Uniqueness of the Level Two Bayesian Network Representing a Probability Distribution
Abstract
Bayesian Networks are graphic probabilistic models through which we can acquire, capitalize on, and exploit knowledge. they are becoming an important tool for research and applications in artificial intelligence and many other fields in the last decade. This paper presents Bayesian networks and discusses the inference problem in such models. It proposes a statement of the problem and the proposed method to compute probability distributions. It also uses D-separation for simplifying the computation of probabilities in Bayesian networks. Given a Bayesian network over a family I of random variables, this paper presents a result on the computation of the probability distribution of a subset S of I using separately a computation algorithm and D-separation properties. It also shows the uniqueness of the obtained result.
1. Introduction
Bayesian networks are graphical models for probabilistic relationships among a set of variables. They have been used in many fields due to their simplicity and soundness. They are used to model, represent, and explain a domain, and they allow us to update our beliefs about some variables when some other variables are observed, which is known as inference in Bayesian networks.
Given a Bayesian network [1] relative to the set XI = (Xi) i∈I of random variables, we are interested in computing the joint probability distribution of a nonempty subset S (called target) of I.
The computation of the probability distribution of XS requires marginalizing out a set of variables of from the joint distribution PI corresponding to the Bayesian network.
In large Bayesian networks, the computation of probability distributions and conditional probability distributions may require summations relative to very large subsets of I. Consequently there is a need to order and segment, if possible, these computations into several computations that are less intensive and more accessible to a parallel treatment. These segmentations are related to the graphic properties of the Bayesian networks.
This paper describes the computation of PS using a specific order described by a proposed algorithm, and the segmentations of PS.
We consider discrete random variables, but the results presented here can be generalized to continuous random variables with the density of Xi relative to a finite measure μi (the summations will be replaced by integrations relative to those measures μi).
The paper is organized as follows. Section 2 introduces Bayesian networks and Level two Bayesian networks. We, then, present in Section 3 the inference problem in Bayesian networks and the proposed computation algorithm. Section 4 outlines D-Separation in Bayesian networks and describes graphical partitions that will allow the segmentations of the computations of probability distributions. Section 5 proves the uniqueness of the results obtained in Sections 3 and 4.
2. Bayesian Networks and level two Bayesian networks
2.1. Bayesian Networks
- (i)
the set I is finite and endowed with a structure of a directed acyclic graph (DAG), G, where, for each i:
- (a)
p(i) is the set of parents of i (p(i) = {j; (j, i) ∈ G})
- (b)
e(i) is the set of children of i (e(i) = {j; (i, j) ∈ G})
- (c)
d(i) is the set of descendants of i (d(i) = {k; ∃(ℓ0, …, ℓn) ℓ0 = i, ℓn = k, ∀ s ∈ {1, …, n} (ℓs−1, ℓs) ∈ G});
- (a)
- (ii)
for each i, Xi is independent of (Xj) j∈I−[p(i)∪d(i)] conditional on Xp(i) (for more details see, e.g., [1–5]).
2.2. level two Bayesian networks
We consider the probability distribution PI of a family (Xi) i∈I of random variables in a finite space ΩI = ∏i∈IΩi. Let ℐ be a partition of I and let us consider a DAG 𝒢 on ℐ.
We say that there is a link from J′ to J′′ (where J′ and J′′ are atoms of the partition ℐ) if (J′, J′′) ∈ 𝒢. If J ∈ ℐ, we denote by p(J) the set of parents of J, that is, the set of J′ such that (J′, J) ∈ 𝒢.
2.3. Close Descendants
We define the set of close descendants of a node J (denoted cd(J)) as the set of vertices containing the children of J and the vertices located on a path between J and one of its children.
2.4. Initial Subset
For each subset S, we denote by S+ the initial subset defined by S, that is, the set consisting of S itself and the S′ such that there is a path in 𝒢 from S′ to S.
