seven herbs—Ginseng Radix et Rhizoma (GRR), Rehmanniae Radix Preparata (RRP), Angelicae Sinensis Radix (ASR), Atractylodis Macrocephala Rhizoma Preparata (ARP), Glycyrrhizae Radix et Rhizoma Preparata cum Melle (GRP), Ziziphi Spinosae Semen (ZSS), and Polygalae Radix Preparata (PRP)—in a ratio of 6 : 9 : 9 : 5 : 3 : 6 : 5 [3]. Qi-Fu-Yin has shown significant effects on Alzheimer’s disease (AD) in clinical studies [4, 5]. Owing to its remarkable therapeutic effects and pharmacological activities, Qi-Fu-Yin has attracted the attention of various researchers. Previous studies showed that Qi-Fu-Yin improves the learning ability and memory of rats injected with advanced glycation end products [6, 7] or β-amyloid protein [8, 9]. Furthermore, 154 chemical components were unambiguously identified or tentatively characterized in Qi-Fu-Yin using ultrahigh-performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry (UHPLC-Q-TOF-MS) [10]. However, it remains unknown which components are absorbed into the plasma and brain after oral administration of Qi-Fu-Yin, which hinders the elucidation of its potentially bioactive constituents and the underlying action mechanisms.
AD is a neurodegenerative disease characterized by the deposition of Aβ and the formation of neurofibrillary tangles in the brain [11]. The ingredients absorbed into blood and that reach a certain concentration can reportedly exert pharmacodynamic effects [12]. The blood-brain barrier (BBB) allows different components to reach the brain and prevents harmful substances from entering the brain. Drugs passing through the BBB can play important roles in brain diseases [13]. Some biotransformed metabolites possess substantial bioactivities and can act as active components [14]. Thus, it is essential to detect components absorbed into blood and elucidate their metabolic profile, which could reveal the pharmacologically active substances and provide potential resources for discovering new drugs from TCM. In this study, a three-step approach based on UHPLC-Q-TOF-MS was implemented to analyze the multicomponent metabolic profiles of Qi-Fu-Yin in rat plasma and cerebrospinal fluid. First, the Qi-Fu-Yin in vitro chemical component database was established by consulting literature on Qi-Fu-Yin and its seven constituent herbs. The components in vitro were identified by their corresponding MS/MS fragment ions in standard solutions and databases. Second, the database of the prototype components was established to characterize the prototypical components in rat plasma and cerebrospinal fluid after oral administration of Qi-Fu-Yin. Under the same LC-MS conditions, the prototype components were identified by comparing the standard solutions, extracts, control, and administered biological samples in parallel. Finally, according to the metabolic pathway and secondary mass spectrometry data of prototype components reported in the literature, the metabolites of Qi-Fu-Yin in plasma and cerebrospinal fluid were tentatively characterized (Figure 1).
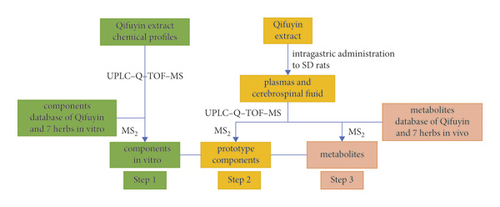
2. Materials and Methods
2.1. Materials and Reagents
GRR, RRP, ASR, ARP, and GRP were purchased from Anxing Traditional Chinese Medicine Co., Ltd. (Anguo, China); ZSS and PRP were purchased from Juyaotang Co., Ltd. (Anguo, China); reference standards of ferulic acid, liquiritin, spinosin, acteoside, 3,6′-disinapoyl sucrose, ginsenoside Rg1 (G-Rg1), ginsenoside Re (G-Re), ginsenoside Rb1 (G-Rb1), tenuifolin, and glycyrrhizic acid were purchased from the National Institute for Food and Drug Control (Beijing, China). Acetonitrile and formic acid were of HPLC grade (Fisher, Carlsbad, CA, USA). Deionized water was prepared using a Milli-Q purification system (Millipore, Bedford, MA, USA). Sodium formate was purchased from Waters (Milford, MA, USA).
2.2. Preparation of Samples of Qi-Fu-Yin and the Seven Herbs
Qi-Fu-Yin was prepared in the laboratory according to the prescribed protocol [3]. Dried pieces of GRR, RRP, ASR, ARP, GRP, ZSS (crushed), and PRP were accurately weighed and immersed in 9 times amount of water for 30 min; then, the samples were serially decocted with 9 times and 7 times amount of water. After mixing and filtering, the extracts were concentrated to a small volume and lyophilized. An appropriate amount of the lyophilized powder was accurately weighed, dissolved in ultrapure water (equivalent to 50 mg crude drug per mL) in a 25 mL volumetric flask, and mixed evenly via ultrasonication for 30 min. Then, the extracts were centrifuged at 13000 rpm and 4°C for 10 min and filtered through a 0.22 µm membrane. The seven herb samples of Qi-Fu-Yin were prepared in the same manner as the prescribed method.
2.3. Animals and Drug Administration
Male SD rats, weighing 200 ± 20 g, were purchased from Beijing Wei Tong Li Hua Experimental Animal Technology Co., Ltd. (Beijing, China). All animal procedures were approved by the Shandong University of Traditional Chinese Medicine Institutional Animal Experimentation Committee (SDUTCM20210119001). All rats were housed at an ambient temperature of 20 ± 1°C with a 12 h light/dark cycle and fed a standard diet and water ad libitum for 3 days before the experiment. The rats were then divided into a control group (orally administered deionized water) and a Qi-Fu-Yin group (orally administered Qi-Fu-Yin) (n = 12). To detect the prototype components and metabolites of Qi-Fu-Yin in the rat plasma and cerebrospinal fluid, an 8-fold clinical dosage (1.72 g crude drug per mL, 10 mL per kg, twice daily) was selected as the oral dose [6, 7]. All groups received intragastric administration twice daily for three consecutive days. Before the experiments, the animals fasted for 12 h, with free access to water.
2.4. Biological Sample Collection and Preparation
After the last intragastric administration, 500 μL aliquots of serial blood samples were collected from the postorbital venous plexus vein of each rat at 0.5, 1.0, 2, and 4 h. Then, approximately 100 μL of cerebrospinal fluid from each rat was collected at 4 h via percutaneous puncture of the cerebellar medulla cistern [15]. The biological samples collected in heparinized polythene tubes were centrifuged at 3000 rpm at 4°C for 15 min. Subsequently, the supernatant was transferred into new tubes and immediately stored at −80°C before preliminary treatment.
After unfreezing the biological samples in an ice-water mixture, plasma or cerebrospinal fluid was mixed at four different times to enrich the biological samples of each group. To each tube containing 1 mL of plasma or cerebrospinal fluid, 4 mL of methanol was added. The mixture was then vortexed for 2 min and centrifuged at 13000 rpm and 4°C for 10 min. Subsequently, the supernatant was transferred to another tube and dried using sanitary nitrogen gas at room temperature. Then, the residue was redissolved in 100 μL of 30% methanol, vortexed for 2 min, and centrifuged at 13000 rpm and 4°C for 10 min.
2.5. UHPLC-Q-TOF-MS Analysis
An ultrahigh-performance liquid chromatography system (ACQUITY H-Class, Waters, Milford, MA, USA) coupled with a Q-TOF (Impact II, Bruker, Bremen, Germany) high-definition mass spectrometer in electrospray ionization mode was used for the chromatographic and mass spectral analyses of all samples. An AMT Halo-C18 column (100 mm × 2.1 mm, 2.7 μm) with a column temperature of 30°C was selected as the separation system. The mobile phase consisted of eluent A (0.1% formic acid in water, v/v) and eluent B (acetonitrile), with a flow rate of 0.30 mL/min. These phases were delivered using a gradient program as follows: 8% B from 0 to 5 min, 8–17% from 5 to 15 min, 17–23% B from 15 to 27 min, 23–35% B from 27 to 43 min, 35–70% B from 43 to 51 min, 70–100% B from 51 to 55 min, and 100% B from 55 to 60 min.
The mass spectra operating parameters were set as follows: capillary voltage of 3.5 kV (ESI+) or −3.0 kV (ESI−), source temperature of 220°C, drying temperature of 220°C, and drying gas flow of 8 L/min. The collision energy was set to range from to 35–75 V for MS/MS acquisition. To ensure mass accuracy and reproducibility, the mass spectrometer was calibrated over a range of 50–1500 Da using a sodium formate solution. All data were processed using Compass Data AnalysisTM (V4.4, Bruker, Bremen, Germany).
3. Results
3.1. In Vitro Chemical Characterization of Qi-Fu-Yin
The base peak chromatograms (BPCs) of Qi-Fu-Yin in the positive and negative ion modes are shown in Figure S1. A total of 180 compounds, including 59 triterpene saponins, 26 flavonoids, 17 organic acids, 16 sucrose esters, 14 oligosaccharide esters, 13 phthalides, 12 phenylethanoid glycosides, 9 alkaloids, 6 xanthones, 3 terpene lactones, 3 ionones, and 2 iridoid glycosides (Table 1), were identified. Twelve compounds were unambiguously identified via comparison with the standard solutions. The structures of other compounds were tentatively characterized based on their retention times, fragmentation pathways, and MS/MS spectra, by referring to the literature.
| No. | tR (min) | Name | Classification | Formula | Theoretical mass (Da) | Measured mass (Da) | Error (ppm) | Precursor ions | Main MS/MS fragment ions | Source | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.99 | Citric acid☆ | Organic acids | C6H8O7 | 191.0197 | 191.0201 | 2.1 | [M − H]− | 129.0196, 111.009 | ZSS, ASR, ARP | [16] |
| 2 | 1.37 | Geniposidic acid | Iridoid glycoside | C16H22O10 | 373.1140 | 373.1143 | 0.8 | [M − H]− | 211.0605, 193.0497, 167.0703, 149.0595, 123.0437 | RRP | [10] |
| 3 | 1.85 | Decaffeoylacteoside☆ | Phenylethanoid glycosides | C20H30O12 | 461.1664 | 461.1669 | 1.1 | [M − H]− | 375.1314, 315.1314, 297.0980, 135.0452 | RRP | [17] |
| 4 | 1.95 | Mussaenosidic acid☆ | Iridoid glycoside | C16H24O10 | 375.1297 | 375.1299 | 0.5 | [M − H]− | 213.0778, 169.0873, 151.0766 | RRP | [18] |
| 5 | 2.05 | 5-Caffeoylquinic acid☆ | Organic acids | C16H18O9 | 353.0878 | 353.0878 | 0.0 | [M − H]− | 191.0563, 179.0352, 161.0245, 155.0350, 111.0088 | ASR | [19] |
| 6 | 2.34 | 3-Caffeoylquinic amide ∗ | Organic acids | C16H19NO8 | 354.1183 | 354.1178 | −1.5 | [M + H]+ | 192.0650, 174.0545, 146.0597 | [10] | |
| 7 | 2.82 | Ferulic acid hexoside☆ | Organic acids | C16H20O9 | 355.1035 | 355.1042 | 2.0 | [M − H]− | 193.0509, 149.0610, 178.0271, 134.0375 | ASR | [20] |
| 8 | 3.01 | 3-Caffeoylquinic amide isomer ∗ | Organic acids | C16H19NO8 | 354.1183 | 354.1177 | −1.8 | [M + H]+ | 192.0650, 174.0545, 146.0597 | [10] | |
| 9 | 3.03 | Hydroxybenzoic acid☆ | Organic acids | C7H6O3 | 137.0244 | 137.0244 | 0.0 | [M − H]− | 136.0170, 108.0215 | ZSS | [16] |
| 10 | 3.21 | p-Hydroxybenzyl malonic acid☆ | Organic acids | C10H10O5 | 209.0455 | 209.0456 | 0.5 | [M − H]− | 419.0982, 165.0562, 121.0662 | GRP | [21] |
| 11 | 3.34 | Sanjoinine IB☆ | Alkaloids | C19H21NO4 | 328.1543 | 328.1534 | −2.7 | [M + H]+ | 265.0855, 251.0665, 237.0902, 223.0712 | ZSS | [22] |
| 12 | 3.53 | Magnocurarine☆ | Alkaloids | C19H24NO3+ | 314.1751 | 314.1748 | −1.0 | [M]+ | 269.1179, 237.0897, 209.0947, 175.0744, 107.0491 | ZSS | [22] |
| 13 | 3.69 | Chlorogenic acid | Organic acids | C16H18O9 | 353.0878 | 353.0885 | 2.0 | [M − H]− | 191.0563, 127.0404 | ASR | [20] |
| 14 | 4.26 | Sibiricose A5 | Sucrose esters | C22H30O14 | 517.1563 | 517.1568 | 1.0 | [M − H]− | 341.1097, 193.0512, 175.0404, 160.0169 | PRP | [23] |
| 15 | 4.55 | 4-Caffeoylquinic acid | Organic acids | C16H18O9 | 353.0878 | 353.0883 | 1.4 | [M − H]− | 191.0562, 179.0350, 173.0457, 161.0243, 111.0453, 93.0346 | ASR | [10] |
| 16 | 4.74 | Vanillic acid | Organic acids | C8H8O4 | 167.0350 | 167.0351 | 0.6 | [M − H]− | 123.0452 | ASR | [24] |
| 17 | 5.32 | Sibiricose A6☆ | Sucrose esters | C23H32O15 | 547.1668 | 547.1678 | 1.8 | [M − H]− | 367.1034, 341.1094, 223.0616, 205.0508, 190.0274 | PRP | [25, 26] |
| 18 | 5.54 | Sanjoinine K | Alkaloids | C17H19NO3 | 286.1438 | 286.1431 | −2.3 | [M + H]+ | 269.1154, 237.0905, 175.0751, 107.0492 | ZSS | [16] |
| 19 | 5.90 | Ferulic acid hexoside isomer☆ | Organic acids | C16H20O9 | 355.1035 | 355.1041 | 1.7 | [M − H]− | 193.0512, 149.0610, 178.0273, 134.0376 | ASR | [20] |
| 20 | 5.99 | Darendoside B☆ | Phenylethanoid glycosides | C21H32O12 | 475.1821 | 475.1830 | 1.9 | [M − H]− | 329.1228, 311.1144, 161.0459, 113.0247 | RRP | [27] |
| 21 | 6.18 | Liquiritigenin-7,4′-di-O-glucoside | Flavonoids | C27H32O14 | 579.1719 | 579.1733 | 2.4 | [M − H]− | 417.1212, 255.0669, 135.0086 | GRP | [28] |
| 22 | 6.38 | Caffeic acid | Organic acids | C9H8O4 | 179.0350 | 179.0352 | 1.1 | [M − H]− | 151.0459, 135.0499 | ASR | [10] |
| 23 | 7.47 | Magnoflorine | Alkaloids | C20H23NO4+ | 342.1700 | 342.1689 | −3.2 | [M + H]+ | 297.1113, 282.0876, 265.0848 | ZSS | [16] |
| 24 | 9.46 | Feruoylquinic acid☆ | Organic acids | C17H20O9 | 367.1035 | 367.1043 | 2.2 | [M − H]− | 191.0563, 173.0461, 111.0453, 93.035 | ASR | [20] |
| 25 | 9.82 | Lotusine☆ | Alkaloids | C19H24NO3+ | 314.1751 | 314.1752 | 0.3 | [M]+ | 269.1162, 237.0912, 209.0949, 107.0485 | ZSS | [22] |
| 26 | 9.94 | Feruoylquinic acid isomer☆ | Organic acids | C17H20O9 | 367.1035 | 367.1034 | −0.3 | [M − H]− | 191.0564, 173.0457, 111.0450, 93.0347 | ASR | [20] |
| 27 | 10.04 | Sibiricose A1 | Sucrose esters | C23H32O15 | 547.1668 | 547.1676 | 1.5 | [M − H]− | 367.1040, 223.016, 190.0275 | PRP | [10, 23] |
| 28 | 10.23 | Vicenin II | Flavonoids | C27H30O15 | 593.1512 | 593.1522 | 1.7 | [M − H]− | 503.1200, 473.1098, 383.0780, 353.0674, 325.0931, | GRP, ZSS | [16, 22] |
| 29 | 10.43 | Ferulic acid isomer ∗☆ | Organic acids | C10H10O4 | 193.0506 | 193.0506 | 0.0 | [M − H]− | 149.0243, 121.0298 | ||
| 30 | 10.81 | Lancerin | Xanthones | C19H18O10 | 405.0827 | 405.0833 | 1.5 | [M − H]− | 285.0410, 257.0456 | PRP | [23] |
| 31 | 11.30 | Rehmaionoside A/B | Ionones | C19H34O8 | 435.2239 | 435.2246 | 1.6 | [M + COOH]− | 389.2223, 179.0591 | RRP | [10, 17] |
| 32 | 11.49 | Ferulic acid | Organic acids | C10H10O4 | 193.0506 | 193.0507 | 0.5 | [M − H]− | 178.0272, 149.0609, 134.0369 | ASR | [20] |
| 33 | 11.97 | Lancerin isomer☆ | Xanthones | C19H18O10 | 405.0827 | 405.0833 | 1.5 | [M − H]− | 285.0413, 315.0518, 257.0458 | PRP | [10] |
| 34 | 12.14 | Caaverine☆ | Alkaloids | C17H17NO2 | 268.1332 | 268.1321 | −4.1 | [M − H]− | 251.1014, 219.0829, 209.0933, 191.0862 | ZSS | [22] |
| 35 | 12.45 | Sibiricaxanthone A/B | Xanthones | C24H26O14 | 537.1250 | 537.1258 | 1.5 | [M − H]− | 405.0832, 387.0730, 327.0524, 315.0514, 297.0412, 285.0410, 267.0303, 243.0302 | PRP | [29] |
| 36 | 12.45 | Echinacoside | Phenylethanoid glycosides | C35H46O20 | 785.2510 | 785.2520 | 1.3 | [M − H]− | 623.2201, 461.1663, 161.0245 | RRP | [30] |
| 37 | 12.74 | Schaftoside | Flavonoids | C26H28O14 | 563.1406 | 563.1408 | 0.4 | [M − H]− | 353.0674, 443.0992, 473.1098, 383.0778, 503.1197, 425.0877, 413.0882 | GRP | [31] |
| 38 | 13.12 | Sibiricose A2 | Sucrose esters | C24H34O15 | 561.1825 | 561.1832 | 1.2 | [M − H]− | 607.1888, 323.0991, 237.0771 | PRP | [10] |
| 39 | 13.41 | Rehmaionoside A/B | Ionones | C19H34O8 | 435.2239 | 435.2239 | 0.0 | [M + COOH]− | 389.2223, 179.0572 | RRP | [29] |
| 40 | 13.79 | Liquiritin | Flavonoids | C21H22O9 | 417.1191 | 417.1194 | 0.7 | [M − H]− | 255.0665, 135.0089, 119.0504 | GRP | [31] |
| 41 | 14.08 | Polygalaxanthone III | Xanthones | C25H28O15 | 567.1355 | 567.1361 | 1.1 | [M − H]− | 447.0945, 435.0932, 417.0839, 357.0621, 345.0620, 327.0518, 315.0515, 297.0408 | PRP | [10] |
| 42 | 14.27 | Jionoside E☆ | Phenylethanoid glycosides | C35H46O19 | 769.2561 | 769.2568 | 0.9 | [M − H]− | 623.2197, 605.2092, 549.1662, 427.1069, 323.0996, 179.0561 | RRP | [27] |
| 43 | 14.37 | Liquiritin apioside | Flavonoids | C26H30O13 | 549.1614 | 549.1616 | 0.4 | [M − H]− | 255.06581, 135.00719, 119.04859, 417.11804 | GRP | [31] |
| 45 | 14.47 | Asimilobine☆ | Alkaloids | C17H17NO2 | 268.1332 | 268.1324 | −3.0 | [M − H]− | 251.1064, 219.0809, 201.0722, 191.0858, 179.0855 | ZSS | [22] |
| 44 | 14.47 | Polygalaxanthone XI☆ | Xanthones | C25H28O15 | 567.1355 | 567.1366 | 1.9 | [M − H]− | 345.0619, 315.0511 | PRP | [32] |
| 46 | 14.56 | Jionoside A1/jionoside A2 | Phenylethanoid glycosides | C36H48O20 | 799.2666 | 799.2672 | 0.8 | [M − H]− | 623.2199, 605.2092, 461.1663, 315.1110, 193.0509, 175.0403 | RRP | [30] |
| 47 | 14.85 | Spinosin | Flavonoids | C28H32O15 | 607.1668 | 607.1674 | 1.0 | [M − H]− | 487.1252, 445.1144, 427.1039, 367.0823, 337.0722, 307.0614 | ZSS | [16] |
| 48 | 15.33 | Swertisin | Flavonoids | C22H22O10 | 445.1140 | 445.1147 | 1.6 | [M − H]− | 355.0839, 325.0721, 297.0409 | ZSS | [16] |
| 49 | 15.33 | Isoviolanthin/violanthin☆ | Flavonoids | C27H30O14 | 577.1563 | 577.1572 | 1.6 | [M − H]− | 383.0777, 353.0670, 413.08783, 457.1145, 487.1248 | GRP | [31] |
| 50 | 15.43 | Tenuifoliside B | Sucrose esters | C30H36O17 | 667.1880 | 667.1894 | 2.1 | [M − H]− | 461.1312, 205.0510, 190.0274, 137.0247, 281.0674 | PRP | [23] |
| 51 | 15.52 | Sibiricose A4☆ | Sucrose esters | C34H42O19 | 753.2248 | 753.2254 | 0.8 | [M − H]− | 547.1678, 529.1574, 461.1306, 367.1041, 223.0615, 205.0509, 190.0274 | PRP | [29] |
| 52 | 15.71 | Tenuifoliside 638☆ | Sucrose esters | C29H34O16 | 637.1774 | 637.1773 | −0.2 | [M − H]− | 461.1309, 443.1208, 175.0402 | PRP | [29] |
| 53 | 16.48 | Acteoside | Phenylethanoid glycosides | C29H36O15 | 623.1981 | 623.1989 | 1.3 | [M − H]− | 461.1667, 443.1555, 315.1083, 179.0349, 161.0243 | RRP | [30] |
| 54 | 17.06 | 6′′′-Vanilloylspinosin☆ | Flavonoids | C36H38O18 | 757.1985 | 757.1993 | 1.1 | [M − H]− | 637.1556, 607.1694, 445.1143, 427.1038, 367.0827, 307.0621 | ZSS | [22] |
| 55 | 17.25 | Senkyunolide I | Phthalides | C12H16O4 | 207.1015 | 207.1009 | −2.9 | [M + H–H2O]+ | 189.0822, 161.0906, 147.0752 | ASR | [33] |
| 56 | 17.44 | Jionoside B1/Jionoside B2 | Phenylethanoid glycosides | C37H50O20 | 813.2823 | 813.2834 | 1.4 | [M − H]− | 637.2359, 619.2254, 491.1780, 193.0507, 175.0402, 160.0167 | RRP | [16] |
| 57 | 17.63 | 6′′′-P-Hydroxyl-benzoyspinosin☆ | Flavonoids | C35H36O17 | 727.1880 | 727.1879 | −0.1 | [M − H]− | 607.1616, 445.1149, 427.1038, 325.0719, 307.0617 | ZSS | [16] |