We can identify this subset with the union of all S′ such that S′ is an ancestor of S
3. Inference in Bayesian Networks
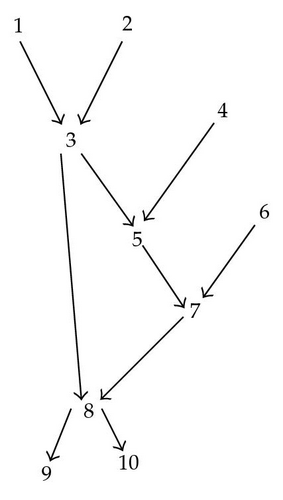
Consider the BN in Figure 1.
Suppose we are interested in computing the distribution of S = I − {3}, in other words all variables are in the target S except X3.
α is a function that depends on X1, X2, X4, X5, and X7 but has nothing to do with P1,2,4,5,7 joint probability distribution of X1, X2, X4, X5, X7.
By doing this we loose the structure of the BN.
In other words α(x1, x2, x4, x5, x7)P7/5,6(x7/x5, x6) = P5,7,8/1,2,4,6(x5, x7, x8/x1, x2, x4, x6).
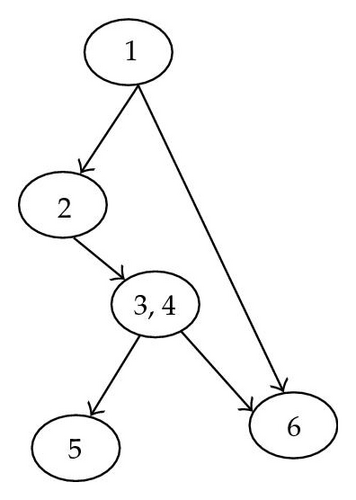
Which provides S with a structure of a level two Bayesian network shown in Figure 3.
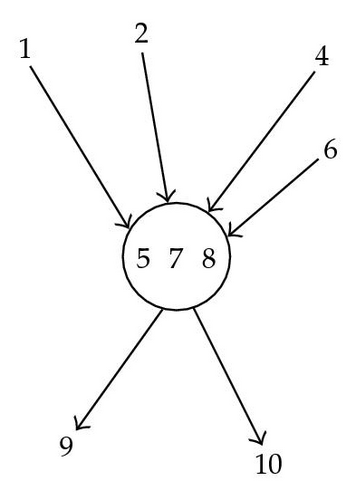
The variables used in the marginalization above, to keep a structure of a BN2, is the the set of close descendants defined above.
More general, if we have to sum out more than one variable there is a need to order the variables first. The aim of the inference will be to find the marginalization, or elimination, ordering for the arbitrary set of variables not in the target. This aim is shared by other node elimination algorithms like “variables elimination” [6], “bucket elimination” [7].
The main idea of all these algorithms is to find the best way to sum over a set of variables from a list of factors one by one. An ordering of these variables is required as an input. The computation depends on the ordering elimination; different elimination ordering produce different factors.
The algorithm we proposed to solve this problem is called the “Successive Restrictions Algorithm” (SRA) [8]. SRA is a goal-oriented algorithm that tries to find an efficient marginalization (elimination) ordering for an arbitrary joint distribution.
The general idea of the algorithm of successive restrictions is to manage the succession of summations on all random variables out of the target S in order to keep on it a structure less constraining than the Bayesian network, but which allows saving in memory; that is the structure of Bayesian Network of Level Two. This was possible using the notion of close descendants.
The principle of the algorithm was presented in details in [8].
4. D-Separation and Computations in a Bayesian Network
We have introduced an algorithm which makes possible the computation of the probability distribution of a subset of random variables (Xs) s∈S of the initial graph. It is also possible to use the SRA to compute any probability distribution of a set of variables XA conditionally to another subset XB (PA∣B).
This algorithm tries to achieve the target distribution by finding a marginalization ordering that takes into account the computational constraints of the application. It may happen that, in certain simple cases, the SRA would be less powerful than the traditional methods [6, 9–13], but it has the advantages of adapting to any subset of nodes of the initial graph, and also to present in each stage interpretable result in terms of conditional probabilities, and thus technically usable.