| 58 | 17.73 | Isoacteoside | Phenylethanoid glycosides | C29H36O15 | 623.1981 | 623.1990 | 1.4 | [M − H]− | 461.1670, 477.1405, 315.1096, 179.0351, 161.0245 | RRP | [30] |
| 59 | 18.69 | Tenuifoliside A isomer☆ | Sucrose esters | C31H38O17 | 681.2036 | 681.2044 | 1.2 | [M − H]− | 443.1208, 281.0672, 237.0774, 223.0616, 205.0510, 137.0246 | PRP | [23] |
| 60 | 18.78 | Senkyunolide H | Phthalides | C12H16O4 | 207.1015 | 207.1008 | −3.4 | [M + H–H2O]+ | 189.0812, 161.0938, 147.0689 | ASR | [33] |
| 61 | 18.98 | 3,6′-Disinapoyl sucrose | Sucrose esters | C34H42O19 | 753.2248 | 753.2249 | 0.1 | [M − H]− | 547.1670, 529.1568, 367.1038, 265.0720, 223.0612, 205.0506, 190.0271 | PRP | [10] |
| 62 | 19.02 | Nornuciferine☆ | Alkaloids | C18H19NO2 | 282.1489 | 282.1481 | −2.7 | [M + H]+ | 265.1214, 250.0979, 121.0280 | ZSS | [22] |
| 63 | 19.05 | 6′′′-Sinapoyl spinosin | Flavonoids | C39H42O19 | 815.2393 | 815.2379 | −1.7 | [M + H]+ | 429.1181, 411.1037, 369.1162, 327.0855, 207.0647, 351.0833, 297.0750, 175.0385 | ZSS | [16] |
| 64 | 19.21 | 6′′′-Dihydrophaseoylspinosin☆ | Flavonoids | C43H52O19 | 873.3176 | 873.3158 | −2.0 | [M + H]+ | 855.2986, 447.1263, 429.1140, 411.1057, 393.0969, 381.0947, 351.0846, 327.0854, 297.0752, 247.1321 | ZSS | [22] |
| 65 | 19.56 | 3,4,5-Trimethoxycinnamic acid☆ | Organic acids | C12H14O5 | 237.0768 | 237.0770 | 0.8 | [M − H]− | 193.0873, 108.0215 | PRP | [34] |
| 66 | 19.60 | 6′′′-p-Coumaroyl spinosin | Flavonoids | C37H38O17 | 755.2182 | 755.2166 | −2.1 | [M + H]+ | 429.1170, 411.1080, 351.0850, 327.0854, 147.0438, 635.1770, 381.0957, 297.0750 | ZSS | [16] |
| 67 | 19.66 | Jionoside D | Phenylethanoid glycosides | C30H38O15 | 637.2138 | 637.2137 | −0.2 | [M − H]− | 161.0242, 461.1660, 267.0660, 175.0401 | RRP | [10] |
| 68 | 19.66 | Arillanin A☆ | Sucrose esters | C33H40O18 | 723.2142 | 723.2149 | 1.0 | [M − H]− | 547.1679, 265.0722, 223.0617, 205.0510, 175.0404, 160.0170 | PRP | [32] |
| 69 | 19.73 | 6′′′-Feruloyl spinosin | Flavonoids | C38H40O18 | 785.2287 | 785.2263 | −3.1 | [M + H]+ | 665.1891, 447.1275, 429.1168, 411.1068, 393.0957, 351.0852, 327.0853, 297.0750, 177.0542 | ZSS | [10] |
| 70 | 19.95 | Tenuifoliside 652☆ | Sucrose esters | C30H36O16 | 651.1931 | 651.1942 | 1.7 | [M − H]− | 443.1199, 281.0671, 207.0668, 175.0403, 137.0244 | PRP | [29] |
| 71 | 20.81 | Ononin☆ | Flavonoids | C22H22O9 | 475.1246 | 475.1249 | 0.6 | [M + COOH]− | 475.1249, 267.0664, 252.0429 | GRP | [31] |
| 72 | 21.00 | Tenuifoliside 652 isomer☆ | Sucrose esters | C30H36O16 | 651.1931 | 651.1941 | 1.5 | [M − H]− | 443.1199, 205.0509, 190.0272, 175.0033, 121.0297 | PRP | [29] |
| 73 | 21.10 | Isoliquiritin apioside | Flavonoids | C26H30O13 | 549.1614 | 549.1620 | 1.1 | [M − H]− | 255.0667, 135.0090, 119.0505, 417.1200 | GRP | [10] |
| 74 | 21.48 | Isoliquiritin | Flavonoids | C21H22O9 | 417.1191 | 417.1197 | 1.4 | [M − H]− | 255.0666, 135.0089, 119.0404 | GRP | [10] |
| 75 | 21.77 | Leucosceptoside A | Phenylethanoid glycosides | C30H38O15 | 637.2138 | 637.2129 | −1.4 | [M − H]− | 461.1661, 175.0400, 265.0722, 161.0239 | RRP | [30] |
| 76 | 21.77 | Tenuifoliside A | Sucrose esters | C31H38O17 | 681.2036 | 681.2038 | 0.3 | [M − H]− | 443.1203, 281.0671, 239.0564, 179.0352, 137.0245 | PRP | [10] |
| 77 | 22.06 | Liquiritigenin | Flavonoids | C15H12O4 | 255.0663 | 255.0665 | 0.8 | [M − H]− | 135.0086, 119.0502 | GRP | [31] |
| 78 | 22.64 | Neoisoliquiritin☆ | Flavonoids | C21H22O9 | 417.1191 | 417.1197 | 1.4 | [M − H]− | 255.0667, 135.0089, 119.0505 | GRP | [28] |
| 79 | 22.64 | Notoginsenoside R1☆ | Saponins | C47H80O18 | 977.5327 | 977.5334 | 0.7 | [M + COOH]− | 931.5284, 637.4332, 475.3809 | GRR | [35] |
| 80 | 22.83 | 6′′′-(-)-Phaseoylspinosin☆ | Flavonoids | C43H50O19 | 869.2874 | 869.2884 | 1.2 | [M − H]− | 839.2765, 607.1683, 589.1575, 427.1045 | ZSS | [22] |
| 81 | 23.70 | Tenuifoliose G | Oligosaccharide esters | C66H84O38 | 1483.4568 | 1483.4582 | 0.9 | [M − H]− | 1337.39795, 1295.38232, 1161.35095, 1119.34119, 997.30548, 851.27283, 753.22705, 631.18640, 452.31161, 307.08231, 175.03891, 163.03868, 145.02803 | PRP | [10] |
| 82 | 23.80 | Senkyunolide D☆ | Phthalides | C12H14O4 | 221.0819 | 221.0820 | 0.5 | [M − H]− | 177.0921, 147.0450 | ASR | [36] |
| 83 | 23.80 | Tenuifoliose M | Oligosaccharide esters | C65H82O37 | 1453.4462 | 1453.4490 | 1.9 | [M − H]− | 1307.3873, 1161.3532, 997.3064, 835.2514, 307.0824, 163.0385, 145.0280 | PRP | [10] |
| 84 | 24.48 | Ginsenoside Rg1 | Saponins | C42H72O14 | 845.4904 | 845.4912 | 0.9 | [M + COOH]− | 799.4877, 637.4342, 475.3809, 161.0458, 179.0565 | GRR | [31] |
| 85 | 24.67 | Licorice glycoside B | Flavonoids | C35H36O15 | 695.1981 | 695.1991 | 1.4 | [M − H]− | 549.1634, 163.0409, 417.1202, 255.0665, 399.1099, 531.1523, 175.0403 | GRP | [31] |
| 86 | 24.77 | Isomartynoside☆ | Phenylethanoid glycosides | C31H40O15 | 651.2294 | 651.2303 | 1.4 | [M − H]− | 505.1703, 475.1826, 193.0511, 175.0403, 160.017, 113.0245 | RRP | [37] |
| 87 | 24.77 | Licorice glycoside A | Flavonoids | C36H38O16 | 725.2087 | 725.2095 | 1.1 | [M − H]− | 549.1639, 255.0668, 193.0508, 135.0086 | GRP | [38] |
| 88 | 24.86 | Ginsenoside Re | Saponins | C48H82O18 | 991.5483 | 991.5501 | 1.8 | [M + COOH]− | 945.5442, 783.4916, 637.4329, 475.3793, 179.0562, 161.0457 | GRR | [31] |
| 89 | 26.11 | Senkyunolide D isomer☆ | Phthalides | C12H14O4 | 221.0819 | 221.0817 | −0.9 | [M − H]− | 177.0920, 147.0453 | ASR | [36] |
| 90 | 26.31 | Tenuifoliside C | Sucrose esters | C35H44O19 | 767.2404 | 767.2416 | 1.6 | [M − H]− | 529.1567, 367.1038, 237.077, 223.0613, 205.0507, 190.0271 | PRP | [10] |
| 91 | 26.69 | Tenuifoliose T☆ | Oligosaccharide esters | C56H70O32 | 1253.3777 | 1253.3792 | 1.2 | [M − H]− | 1223.3637, 1077.3279, 955.2908, 647.1988, 451.1232, 307.0810, 287.0549, 257.0444 | PRP | [23] |
| 92 | 26.88 | Martynoside☆ | Phenylethanoid glycosides | C31H40O15 | 651.2294 | 651.2299 | 0.8 | [M − H]− | 505.172, 475.1829, 193.0508, 175.0403, 160.0169, 113.0244 | RRP | [17] |
| 93 | 26.90 | (hydroxy benzoyl)-(hydroxy cinnamoyl)-trihydroxyphenyl sucrose | Sucrose esters | C34H42O18 | 783.2353 | 783.2365 | 1.5 | [M + COOH]− | 737.2325, 615.1934, 467.1415, 323.0980, 179.0547, 161.0458, 147.0453, 121.0296 | PRP | [10] |
| 94 | 27.75 | Tenuifoliose L | Oligosaccharide esters | C67H84O38 | 1495.4569 | 1495.4569 | 0.0 | [M − H]− | 1349.3923, 1307.3988, 163.0410, 145.0294 | PRP | [10] |
| 95 | 28.14 | Tenuifoliose K | Oligosaccharide esters | C57H70O32 | 1265.3777 | 1265.3801 | 1.9 | [M − H]− | 1119.3395, 1077.3346, 997.3037, 163.0403, 145.0294 | PRP | [10] |
| 96 | 29.00 | Tenuifoliose C | Oligosaccharide esters | C58H72O33 | 1295.3883 | 1295.3903 | 1.5 | [M − H]− | 1173.3653, 1119.3401, 1077.3265, 997.3061, 145.0296, 175.0404 | PRP | [10] |
| 97 | 29.78 | Amphibine D☆ | Alkaloids | C36H49N5O5 | 632.3806 | 632.3805 | −0.2 | [M + H]+ | 289.1874, 148.1111 | ZSS | [16] |
| 98 | 30.35 | Desacylsenegasaponin B☆ | Saponins | C57H70O32 | 1265.5808 | 1265.5831 | 1.8 | [M − H]− | 455.3179, 425.3077 | PRP | [29] |
| 99 | 30.44 | Uralsaponin C | Saponins | C42H64O16 | 823.4122 | 823.4133 | 1.3 | [M − H]− | 647.3829, 351.0580, 193.0357 | GRP | [28] |
| 100 | 30.44 | Tenuifoliose I | Oligosaccharide esters | C59H72O33 | 1307.3883 | 1307.3904 | 1.6 | [M − H]− | 1161.3529, 1119.3479, 1101.3331, 997.3023, 631.1891, 163.0400, 145.0299 | PRP | [10] |
| 101 | 30.70 | Aeginetic acid☆ | Ionones | C15H24O4 | 267.1602 | 267.1610 | 3.0 | [M − H]− | 223.1780, 205.1615, 178.9208, 153.0924 | RRP | [39] |
| 102 | 30.81 | Methoxyl benzoyl-trimethoxyl cinnamoyl sucrose | Sucrose esters | C32H40O17 | 741.2248 | 741.2255 | 0.9 | [M + COOH]− | 237.0773, 151.0402 | PRP | [10] |
| 103 | 31.12 | Tenuifoliose D | Oligosaccharide esters | C60H74O34 | 1337.3989 | 1337.4007 | 1.3 | [M − H]− | 1161.3546, 1119.3412, 1039.3161, 997.3030, 175.0404 | PRP | [10] |
| 104 | 31.41 | Notoginsenoside R2 | Saponins | C41H70O13 | 815.4834 | 815.4834 | 0.0 | [M + COOH]− | 769.4745, 637.4342, 475.3791, 161.0462 | GRR | [40] |
| 105 | 31.41 | Tenuifoliose E☆ | Oligosaccharide esters | C58H72O33 | 1295.3883 | 1295.3933 | 3.9 | [M − H]− | 1173.3506, 1119.3442, 795.2398, 175.0404, 145.0300 | PRP | [29] |
| 106 | 31.79 | Polygalasaponin XXIII☆ | Saponins | C53H82O24 | 1101.5123 | 1101.5164 | 3.7 | [M − H]− | 423.2925, 453.3029 | PRP | [29] |
| 107 | 32.08 | Polygalasaponin XXVIII | Saponins | C53H84O24 | 1103.5380 | 1103.5328 | −4.7 | [M − H]− | 455.3185, 425.3075 | PRP | [23] |
| 108 | 32.28 | 24-Hydroxyl-licorice-saponin A3 | Saponins | C48H72O22 | 999.4442 | 999.4488 | 4.6 | [M − H]− | 837.3942, 351.0584, 193.0359 | GRP | [10] |
| 109 | 32.57 | Tenuifoliose J☆ | Oligosaccharide esters | C59H72O33 | 1307.3883 | 1307.3898 | 1.1 | [M − H]− | 1161.3549, 1039.3096, 163.0408, 145.0304 | PRP | [29, 32] |
| 110 | 32.81 | Butylidenephthalide | Phthalides | C12H12O2 | 189.0910 | 189.0904 | −3.2 | [M + H]+ | 171.0799, 161.0954, 143.0852, 117.0694 | ASR | [20] |
| 111 | 32.85 | Senkyunolide F☆ | Phthalides | C12H14O3 | 205.0870 | 205.0880 | 4.9 | [M − H]− | 161.0975 | ASR | [20] |