In addition to the SRA we propose, especially for large Bayesian networks, to segment the computations into several less heavier computations that could be carried independently. These segmentations are possible using the D-separation.
4.1. D-Separations and Classical Results
Consider a DAG (I, G). A chain is a sequence (x0, …, xn) of elements of I such that for all i ≥ 1, (xi−1, xi) ∈ G or (xi, xi−1) ∈ G.
x1, …, xn−1 are called intermediate nodes on this chain.
On an intermediate node xi a chain can have three connexions as illustrated in Figure 4.
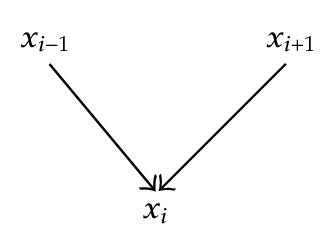
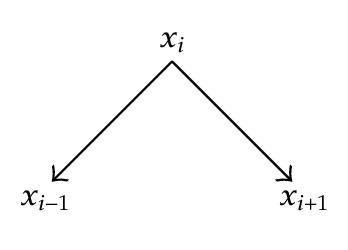
- (i)
x ∈ S and the connection is, on x, serial or diverging,
- (ii)
x ∉ S+ and the connection is converging on x.
In other words, A chain is not d-separated by S if it is in a converging connection at each intermediate node of S, and in a serial or a diverging connection at each intermediate node that has no descendants in S.
Classic Result 1 If A and B are d-separated by S. then the variables XA and XB are independent conditional on XS.
4.2. Notions
-
Markov Blanket. F(C) of a subset C is made of the parents of C, the children of C and the variables sharing a child with C.
-
Markov Boundary of C. M(C) = C ∪ F(C)
-
Close Descent of C. T(C) is the set of all close descendants of the elements in C other that those in C itself.
-
Exterior Roots of C. R(C) is the set of the parents of the elements of C ∪ T(C) other that those in C ∪ T(C) itself.
As we can see on the following example:
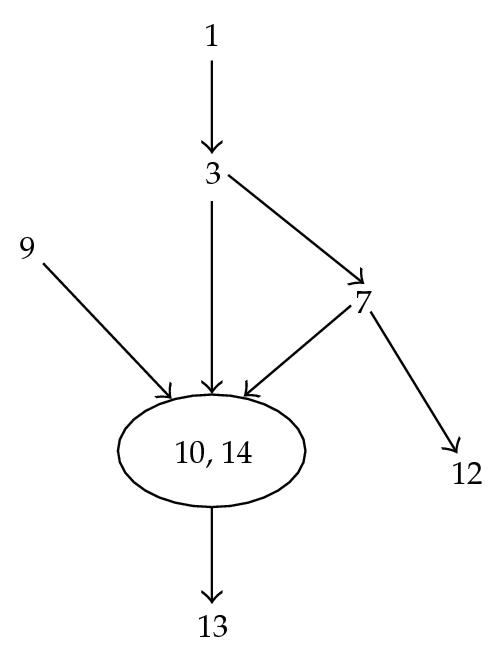
F(C) = {1,10,14,12,9}, M(C) = {3,7, 1,10,14,12,9}, T(3,7) = {10,14,12}, R(3,7) = {1,9}.
Classic Result 2 XC is d-separated from the rest of the variables conditional on the Markov blanket of C. The proof of these two results and other results related to D-separation can be found in [11].
4.3. Moral and Hypermoral Graphs
Another classic graphic property used with some inference algorithms that we can find in the literature is the notion of the Moral graph.
Moral Graph Given a DAG (I, G)
its associated moral graph is the undirected graph (I, Gm), where Gm is the set
- (i)
of pairs {i, j} such that (i, j) ∈ G or (j, i) ∈ G,
- (ii)
of pairs {i, j} such that i and j have a child in common.