| 112 | 32.92 | Uralsaponin F | Saponins | C44H64O19 | 895.3969 | 895.3995 | 2.9 | [M − H]− | 719.3703, 351.0586, 193.0363 | GRP | [31] |
| 113 | 32.95 | Onjisaponin TF | Saponins | C59H94O28 | 1249.5859 | 1249.5880 | 1.7 | [M − H]− | 1025.5362, 455.3185, 425.3077 | PRP | [23] |
| 114 | 33.05 | Licorice saponin H2/K2☆ | Saponins | C42H62O16 | 821.3965 | 821.3981 | 1.9 | [M − H]− | 351.0583, 193.0364, 175.0255 | GRP | [28, 41] |
| 115 | 33.05 | 22-Hydroxyl-licorice-saponin G2 | Saponins | C42H62O18 | 853.3863 | 853.3882 | 2.2 | [M − H]− | 677.3568, 351.0583, 193.0365 | GRP | [28] |
| 116 | 33.22 | Butylphthalide | Phthalides | C12H14O2 | 191.1067 | 191.1062 | −2.6 | [M + H]+ | 173.0959, 155.0842, 145.1008, 117.0698 | ASR | [20] |
| 117 | 33.34 | Tenuifoliose B | Oligosaccharide esters | C60H74O34 | 1337.3989 | 1337.4027 | 2.8 | [M − H]− | 1161.3551, 1119.342, 1101.3324, 1039.3156, 175.0410, 145.0306 | PRP | [10] |
| 118 | 33.92 | Ginsenoside Rf | Saponins | C42H72O14 | 845.4904 | 845.4928 | 2.8 | [M + COOH]− | 799.4880, 637.4349, 475.3820, 179.0574, 161.0466 | GRR | [35] |
| 119 | 33.92 | Tenuifoliose H | Oligosaccharide esters | C61H74O34 | 1349.3989 | 1349.4019 | 2.2 | [M − H]− | 1307.3907, 1161.3503, 731.2194, 145.0304 | PRP | [10] |
| 120 | 34.40 | Senkyunolide A | Phthalides | C12H16O2 | 193.1223 | 193.1218 | −2.6 | [M + H]+ | 147.1170, 175.1113, 137.0593 | ASR | [20] |
| 121 | 34.59 | Tenuifoliose A | Oligosaccharide esters | C62H76O35 | 1379.4094 | 1379.4131 | 2.7 | [M − H]− | 1203.3649, 1161.3529, 175.041, 145.0303 | PRP | [10] |
| 122 | 35.08 | Tenuifoliose N☆ | Oligosaccharide esters | C63H78O36 | 1409.4200 | 1409.4234 | 2.4 | [M − H]− | 1233.3879, 175.0410 | PRP | [23] |
| 123 | 35.37 | Ginsenoside F5☆ | Saponins | C41H70O13 | 815.4834 | 815.4821 | −1.6 | [M + COOH]− | 769.4765, 637.4337, 475.3807 | GRR | [42] |
| 124 | 35.41 | Licorice saponin A3 | Saponins | C48H72O21 | 983.4493 | 983.4518 | 2.5 | [M − H]− | 821.3988, 645.3687, 351.0584, 193.0366 | GRP | [31] |
| 125 | 35.79 | 24-Hydroxyl-licorice-saponin E2 | Saponins | C42H60O17 | 835.3793 | 835.3785 | −1.0 | [M − H]− | 659.3446, 351.0582, 193.0362 | GRP | [28] |
| 126 | 35.84 | Isoliquiritigenin ∗☆ | Flavonoids | C15H12O4 | 255.0663 | 255.0674 | 4.3 | [M − H]− | 135.0094, 119.0510 | [28] | |
| 127 | 36.04 | Formononetin ∗☆ | Flavonoids | C16H12O4 | 267.0663 | 267.0671 | 3.0 | [M − H]− | 252.0458, 195.0458 | [31] | |
| 128 | 36.32 | Senkyunolide F isomer☆ | Phthalides | C12H14O3 | 205.0870 | 205.0879 | 4.4 | [M − H]− | 161.0993 | ASR | [20] |
| 129 | 36.42 | 22β-Acetoxyl-glycyrrhizin | Saponins | C44H64O18 | 879.4020 | 879.4034 | 1.6 | [M − H]− | 351.0583, 193.0362 | GRP | [31] |
| 130 | 36.61 | Tenuifolin | Saponins | C36H56O12 | 679.3699 | 679.3718 | 2.8 | [M − H]− | 455.3180, 425.3074 | PRP | [10] |
| 131 | 36.71 | Ginsenoside F3☆ | Saponins | C41H70O13 | 815.4834 | 815.4818 | −2.0 | [M + COOH]− | 769.4761, 637.4332, 475.3810, 161.0463 | GRR | [42] |
| 132 | 36.90 | 20(S)-Ginsenoside Rh1 | Saponins | C36H62O9 | 683.4376 | 683.4390 | 2.0 | [M + COOH]− | 637.4335, 475.3806, 161.0462 | GRR | [10] |
| 133 | 36.90 | 20(S)-Ginsenoside Rg2 | Saponins | C42H72O13 | 829.4955 | 829.4969 | 1.7 | [M + COOH]− | 783.4911, 637.4334, 475.3807, 161.0461 | GRR | [35] |
| 134 | 36.90 | 22-Hydroxyl-glycyrrhizin | Saponins | C42H62O17 | 837.3914 | 837.3929 | 1.8 | [M − H]− | 661.3603, 485.3294, 351.0583, 193.0362 | GRP | [28] |
| 135 | 37.35 | Senkyunolide A isomer☆ | Phthalides | C12H16O2 | 193.1223 | 193.1217 | −3.1 | [M + H]+ | 147.1163, 175.1113, 137.0594 | ASR | [20] |
| 136 | 37.39 | 20(R)-Ginsenoside Rg2 | Saponins | C42H72O13 | 829.4955 | 829.4972 | 2.0 | [M + COOH]− | 783.4913, 637.4332, 475.3808, 161.0462 | GRR | [42] |
| 137 | 37.68 | 20(R)-Ginsenoside Rh1 | Saponins | C36H62O9 | 683.4376 | 683.4393 | 2.5 | [M + COOH]− | 637.4336, 475.3807, 161.0463 | GRR | [40] |
| 138 | 37.89 | Jujuboside A | Saponins | C58H94O26 | 1251.6015 | 1251.6036 | 1.7 | [M + COOH]− | 1205.5983, 1073.5549, 749.4461, 455.1431, 179.0564, 161.0463 | ZSS | [16] |
| 139 | 38.73 | Ginsenoside Rb1 | Saponins | C54H92O23 | 1153.6011 | 1153.6033 | 1.9 | [M + COOH]− | 1107.5962, 945.5427, 783.4889, 621.4396, 459.3908 | GRR | [31] |
| 140 | 39.41 | Licorice saponin E2 | Saponins | C42H60O16 | 819.3809 | 819.3819 | 1.2 | [M − H]− | 645.3648, 351.0581, 193.0362 | GRP | [28] |
| 141 | 39.70 | Ginsenoside Ro | Saponins | C48H76O19 | 955.4908 | 955.4918 | 1.0 | [M − H]− | 793.4382, 775.4275, 749.451, 731.4392, 523.3806, 455.3537, 613.3755, 569.3857, 179.0569, 119.0355 | GRR | [31] |
| 142 | 39.70 | Ginsenoside Rc | Saponins | C53H90O22 | 1123.5906 | 1123.5918 | 1.1 | [M + COOH]− | 1077.5854, 915.5348, 459.3809, 149.0451, 191.0563 | GRR | [35] |
| 143 | 39.79 | Licorice saponin G2 | Saponins | C42H62O17 | 837.3914 | 837.3921 | 0.8 | [M − H]− | 775.3927, 661.3593, 485.3277, 351.0576, 193.0359 | GRP | [28] |
| 144 | 40.75 | Ginsenoside Rb2 | Saponins | C53H90O22 | 1123.5906 | 1123.5908 | 0.2 | [M + COOH]− | 1077.5865, 783.4945, 621.4307, 459.3789 | GRR | [35] |
| 145 | 41.14 | Ginsenoside Rb3 | Saponins | C53H90O22 | 1123.5906 | 1123.5907 | 0.1 | [M + COOH]− | 1077.5871, 783.4955, 621.4311, 459.3792 | GRR | [43] |
| 146 | 41.33 | Rhaoglycyrrhizin | Saponins | C48H72O20 | 967.4544 | 967.4567 | 2.4 | [M − H]− | 497.1159, 321.0841, 339.0941 | GRP | [10] |
| 147 | 41.33 | Jujuboside B | Saponins | C52H84O21 | 1045.5578 | 1045.5582 | 0.4 | [M + H]+ | 733.4491, 587.39348, 533.3637, 455.3536, 437.3432, 369.2802 | ZSS | [16] |
| 148 | 42.59 | Chikusetsusaponin IVa | Saponins | C42H66O14 | 793.4380 | 793.4389 | 1.1 | [M − H]− | 631.3854, 455.3525, 569.3834 | GRR | [31] |
| 149 | 42.68 | Ginsenoside Rd | Saponins | C48H82O18 | 991.5483 | 991.5496 | 1.3 | [M + COOH]− | 945.5438, 783.4892, 621.438, 459.3857, 179.0563, 161.0457 | GRR | [35] |
| 150 | 42.78 | Glycyrrhizic acid | Saponins | C42H62O16 | 821.3965 | 821.3972 | 0.9 | [M − H]− | 759.3961, 645.3648, 469.3324, 351.0572, 193.0356 | GRP | [31] |
| 151 | 43.19 | Senkyunolide A isomer☆ | Phthalides | C12H16O2 | 193.1223 | 193.1220 | −1.6 | [M + H]+ | 147.1166, 175.1117, 137.0599 | ASR | [20] |
| 152 | 44.03 | 6,8-Dihydroxy-1,2,4-trimethoxyxanthone ∗☆ | Xanthones | C16H14O7 | 317.0667 | 317.0675 | 2.5 | [M − H]− | 302.0444, 287.0203, 259.0254, 231.0297 | [23] | |
| 153 | 44.61 | Licorice saponin B2☆ | Saponins | C42H64O15 | 807.4172 | 807.4178 | 0.7 | [M − H]− | 631.3870, 351.0572, 193.0356 | GRP | [31] |
| 154 | 44.62 | Atractylenolide I | Terpene lactones | C15H18O2 | 231.1379 | 231.1373 | −2.6 | [M + H]+ | 213.1266, 203.1427, 189.0913, 185.1314, 157.1007 | ARP | [10] |
| 155 | 44.70 | Atractylenolide III | Terpene lactones | C15H20O3 | 249.1485 | 249.1485 | −0.1 | [M + H]+ | 231.1405, 213.1207, 185.1277, 175.0688 | ARP | [10] |
| 156 | 45.19 | Uralsaponin B | Saponins | C42H62O16 | 821.3965 | 821.3972 | 0.9 | [M − H]− | 759.3961, 645.3648, 469.3324, 351.0572, 193.0356 | GRP | [44] |
| 157 | 46.15 | Licorice saponin J2 | Saponins | C42H64O16 | 823.4122 | 823.4131 | 1.1 | [M − H]− | 351.0573, 193.0357 | GRP | [41] |
| 158 | 46.25 | Ginsenoside Rg6 | Saponins | C42H70O12 | 811.4849 | 811.4852 | 0.4 | [M + COOH]− | 765.4808, 619.4225, 205.0721, 161.0459 | GRR | [31] |
| 159 | 46.25 | Senegasaponin B☆ | Saponins | C69H102O31 | 1425.6332 | 1425.6381 | 3.4 | [M − H]− | 1395.6243, 1201.5864, 455.3163, 425.3061 | PRP | [29] |
| 160 | 46.25 | Onjisaponin Z☆ | Saponins | C71H106O32 | 1469.6594 | 1469.6600 | 0.4 | [M − H]− | 1245.6054, 1439.6517, 425.3061, 405.1400, 455.3165 | PRP | [29] |
| 161 | 46.34 | Onjisaponin E | Saponins | C71H106O33 | 1485.6544 | 1485.6545 | 0.1 | [M − H]− | 455.3187, 425.3029 | PRP | [23] |
| 162 | 46.53 | Onjisaponin Y☆ | Saponins | C69H102O30 | 1409.6383 | 1409.6376 | −0.5 | [M − H]− | 1379.6184, 1185.5881, 425.3062, 455.3166 | PRP | [29] |
| 163 | 46.53 | Onjisaponin G☆ | Saponins | C70H104O32 | 1455.6438 | 1455.6447 | 0.6 | [M − H]− | 1425.6341, 993.5078, 425.3062, 455.3166 | PRP | [23] |
| 164 | 46.63 | Ginsenoside Rg4☆ | Saponins | C42H70O12 | 811.4849 | 811.4854 | 0.6 | [M + COOH]− | 765.4798, 619.4212, 161.0456 | GRR | [42] |
| 165 | 46.82 | Ginsenoside Rk3 | Saponins | C36H60O8 | 665.4270 | 665.4271 | 0.2 | [M + COOH]− | 619.4211, 457.3698, 161.0458 | GRR | [31] |
| 166 | 46.82 | Licorice saponin C2☆ | Saponins | C42H62O15 | 805.4016 | 805.4020 | 0.5 | [M − H]− | 645.3637, 351.0575, 193.0356 | GRP | [41] |
| 167 | 46.92 | Onjisaponin TH | Saponins | C65H96O28 | 1323.6015 | 1323.5991 | −1.8 | [M − H]− | 455.3171, 425.3048 | PRP | [23] |
| 168 | 47.11 | Ginsenoside Rh4 | Saponins | C36H60O8 | 665.4270 | 665.4277 | 1.1 | [M + COOH]− | 619.4218, 457.3679, 161.0459 | GRR | [31] |
| 169 | 47.40 | Zingibroside R1 | Saponins | C42H66O14 | 793.4380 | 793.4386 | 0.8 | [M − H]− | 731.4390, 631.3853, 613.3751, 569.3853, 455.3538 | GRR | [42] |
| 170 | 47.88 | Ginsenoside Rg3 | Saponins | C42H72O13 | 829.4955 | 829.4953 | −0.2 | [M + COOH]− | 783.4894, 621.4369, 459.3844, 161.0456 | GRR | [31] |
| 171 | 48.10 | E-Ligustilide | Phthalides | C12H14O2 | 191.1067 | 191.1060 | −3.7 | [M + H]+ | 173.0959, 163.1111, 155.0845, 145.1010 | ASR | [20, 33] |
| 172 | 48.17 | Licochalcone A ∗☆ | Flavonoids | C21H22O4 | 337.1445 | 337.1445 | 0.0 | [M − H]− | 307.0978, 281.082, 243.104 | [31] | |
| 173 | 48.56 | Isoglycyrol ∗☆ | Flavonoids | C21H18O6 | 365.1031 | 365.1029 | −0.5 | [M − H]− | 335.0561, 307.0248, 295.0251 | [31] | |
| 174 | 49.13 | 20(S)-Ginsenoside Rs3 ∗ | Saponins | C44H74O14 | 871.5061 | 871.5056 | −0.6 | [M + COOH]− | 825.5012, 783.4903, 621.4387, 459.3845, 765.4792 | [35] | |
| 175 | 49.26 | Atractylenolide II | Terpene lactones | C15H20O2 | 233.1536 | 233.1532 | −1.7 | [M + H]+ | 215.1431, 187.1473, 169.1047, 151.0747, 145.1009 | ARP | [10] |
| 176 | 49.33 | 20(R)-Ginsenoside Rs3 ∗☆ | Saponins | C44H74O14 | 871.5061 | 871.5074 | 1.5 | [M + COOH]− | 825.5021, 783.4910, 621.4384, 459.3875, 765.4807 | [35] | |
| 177 | 49.39 | Z-Ligustilide | Phthalides | C12H14O2 | 191.1067 | 191.1062 | −2.6 | [M + H]+ | 173.0956, 163.1112, 155.0847, 145.1010 | ASR | [20, 33] |
| 178 | 50.00 | Ginsenoside Rk1 ∗ | Saponins | C42H70O12 | 811.4849 | 811.4850 | 0.1 | [M + COOH]− | 765.4802, 603.4275, 161.0458 | [31] | |
| 179 | 50.19 | Ginsenoside Rg5 ∗ | Saponins | C42H70O12 | 811.4849 | 811.4856 | 0.9 | [M + COOH]− | 765.4800, 603.4263, 161.0458 | [40] | |
| 180 | 52.21 | Glycyrrhetinic acid ∗☆ | Saponins | C30H46O4 | 469.3323 | 469.3327 | 0.9 | [M − H]− | 425.3406 | [31] |
- ∗Only detected in Qi-Fu-Yin prescription, not detected in herbs; ☆detected in Qi-Fu-Yin prescription for the first time.
3.1.1. GRR
Triterpene saponins are the main components of GRR [45]. Ginsenosides can be divided into protopanaxatriol (PPT), protopanaxadiol (PPD), and oleanolic acid (OA) according to their mother skeleton. The diagnostic ions at m/z 475.38, 459.38, and 455.35 corresponded to the PPT, PPD, and OA-type aglycones, respectively. Some special PPT-type ginsenosides were detected at m/z 457.37 owing to dehydration between the 20(21) or 20(22) bonds (Table 1). Continuous or simultaneous loss of different types of glycosyl moieties is another characteristic fragment distribution of ginsenosides. The 132, 146, 162, and 176 Da values indicated the presence of an Ara or Xyl, Rha, Glc, and GlcA glycosyl moiety, respectively. Based on the fragmentation rules, 28 saponins were identified.
Compound 142 produced the adduct ion [M + COOH]− (m/z 1123.5918) and deprotonated molecular ion [M − H]− (m/z 1077.5854), indicating a molecular formula of C53H90O22. Diagnostic ions at m/z 915.5348, 783.4945, 621.4401, and 459.3809 revealed that it was a PPD-type ginsenoside with continuous or simultaneous elimination of Glc and Ara moieties. Thus, compound 142 was assigned to ginsenoside Rc (Table 1). Analogously, PPT-type compounds 79, 84, 88, 104, 118, 123, 131–133, 136, and 137 and PPD-type compounds 139, 142, 144, 145, 149, 170, 174, 176, 178, and 179 were also preliminarily characterized according to their fragmentation pathways and retention times (Table 1). Compounds 158, 164, 165, and 168 had characteristic fragments at m/z 457.37 and were characterized as special PPT-type ginsenosides (Table 1).
Compound 141 only produced a deprotonated molecular ion [M − H]− and diagnostic ions at m/z 455.3527, which indicated that it was an OA-type ginsenoside. Fragmentation ions at m/z 793.4382, 731.4392, 613.3755, and 569.3857 indicated the continuous or simultaneous loss of Glc, GlcA, and CO2. Similarly, compounds 148 and 169 were tentatively assigned (Table 1).
3.1.2. RRP
Iridoid glycosides are considered the main components of RRP. The negative ion mode was selected to characterize the RRP components because the fragmentation pathway of glycosyl was easier to detect in the negative ion mode (Figure S1). According to the fragmentation rules, 12 phenylethanoid glycosides, 2 iridoid glycosides, 3 ionone glycosides, and 1 organic acid were identified.
The loss of acyl residues is a characteristic fragmentation pattern of phenylethanoid glycosides. Compound 53 produced a deprotonated molecular ion [M − H]− (m/z 623.1989) in the negative ion mode, which indicated a molecular formula of C29H36O15. The detection of fragmentation ions at m/z 461.1667, 443.1555, and 315.1083 suggested the continuous neutral loss of caffeoyl, H2O, and Rha; therefore, compound 53 was identified as acteoside (Figure S2A). Compounds 86 and 92 produced deprotonated molecular ions [M − H]− (m/z 651.23), indicating a molecular formula of C31H40O15. Fragmentation ions at m/z 505.17 and 475.18 corresponded to their neutral loss of Rha and feruloyl. Compounds 86 and 92 were identified as isomartynoside and martynoside, respectively, based on their retention times (Table 1). Other compounds were also preliminarily characterized according to MS1/MS2 data and retention times available in the literature.
3.1.3. ASR
Organic acids and phthalides are the primary components of ASR, and both can be detected in the positive as well as negative ion modes. The loss of acyl residues in the negative ion mode is characteristic of the fragmentation pattern of organic acids. Phthalides were easily detected by the loss of H2O and CO through ring opening in the positive ion mode. According to the fragmentation rules, 14 organic acids and 13 phthalides were identified.
Compound 5 produced a deprotonated molecular ion [M − H]− (m/z 353.0878) in the negative ion mode, indicating a molecular formula of C16H18O9. Fragmentation ions at m/z 191.0563 and 161.0245 indicated the presence of caffeoyl, and the m/z values 155.0350 and 127.0400 indicated the continuous loss of CO and CO2. Compounds 13 and 15 were isomers of compound 5. Compounds 5, 13, and 15 were identified as 5-caffeoylquinic acid, chlorogenic acid, and 4-caffeoylquinic acid, respectively, according to the retention time (Table 1).
Alkyl phthalides, such as compound 116 (3-n-butylphthalide), showed abundant protonated molecular ions [M + H]+ in the positive ion mode (Table 1). Characteristic fragmentation ions were produced at m/z 173, 155, and 145 because of the continuous or simultaneous neutral loss of H2O and CO, while hydroxylated phthalides such as compound 55 (senkyunolide I) showed higher intensities at [M + H−H2O]+ (Table 1).
3.1.4. ARP
Terpenoids and their lactones are the main components of ARP. Terpene lactones were easily detected by the loss of H2O, CO, and CnH2n in the positive ion mode. One organic acid and three terpene lactones were identified according to the fragmentation rules.
Compound 175 presented a deprotonated molecular ion [M − H]− (m/z 233.1532) in the positive ion mode, indicating a molecular formula of C16H18O9. Fragmentation ions at m/z 215.1431 and 187.1473 indicated the continuous neutral loss of H2O and CO, whereas the m/z values 159.0795, 145.1009, and 131.0848 indicated the continuous neutral loss of CnH2n; thus, compound 175 was identified as atractylenolide II (Table 1).
3.1.5. GRP
Flavonoids and saponins are the primary components of GRP. Flavonoids have a cyclohexene structure, which readily occurred owing to reverse Diels–Alder (RDA) cleavage in the negative ion mode. Except for the aglycones of compounds 77 and 127, all flavonoids were flavonoid glycosides, which were subdivided into O-glycosides and C-glycosides owing to the different bonding types between glycosyl and aglycones (Table 1). The former can only be detected by the loss of different types of glycosyl groups (Glc, Api, and others), whereas the latter can also be detected by the fragments of CnH2nOn generated from cross-ring cleavage reactions. Saponins can be easily detected by the characteristic fragments of glucuronic acid residues (GlcA) at m/z 351.05 and 193.03 in the negative ion mode. Seventeen flavonoids, 18 saponins, and 1 organic acid were identified according to the fragmentation rules.
Compound 37 presented an [M − H]− peak at m/z 563.1408, indicating a molecular formula of C26H28O14. Fragmentation ions at the m/z values 503.1197, 473.1098, 443.0992, 413.0882, 383.0778, and 353.0674 indicated the continuous neutral loss of CH2O (30 Da); therefore, compound 37 was identified as schaftoside, as shown in Figure S2B. Compound 40 was identified as liquiritin using standard solutions, which presented an [M − H]− peak at m/z 417.1194 and characteristic product ions at m/z 255.0665 with the loss of Glc, and m/z values of 135.0089 and 119.0504 due to RDA cleavage (Table 1). Other flavonoids were identified using data from the literature.
According to the standard solutions, compound 150 was identified as glycyrrhizic acid, which showed [M − H]− at m/z 821.3972, and m/z 803.3855, 777.4059, and 759.3961 due to the simultaneous loss of CO2 and H2O. Fragmentation ions at m/z 645.3648, 469.3324, 351.0572, and 193.0356 indicated that the mother skeleton was connected to two GlcA groups (Table 1). There were some isomers at m/z 821.39, 823.41, and 837.39 that were preliminarily characterized according to their fragmentation rules and retention times in the literature.
3.1.6. ZSS
Flavonoids and saponins are the main components of ZSS. A total of 10 flavonoids, 2 saponins, 9 alkaloids, and 2 organic acids were identified.