In a similar way, we define what we call the hypermoral graph defined as follow:
Hypermoral Graph Given a DAG (I, G)
its associated hypermoral graph is the undirected graph (I, Ghm), where Ghm is the set:
- (i)
of pairs {i, j} such that (i, j) ∈ G or (j, i) ∈ G,
- (ii)
of pairs {i, j} such that i and j have a close descendant in common.
In Figure 5, there is a link between 4 and 7 in the hypermoral graph because they share 9 as a close descendant.
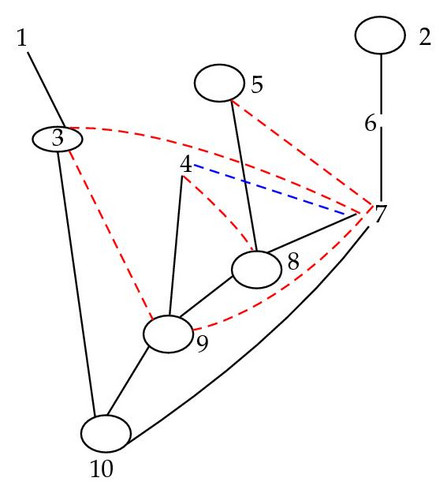
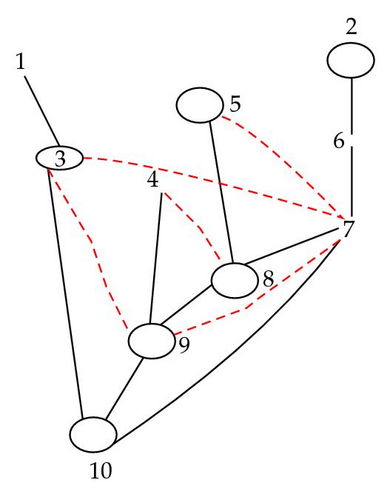
4.4. Moral and Hypermoral Partitions
The moral graph helps defining the moral partition as follow.
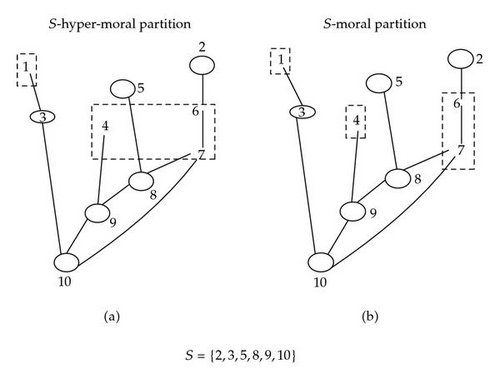
(i) We call a S-moral partition the partition of S+ − S, denoted , defined by the equivalence relation , where means “ there exists a chain from x to y, in S+ − S, not blocked by S.”
In an equivalent way “there exists a chain, in the moral graph Gm, connecting x to y without an intermediate node in S.”
In a similar way we define the hypermoral partition.
(ii) We call an S-hypermoral the partition of S+ − S, denoted , defined by the equivalence relation , where means “there exists a chain, in the hypermoral graph Ghm, connecting x to y without an intermediate node in S”. As Illustrated in Figure 6.
4.5. Results
The following results show the possibility of segmenting the computation of the probability distribution PS.
Theorem 4.1. Let (I, G, PI) be a BN, and let S be a subset of I. Let be the S-hypermoral partition of S+ − S and let K be the set of elements of S which are not close descendants of any element of S+ − S, that is, K = {k ∈ S, ∀ y ∈ S+ − S, k ∉ cd(y)}. Then,
The proof of the theorem can be found in [14].
Theorem 4.2. The set Qs of singletons {k}, where k ∈ K (if K ≠ ∅), and of subsets T(C)∩S+, where , constitutes a partition of S. As Illustrated in Figure 7.