Most of the identified flavonoids contained a structure nucleus of spinosin, and a few of them were the common C-glycosyl flavonoids. Fragmentation ions at m/z 327.08 represented the flavonoid base peak of spinosin in the positive ion mode, and m/z 445.11, 427.10, 325.07, and 307.06 were detected in the negative ion mode (Table 1). Compound 47 was identified as spinosin based on a comparison of standard solutions and presented [M − H]− at m/z 607.1674. Owing to the cross-ring cleavage reaction, characteristic product ions at m/z 487.1252, 367.0823, 337.0722, and 307.0614 were readily observed. In addition, m/z 445.1144 and 427.1039 indicated the neutral loss of Glc and H2O, as shown in Figure S2C. Other spinosin flavonoids were identified in the same manner. Common C-glycosyl flavonoids also displayed a neutral loss of CnH2nOn due to the cross-ring cleavage reaction. Combined with the [M − H]− peak, compounds 28 and 48 were identified as vicenin II and swertisin, respectively (Table 1).
A large number of dammarane-type triterpene glycosides, including inner and outer sugar, were detected in ZSS. The inner sugar was usually Ara (132 Da), whereas the outer sugar generally included Xyl (132 Da), Rha (146 Da), or Glc (162 Da). The characteristic aglycone ions and dehydration products of saponin were easily observed at m/z 455.35 and 437.34, respectively.
Alkaloids can only be detected in the positive ion mode. Compounds 12, 23, and 25 yielded [M]+, whereas others produced [M + H]+ peaks (Table 1). According to the MS1/MS2 data, eight isoquinoline alkaloids and one cyclopeptide alkaloid were identified.
3.1.7. PRP
The main components of PRP are xanthones, sucrose esters, oligosaccharide esters, and saponins. Both sucrose esters and xanthones have low molecular weights, whereas oligosaccharide esters and saponins are larger. Based on the fragmentation characteristics of the different types of components, 16 sucrose esters, 14 oligosaccharide esters, 11 saponins, 6 xanthones, and 2 organic acids were identified.
The main characteristic of sugar esters in the negative mode is the neutral loss of acyl (acetyl, feruloyl, p-coumaroyl, sinapoyl, and p-hydroxy benzoyl) residues. For example, compound 90 produced an [M − H]− ion at m/z 767.2416, which corresponds to the molecular formula of C35H44O19. In the MS/MS spectrum, Z2− (m/z 529.1567), Z1− (m/z 367.1038), 0,4X− (m/z 325.0935), 0,2X− (m/z 265.0721), Y2− (m/z 237.0770), Z0− (m/z 205.0507), Y0− (m/z 223.0613), and Z0−−CH3 (m/z 190.0271) ions were formed. The presence of Z2−, Y2− and Y0−, Z0− ions indicated the existence of 3,4,5-trimethoxycinnamic acid and sinapoyl, respectively. The presence of Z2−, Z1− and Z0− ions indicated that 3,4,5-trimethoxycinnamic acid and sinapoyl moieties were situated on the glucose and fructose residues, respectively. Therefore, compound 90 was deduced to be tenuifoliside C, as shown in Figure S3. The fragmentation rule of oligosaccharide esters was similar to that of sucrose esters. Compound 119 produced an [M − H]− ion at m/z 1349.4019, corresponding to the molecular formula of C61H74O34, whereas the m/z values 1307.3907 and 163.0409, 145.0304 indicated the presence of acetyl and p-coumaroyl, respectively; thus, it was identified as tenuifoliose H (Table 1). The remaining 15 sucrose esters and 13 oligosaccharide esters were characterized on the basis of fragmentation rules and the literature.
The basic structure of saponins in PRP mainly comprised an aglycone substituted at C-3 with a mono-glucosyl saccharide (A-chain) and at C-28 with a second complex oligosaccharide (B-chain). Saponins produced characteristic fragments at m/z 455 and 425 in the negative ion mode because of the easy elimination of CH2OH (30 Da) on C-14. For example, compound 107 produced a deprotonated molecular ion [M − H]− (m/z 1103.5328) in the negative ion mode, indicating a molecular formula of C53H84O24. Characteristic fragments were easily observed at m/z 455.3185 [M − H−Glc−H2O−CO2−Fuc−Rha−Xyl]− and m/z 425.3075 [M−H−Glc−H2O−CO2−Fuc−Rha−Xyl−CH2O]− in the MS/MS spectrum. Therefore, compound 107 was deduced to be polygalasaponin XXVIII (Table 1). According to the fragmentation rules, the remaining 10 saponins were preliminarily characterized.
Characteristic fragments of CnH2nOn were found for xanthones due to cross-ring cleavage. Compound 41 showed a deprotonated molecular [M − H]− ion at m/z 567.1361, indicating a molecular formula of C25H28O15. In the MS/MS spectrum, fragment ions at m/z 435.0932, 417.0839, 375.0736, 357.0621, 345.0620, 327.0518, 315.0515, and 297.0408 corresponded to Y1−, Y1−−H2O, 0,4X−, 0,4X−−H2O, 0,3X−, 0,3X−−H2O, 0,2X−, and 0,2X−−H2O, respectively. The Y1− ions were generated by the loss of Api. The 0,2X−, 0,3X−, and 0,4X− ions were observed in the MS/MS spectrum, mainly via the cross-ring cleavage reactions in the Glc residue. Therefore, compound 10 was identified as polygalaxanthone III, as shown in Figure 2.
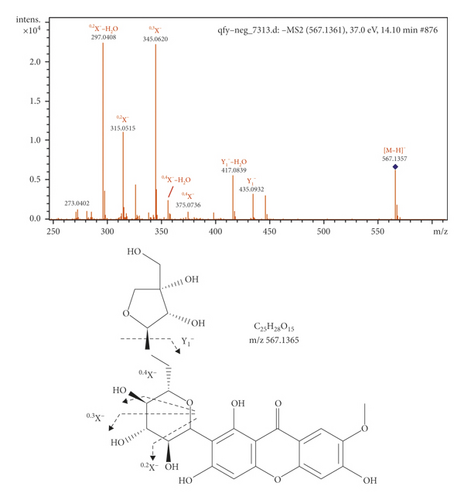
3.2. Characterizing the Prototype Components in Plasma after Oral Administration of Qi-Fu-Yin
The identification process for the prototype components was similar to that used in vitro. Using the same UPLC-Q-TOF-MS conditions, 51 prototype components were preliminarily identified by comparing the components of Qi-Fu-Yin in vitro, including 24 triterpene saponins, 10 phthalides, 8 flavonoids, 4 sucrose esters, 1 organic acid, 1 alkaloid, 1 xanthone, 1 terpene lactone, and 1 ionone. Among them, 10 components were compared with the reference standards, and others were identified by comparing the retention times, fragmentation pathways, and MS/MS spectra (Table 2, Figure 3).
| No. | tR (min) | Name | Formula | Theoretical mass (Da) | Measured mass (Da) | Error (ppm) | Precursor ions | Main MS/MS fragment ions | P | CSF |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 4.17 | Sibiricose A5 | C22H30O14 | 517.1563 | 517.1566 | 0.6 | [M − H]− | 193.0514, 175.0405, 160.0170 | + | |
| P2 | 5.13 | Sibiricose A1 | C23H32O15 | 547.1668 | 547.1658 | −1.8 | [M − H]− | 367.1030, 223.0627, 205.0508, 190.0274 | + | |
| P3 | 7.31 | Magnoflorine | C20H23NO4+ | 342.1700 | 342.1697 | −0.8 | [M + H]+ | 297.1119, 282.0888, 265.0843 | + | |
| P4 | 13.59 | Liquiritin | C21H22O9 | 417.1191 | 417.1189 | −0.5 | [M − H]− | 255.0666, 135.0091, 119.0508 | + | |
| P5 | 14.07 | Polygalaxanthone III | C25H28O15 | 567.1355 | 567.1352 | −0.5 | [M − H]− | 435.0944, 357.0600, 345.0606, 315.0522, 297.0395 | + | |
| P6 | 14.16 | Liquiritin apioside | C26H30O13 | 549.1614 | 549.1609 | −0.9 | [M − H]− | 255.0662, 417.1186, 175.02373, 135.0086, 113.0248 | + | |
| P7 | 14.85 | Spinosin | C28H32O15 | 607.1668 | 607.1665 | −0.5 | [M − H]− | 487.1252, 445.1177, 367.0823, 337.0722, 307.0614 | + | |
| P8 | 17.24 | Senkyunolide I | C12H16O4 | 207.1015 | 207.1012 | −1.4 | [M + H–H2O]+ | 189.0910, 161.1026, 147.0814 | + | + |
| P9 | 18.77 | Senkyunolide H | C12H16O4 | 207.1015 | 207.1013 | −1.0 | [M + H–H2O]+ | − | + | + |
| P10 | 18.88 | 3,6′-Disinapoyl sucrose | C34H42O19 | 753.2248 | 753.2251 | 0.4 | [M − H]− | 547.1668, 529.1565, 265.0748, 223.0595, 205.0540 | + | + |
| P11 | 19.46 | 3,4,5-Trimethoxycinnamic acid | C12H14O5 | 237.0768 | 237.0766 | −0.8 | [M − H]− | 193.0870, 161.0609, 108.0217 | + | |
| P12 | 20.90 | Isoliquiritin apioside | C26H30O13 | 549.1614 | 549.1628 | 2.5 | [M − H]− | 255.0664, 135.0077, 119.0515 | + | |
| P13 | 21.29 | Isoliquiritin | C21H22O9 | 417.1191 | 417.1195 | 1.0 | [M − H]− | 255.0659, 135.0089, 119.0499 | + | |
| P14 | 21.58 | Tenuifoliside A | C31H38O17 | 681.2036 | 681.2002 | −5.0 | [M − H]− | 179.0327, 137.0244 | + | |
| P15 | 22.35 | Liquiritigenin | C15H12O4 | 255.0663 | 255.0665 | 0.8 | [M − H]− | 135.0087, 119.0503 | + | |
| P16 | 23.69 | Senkyunolide D or isomer | C12H14O4 | 221.0819 | 221.0823 | 1.8 | [M − H]− | 177.0927, 147.0459 | + | |
| P17 | 24.48 | Ginsenoside Rg1 | C42H72O14 | 845.4904 | 845.4900 | −0.5 | [M + COOH]− | 475.3815, 179.0564, 161.0454 | + | |
| P18 | 24.85 | Ginsenoside Re | C48H82O18 | 991.5483 | 991.5436 | −4.7 | [M + COOH]− | 783.4934, 475.3719, 179.0566, 161.0460 | + | |
| P19 | 26.01 | Senkyunolide D or isomer | C12H14O4 | 221.0819 | 221.0821 | 0.9 | [M − H]− | 177.0925, 147.0453, 134.0374 | + | |
| P20 | 30.70 | Aeginetic acid | C15H24O4 | 267.1602 | 267.1608 | 2.2 | [M − H]− | 178.9213, 153.0928 | + | |
| P21 | 32.08 | Polygalasaponin XXVIII | C53H84O24 | 1103.5280 | 1103.5280 | 0.0 | [M − H]− | 455.3189, 425.3078 | + | |
| P22 | 32.66 | Senkyunolide F or isomer | C12H14O3 | 205.0870 | 205.0872 | 1.0 | [M − H]− | 161.0977, 187.9911, 149.0043 | + | |
| P23 | 32.90 | Butylidenephthalide | C12H12O2 | 189.0910 | 189.0913 | 1.6 | [M + H]+ | 171.0768, 161.0935, 143.0845, 117.0676 | + | + |
| P24 | 33.22 | Butylphthalide | C12H14O2 | 191.1066 | 191.1064 | −1.0 | [M + H]+ | − | + | + |
| P25 | 34.01 | Ginsenoside Rf | C42H72O14 | 845.4904 | 845.4887 | −2.0 | [M + COOH]− | 179.0575, 161.0465 | + | |
| P26 | 34.38 | Senkyunolide A or isomer | C12H16O2 | 193.1223 | 193.1228 | 2.6 | [M + H]+ | 147.1167, 175.1169, 137.0591 | + | |
| P27 | 35.35 | Licorice saponin A3 | C48H72O21 | 983.4493 | 983.4463 | −3.1 | [M − H]− | 351.0583, 193.0364 | + | |
| P28 | 35.64 | Isoliquiritigenin | C15H12O4 | 255.0663 | 255.0658 | −2.0 | [M − H]− | 135.0083, 119.0498 | + | |
| P29 | 35.83 | Formononetin | C16H12O4 | 267.0663 | 267.0657 | −2.2 | [M − H]− | − | + | |
| P30 | 36.61 | Tenuifolin | C36H56O12 | 679.3699 | 679.3718 | 2.8 | [M − H]− | 455.3136, 425.3101 | + | + |
| P31 | 36.80 | 22-Hydroxyl-glycyrrhizin | C42H62O17 | 837.3914 | 837.3894 | −2.4 | [M − H]− | 351.0584, 193.0366 | + | |
| P32 | 36.89 | 20(S)-Ginsenoside Rh1 | C36H62O9 | 683.4376 | 683.4367 | −1.3 | [M + COOH]− | 637.4335, 475.3806, 161.0462 | + | + |
| P33 | 37.37 | Senkyunolide A or isomer | C12H16O2 | 193.1223 | 193.1224 | 0.5 | [M + H]+ | 175.1158, 147.1162, 137.0595 | + | |
| P34 | 37.66 | 20(R)-Ginsenoside Rh1 | C36H62O9 | 683.4376 | 683.4367 | −1.3 | [M + COOH]− | 161.0463 | + | + |
| P35 | 37.85 | Jujuboside A | C58H94O26 | 1251.6015 | 1251.5971 | −3.5 | [M + COOH]− | 179.0566, 161.0465 | + | |
| P36 | 38.72 | Ginsenoside Rb1 | C54H92O23 | 1153.6011 | 1153.5980 | −2.7 | [M + COOH]− | 1107.5959 | + | |
| P37 | 39.69 | Ginsenoside Ro | C48H76O19 | 955.4908 | 955.4899 | −0.9 | [M − H]− | 793.4379, 179.0563, 119.0352 | + | |
| P38 | 39.69 | Ginsenoside Rc | C53H90O22 | 1123.5906 | 1123.5856 | −4.5 | [M + COOH]− | 459.3809, 149.0451, 191.0563 | + | |
| P39 | 39.78 | Licorice saponin G2 | C42H62O17 | 837.3914 | 837.3891 | −2.7 | [M − H]− | 351.056, 193.0351 | + | |
| P40 | 40.75 | Ginsenoside Rb2 | C53H90O22 | 1123.5906 | 1123.5908 | 0.2 | [M + COOH]− | 1077.5866 | + | |
| P41 | 41.32 | Rhaoglycyrrhizin | C48H72O20 | 967.4544 | 967.4506 | −3.9 | [M − H]− | 1077.5859 | + | |
| P42 | 42.76 | Glycyrrhizic acid | C42H62O16 | 821.3965 | 821.3942 | −2.8 | [M − H]− | 645.3641, 351.0564, 193.0351, 175.0249 | + | |
| P43 | 42.76 | Ginsenoside Rd | C48H82O18 | 991.5483 | 991.5474 | −0.9 | [M + COOH]− | 179.0564, 161.0456 | + | |
| P44 | 44.63 | Atractylenolide I | C15H18O2 | 231.1379 | 231.1378 | −0.4 | [M + H]+ | − | + | |
| P45 | 46.13 | Licorice saponin J2 | C42H64O16 | 823.4122 | 823.4091 | −3.8 | [M − H]− | 351.0573, 193.0357 | + | |
| P46 | 46.90 | Ginsenoside Rk3 | C36H60O8 | 665.4270 | 665.4248 | −3.3 | [M + COOH]− | 161.0449 | + | |
| P47 | 47.09 | Ginsenoside Rh4 | C36H60O8 | 665.4270 | 665.4258 | −1.8 | [M + COOH]− | 161.0450 | + | |
| P48 | 47.38 | Zingibroside R1 | C42H66O14 | 793.4380 | 793.4374 | −0.8 | [M − H]− | 731.4388, 631.3849 | + | + |
| P49 | 47.86 | Ginsenoside Rg3 | C42H72O13 | 829.4955 | 829.4934 | −2.5 | [M − H]− | 783.4886, 621.4365, 459.3812, 161.0454 | + | |
| P50 | 49.40 | Z-Ligustilide | C12H14O2 | 191.1066 | 191.1070 | 2.1 | [M + H]+ | − | + | |
| P51 | 52.30 | Glycyrrhetinic acid | C30H46O4 | 469.3323 | 469.3316 | −1.5 | [M − H]− | 425.3414 | + | + |
| M1 | 8.78 | Ferulic acid-4-sulfate | C10H10O7S | 273.0074 | 273.0074 | 0.0 | [M − H]− | 193.0507, 149.0246 | + | |
| M2 | 9.45 | Ferulic acid-4-sulfate isomer | C10H10O7S | 273.0074 | 273.0073 | −0.4 | [M − H]− | 193.0504, 149.0245 | + | |
| M3 | 13.59 | Liquiritigenin-7-O-glucuronide | C21H20O10 | 431.0984 | 431.0977 | −1.6 | [M − H]− | 255.0662, 175.0250, 135.0088 | + | + |
| M4 | 13.97 | Liquiritigenin-4′-O-glucuronide | C21H20O10 | 431.0984 | 431.0982 | −0.5 | [M − H]− | 255.0662, 175.025, 135.0088 | + | + |
| M5 | 15.70 | Liquiritigenin+2H + sulfate | C15H14O7S | 337.0382 | 337.0380 | −0.6 | [M − H]− | 257.0824 | + | |
| M6 | 17.83 | Liquiritigenin-4′-O-sulfate | C15H12O7S | 335.0231 | 335.0225 | −1.8 | [M − H]− | 255.0664, 135.0088, 119.0503 | + | |
| M7 | 19.36 | (Iso)Liquiritigenin+2H + sulfate | C15H14O7S | 337.0382 | 337.0383 | 0.3 | [M − H]− | 257.0823, 151.0401 | + | |
| M8 | 20.81 | (Iso)Liquiritigenin+2H + sulfate | C15H14O7S | 337.0382 | 337.0385 | 0.9 | [M − H]− | 257.0820, 151.0398 | + | |
| M9 | 21.10 | Formononetin-7-O-glucuronide | C22H20O10 | 443.0984 | 443.0984 | 0.0 | [M − H]− | 267.0661, 175.0249, 135.0453 | + | |
| M10 | 23.12 | Isoliquiritigenin-4′-O-glucuronide | C21H20O10 | 431.0984 | 431.0978 | −1.4 | [M − H]− | 255.0662, 175.0247, 135.0088 | + | + |
| M11 | 27.07 | Isoliquiritigenin+2H + sulfate | C15H14O7S | 337.0382 | 337.0390 | 2.4 | [M − H]− | 257.0821 | + | |
| M12 | 28.20 | Acetylcysteine conjugate of senkyunolide I or senkyunolide H | C17H23NO6S | 370.1324 | 370.1316 | −2.2 | [M + H]+ | 207.1024, 189.0925, 161.0957 | + | |
| M13 | 29.38 | Formononetin-7-O-sulfate | C16H12O7S | 347.0231 | 347.0230 | −0.3 | [M − H]− | 267.0664, 252.0429 | + | |
| M14 | 29.67 | Isoliquiritigenin-6′-O-sulfate | C15H12O7S | 335.0231 | 335.0236 | 1.5 | [M − H]− | 255.0666, 135.009, 119.0508 | + | |
| M15 | 38.72 | Compound K-H2 | C36H60O8 | 619.4215 | 619.4193 | −3.6 | [M − H]− | 457.3683, 439.3216 | + | |
| M16 | 45.27 | Compound K | C36H62O8 | 621.4372 | 621.4355 | −2.7 | [M − H]− | 459.3846, 179.0559, 161.0453 | + | |
| M17 | 45.94 | Compound K+2O-2H2 | C36H58O10 | 665.3906 | 665.3883 | −3.5 | [M − H]− | 651.4118, 409.2751, 375.2533 | + | |
| M18 | 46.42 | Compound K+3O-H2 | C36H59O11 | 667.4063 | 667.4047 | −2.4 | [M − H]− | 605.4042, 491.3720, 175.0237, 113.0242 | + | |
| M19 | 46.90 | Compound K+3O-H2 | C36H59O11 | 667.4063 | 667.4042 | −3.1 | [M − H]− | 605.4029, 491.3724, 175.0241, 113.0242 | + | |
| M20 | 46.99 | Compound K+2O-2H2 | C36H58O10 | 665.3906 | 665.3893 | −2.0 | [M − H]− | 651.4113, 409.2746, 375.2527 | + | |
| M21 | 47.76 | Compound K+2O-2H2 | C36H58O10 | 665.3906 | 665.3897 | −1.4 | [M − H]− | 651.4119, 409.2752, 375.2535 | + | |
| M22 | 48.13 | Glycyrrhetinic acid-2H | C30H44O4 | 469.3318 | 469.3312 | −1.3 | [M+H]+ | 451.3203, 423.3243 | + | |
| M23 | 48.15 | Glycyrrhetinic acid + O | C30H46O5 | 485.3272 | 485.3263 | −1.9 | [M − H]− | 441.3357 | + | + |
| M24 | 48.34 | Compound K+2O-2H2 | C36H58O10 | 665.3906 | 665.3904 | −0.3 | [M − H]− | 491.3368, 473.3269, 443.3161, 193.0352, 175.0246, 113.0242 | + | |
| M25 | 48.92 | Glycyrrhetinic acid + O | C30H46O5 | 485.3272 | 485.3256 | −3.3 | [M − H]− | 441.3361 | + | + |
| M26 | 49.59 | Protopanaxadiol+2O + H2 | C30H50O5 | 489.3585 | 489.3575 | −2.0 | [M − H]− | 473.3261, 445.3677, 375.2896 | + | |
| M27 | 45.36 | Glycyrrhetinic acid + O | C30H46O5 | 485.3272 | 485.3279 | 1.4 | [M − H]− | 441.3383 | + |
- P, plasma; CSF, cerebrospinal fluid; −, not detected +, detected.
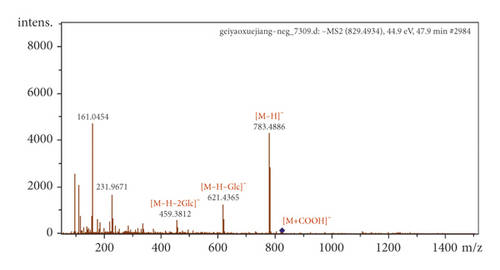
Some saponins with low molecular weights can be directly absorbed into blood. For example, P53 produced the adduct ion [M + COOH]− (m/z 829.4934) and deprotonated molecular ion [M − H]− (m/z 783.4886), indicating a molecular formula of C42H72O13. Diagnostic ions at m/z 621.4365, 459.3812, and 161.0454 suggested that it was a PPD-type ginsenoside with continuous or simultaneous elimination of Glc moieties. Thus, P53 was assigned to ginsenoside Rg3 (Figure 4(a)). P41 produced an [M − H]− peak at m/z 837.3891, indicating a molecular formula of C42H62O17. Furthermore, P41 was identified as glycyrrhizin G2 because of the characteristic fragments of glucuronic acid residues, which were readily detected at m/z 351.056 and 193.0351 in the negative ion mode (Figure 4(b)).
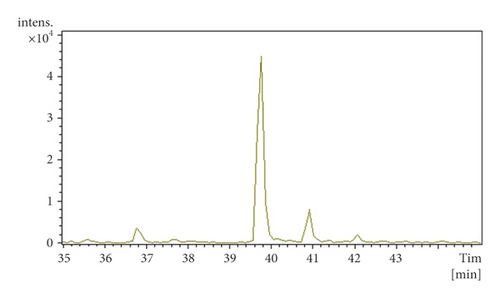
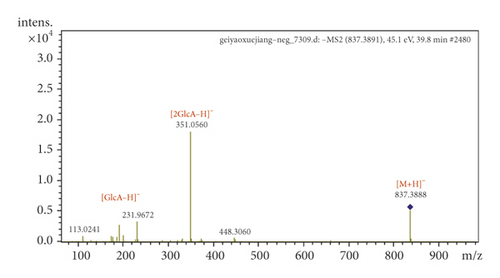
Hydroxylated phthalides showed a higher intensity at [M + H−H2O]+ and were detected by the loss of H2O, CO, and CnH2n through ring opening in the positive ion mode. For example, P10 and P11 produced [M + H–H2O]+ at m/z 207.10, and the characteristic fragmentation ions at m/z 189.09, 161.10, and 147.08 indicated neutral loss of H2O, CO, and C3H6. P10 and P11 were identified as senkyunolides I and H, respectively, according to the retention time (Figure S4).
3.3. Characterization of Metabolites in Plasma after Oral Administration of Qi-Fu-Yin
Twenty-six metabolites were preliminarily identified by comparing with data from the metabolite database, mainly including oxidation, reduction, glucuronidation, and sulfation (Table 2, Figure 5). The pathways of some metabolites are shown in Figure 6.

The [M–H]– ions of M1 and M2 were at m/z 273.00, which showed a mass shift of 79.96 Da (SO3) from 193.05 [ferulic acid–H]– and provided the fragment ions at m/z 149.02 [ferulic acid–H–CO2]–. Combined with the predicted chemical formula of C10H10O7S, M1 and M2 were tentatively deduced to be sulfate conjugates of ferulic acid [36] (Figure 6).
M3, M4, and M10 showed the [M–H]– ion at m/z 431.10, which was 176.03 Da more than that of isoliquiritigenin. The MS2 spectra of M3, M4, and M10 all provided fragment ions at m/z 255.07, 175.02, and 135.01, respectively, which suggested the presence of an isoliquiritigenin group. Combining these data with the retention times [46], M3, M4, and M10 were tentatively deduced to be liquiritigenin-7-O-glucuronide, liquiritigenin-4′-O-glucuronide, and isoliquiritigenin-4′-O-glucuronide, respectively (Figure 6).
M6 and M14 showed the [M–H]– ion at m/z 335.02 (C15H12O7S), which was 79.96 Da (SO3) more than that at m/z 255.07. Upon combining data from the retention time and characteristic fragmentation ions at m/z 255.07 and 135.01, M6 and M14 were identified as liquiritigenin-4′-O-sulfate and isoliquiritigenin-6′-O-sulfate, respectively (Figure 6). Similarly, the [M–H]– ion of M5, M7, M8, and M11 at m/z 337.04 was approximately 2 Da more than that of M6 and M14. The product ions at m/z 257.08 were also approximately 2 Da more than those at 255.07. Combining these data with the retention time, M5, M7, M8, and M11 were deduced to be hydrogenation and sulfate conjugates of (iso)liquiritigenin (Figure 6).
M9 and M13 produced the same fragment ions at m/z 267.07, which were believed to be metabolites of formononetin; according to the adduct ions of m/z 443.0984 and 347.0230, they were identified as formononetin-7-O-glucuronide and formononetin-7-O-sulfate, respectively (Figure 6).
M12 produced fragmentation ions at m/z 207.1024 [M + H−145−H2O]+ and 189.0925 [M + H−145−2H2O]+, which suggested the presence of a phthalide group. Combining these data with the [M + H]+ ion at m/z 370.1316 (C17H23NO6S), M12 was identified as an acetylcysteine conjugate of ligustilide I or H (Table 2).
The fragment ions at m/z 459.3846, 179.0559, and 161.0453 suggested that M16 was a PPD-type ginsenoside. Combining the predicted chemical formula of C36H62O8 and literature [29], M15, M17-21, and M24 were identified as related metabolites of compound K, according to their retention times and chemical formulae [29] (Table 2).
M22 produced fragments of m/z 423.3243 [M + H−CO2]+ in the positive ion mode, which is in accordance with the fragmentation rules of glycyrrhetinic acid. Furthermore, M22 exhibited [M + H]+ at m/z 469.3312, which was determined to be C30H44O4; therefore, M22 was identified as the dehydrogenization of glycyrrhetinic acid. Likewise, M23 and M25 produced [M–H]– ions at m/z 485.3263 and fragments of m/z 441.3357 in the negative ion mode, which represented a neutral loss of CO2 (44 Da), and were identified as hydroxylate conjugates of glycyrrhetinic acid (Table 2).
3.4. Characterization of Prototypical Components and Metabolites in the Cerebrospinal Fluid after Oral Administration of Qi-Fu-Yin
Using the same UPLC-Q-TOF-MS conditions, 10 prototype components (P8-P10, 23, 24, 30, 32, 34, 48, and 51) and 6 metabolites (M3, 4, 10, 23, 25, and 27) were preliminarily identified by comparing the components of the drugged rat plasma, among which two components were compared with the reference standards, and others were identified by comparing the retention times, fragmentation pathways, and MS/MS spectra (Table 2 and Figure 7).
4. Discussion
In recent years, LC-MS technology has been widely used in the analysis of components of TCM, combining the high separation ability of liquid chromatography with the high sensitivity of mass spectrometry [47, 48]. Up to now, the only research on the identification of components in Qi-Fu-Yin was based on UPLC-Q-TOF-MS in vitro [10]. In this present study, the same 110 components were detected consistent with previous studies [10], and 70 components were preliminarily identified for the first time in vitro (Table 1, Table S1). Among them, forty-four reported components [10] were undetected, and 18 of them were lost due to different scanning ranges (Table S1).
Qi-Fu-Yin consists of seven herbs, but there is no research on the similarities and differences of components between them after decocting. For the first time, upon comparing Qi-Fu-Yin with the seven herbs, the categories of chemical components were found to be unanimous, and the number of flavonoids and organic acids in Qi-Fu-Yin was more than the sum of seven herbs; however, the opposite was true for phenylethanoid glycosides (Figure S5). Most of the chemical components could be detected in both, but 9 and 13 chemical components were only detected in the seven herbs and Qi-Fu-Yin, respectively, and the configuration of some components changed (Figure S5, Table 1). This showed that the chemical composition of Qi-Fu-Yin is not a simple addition of compounds in its single herbs.
As far as we know, the prototype components and metabolites of the seven herbs, not Qi-Fu-Yin, in the plasma after oral administration have been reported. For example, saponins in GRR [49], GRP [46], ZSS [50], flavonoids in GRP [51], ZSS [50], phthalides in ASR [36, 52], sugar esters in PRP [53], phenylethanoid glycosides, and iridoid glycoside in RRP [54] are the main components in plasma after oral administration of herbs. In this research, 51 prototypical components and 26 metabolites of Qi-Fu-Yin, including saponins, phthalides, flavonoids, sucrose esters, organic acids, alkaloids, ionones, terpene lactones, iridoid glycoside, and their derivatives have been tentatively identified in the plasma for the first time.
Similarly, the prototype components and metabolites in the cerebrospinal fluid after oral administration of Qi-Fu-Yin have not been reported. Several research showed that some saponins in GRR [55, 56], GRP [57], and phthalides in ASR [58, 59] can be absorbed into the cerebrospinal fluid. In addition, saponins in GRR [60] and GRP [61], flavonoids in ZSS [62], and source esters in PRP [53] have been determined in the brain tissue homogenate. In this research, 10 prototypical components and 6 metabolites were preliminarily characterized in the rat cerebrospinal fluid after oral administration of Qi-Fu-Yin. Among them, butylidenephthalide, butylphthalide, 20(S)-ginsenoside Rh1, 20(R)-ginsenoside Rh1, zingibroside R1, and six other metabolites were detected in the cerebrospinal fluid for the first time. Some prototype components, as saponins, phthalides, and sucrose esters, could be directly absorbed into plasma and cerebrospinal fluid, and phthalides had a higher absorption rate (Figure 8). Some flavonoids, organic acids, alkaloids, xanthones, terpene lactones, and iridoid glycosides could be absorbed into the plasma, whereas other categories of chemical components were not detected in the plasma and cerebrospinal fluid.

Studies have shown that glycyrrhetinic acid [57], 3,6′-disinapoyl sucrose [63], tenuifolin [64], and senkyunolide I and H [65] can be absorbed into cerebrospinal fluid. Some components have been determined in the brain tissue homogenate [66–68], but whether these components can penetrate the BBB is unknown, and they may only exist in the astrocytes and/or vascular endothelial cells constituting the BBB. In this study, 3,6′-disinapoyl sucrose, ginsenoside Rh1, butylphthalide, glycyrrhetinic acid, tenuifolin, and senkyunolide I and H were detected in cerebrospinal fluid. Many studies showed that they had promising effects on neuroprotection, antiapoptosis, anti-inflammation, or antioxidative stress (Table 3). This suggested that these compounds might be potentially active components of Qi-Fu-Yin for treating AD.
| Compound | Samples | Biomarkers | Effects | References |
|---|---|---|---|---|
| 3,6′-Disinapoyl sucrose | Glutamate and H2O2-induced SHSY5Y cells |
|
Neuroprotection | [69] |
| Glutamate-induced SHSY5Y cells |
|
Antiapoptosis | [70] | |
| Ginsenoside Rh1 | Mice (6-month-old) |
|
Neuroprotection | [71] |
| IFN-γ-stimulated BV2 cells | Amounts of NO, ROS, and TNF-α↓ | Anti-inflammation | [72] | |
| Scopolamine-induced amnesic mice |
|
Antioxidative stress | [73] | |
| Butylphthalide | APP/PS1 mice | Escape latency↓, the time spent and travel distance in the target quadrant↑ in the Morris water maze test | Neuroprotection | [74] |
| Aβ1-42-induced SD rats | Protein expression of MAPK↓ | Antiapoptosis | [75] | |
| Senkyunolide H |
|
|
Antioxidative stress | [76] |
| Protein expression of Bax and caspase-3↓ | Antiapoptosis | [76] | ||
| Tenuifolin | Aβ1-42-induced BV2 cells | Amounts of TNF-α, IL-6, and IL-1β↓ | Anti-inflammation | [77] |
|
Antioxidative stress | [77] | ||
| Senkyunolide I | Glutamate-induced Neuro2a cells | Amount of caspase-3↓ | Antiapoptosis | [78] |
| Glycyrrhetinic acid | BACE1 FRET assay | Activity of BACE1↓ | Neuroprotection | [79] |
- ↓, decrease; ↑, increase; Aβ, amyloid-β; CREB, cyclic AMP response element binding protein; BDNF, brain-derived neurotrophic factor; Bax, Bcl-2 associated X protein; Bcl-2, B cell lymphoma/leukemia-2; NO, nitric oxide; ROS, reactive oxygen species; TNF-α, tumor necrosis factor-α; MDA, malondialdehyde; SOD, superoxide dismutase; CAT, catalase; GSH-Px, glutathione peroxidase; IL-6, interleukin 6; IL-1β, interleukin 1β; iNOS, inducible nitric oxide synthase; COX-2, cyclooxygenase-2; MAPK, mitogen-activated protein kinase; BACE1: β-site APP cleaving enzyme 1.
5. Conclusions
In this study, the chemical components of Qi-Fu-Yin in the plasma and cerebrospinal fluid after oral administration of Qi-Fu-Yin were preliminarily characterized using UPLC-Q-TOF-MS. To our knowledge, this is the first systematic investigation of the metabolic profiles of the constituents of Qi-Fu-Yin. In total, 51 prototypical components and 26 metabolites were tentatively identified in plasma. The major phase I metabolic pathway of Qi-Fu-Yin involved hydrogenation and oxidation, whereas that of phase II reactions included sulfate and glucuronic acid conjugation. Furthermore, 10 prototypical components and 6 metabolites, which might be responsible for the potential activity of Qi-Fu-Yin, were preliminarily characterized in the cerebrospinal fluid. This study provides a chemical basis for elucidating the active components of Qi-Fu-Yin that play roles in the treatment of AD and should further motivate research on the mechanisms underlying the anti-AD activity of Qi-Fu-Yin.
Ethical Approval
All animal procedures were approved by the Shandong University of Traditional Chinese Medicine Institutional Animal Experimentation Committee (SDUTCM20210119001).
Disclosure
Hengyu Li and Hongwei Zhao are co-first authors. Xiaorui Cheng and Jiafeng Wang are conjointly designated as corresponding authors.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
Authors’ Contributions
Xiaorui Cheng, Jiafeng Wang initiated and designed the study. Hengyu Li, Hongwei Zhao, and Xiaorui Cheng developed the method and drafted the manuscript. Dongmei Qi and Yong Yang provided experimental platform and equipment. All authors read and approved the final manuscript.
Acknowledgments
The authors would like to thank Shandong Academy of Sciences and Shandong University of Traditional Chinese Medicine Experimental Center for providing experimental platform and equipment and thanks to Xiaoming Wang for guiding this research. This study was supported by Major Basic Research Projects of Natural Science Foundation of Shandong Province (ZR2020ZD17) and Natural Science Foundation of Shandong Province (ZR202103040693, ZR2021QH271).
Open Research
Data Availability
The data used to support the findings of this study are included within the article and are available from the corresponding author upon request.