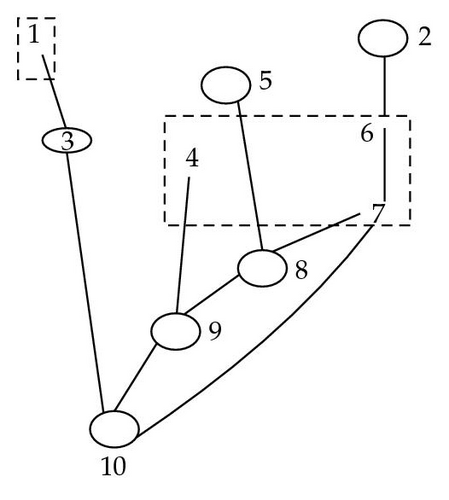
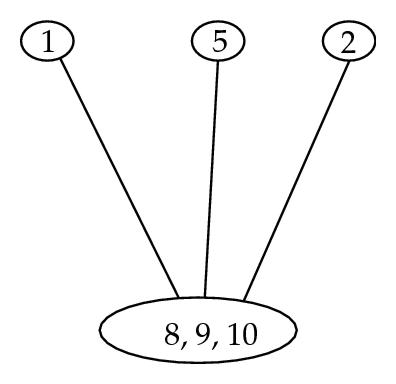
5. Unique Partition
We have seen in the last two sections that the application of the SRA for the computation of PS provides a structure of BN2, and that the use of D-Separations properties allows the segmentation of the computation of PS and provides also a structure of a level two Bayesian network on S. In fact the two obtained structures are same, this results is giving by the following theorem.
Theorem 5.1. The following two sets.
- (1)
The subsets T(C)∩S+, where .
- (2)
The set Qs of singletons {k}, where k ∈ K (if K ≠ ∅)
Interpretation This theorem indicates that the level two Bayesian network, characterizing the probability distribution PS, obtained by application of the SRA, is unique independently of the choices done while running the algorithm. This unique partition is constituted of sets of the two types 1 and 2 mentioned above.
Proof. Let us show that the partition of the target S consists of the subsets of types 1 and 2 as mentioned above in the theorem, in other words consists of T(C)∩S+ where , and {k} for all k ∈ K.
As S+ is a BN-containing S, without a loss of generality, we can limit ourselves to the case where S+ = I.
The application of the SRA for the computation of PS requires marginalizing out the set of variables of following a specific order. Let try to show that the obtained partition by application of the SRA is same mentioned above.
Let us proof this result by induction on the cardinality of .
Let us assume that Card. In this case has only one element, .
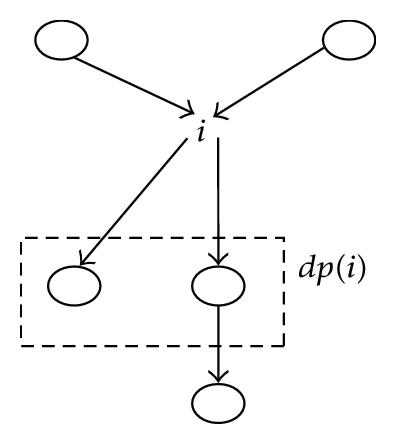
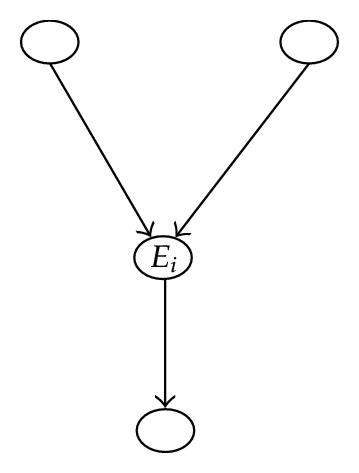
On one side, marginalizing out the variable (á) i, by application of the SRA, creates a new node Ei that contains the close descendants of i (i.e., Ei = cd(i)).
The BN2 resulting from this marginalization is formed of the new node Ei along with all other remaining nodes, in other words all the {k} such that k ∈ I − (i1 ∪ cd(i1)) = S − Ei, which is shown in Figure 8.
On the other side, since Card, is constituted of a unique equivalent class, C = {i}, so, by definition, the partition of S is constituted of
- (1)
T(C)∩S+ = cd(i)∩I = cd(i),
- (2)
all other nodes, in other words the nodes k ∈ K such that
(5.1)
Let us suppose now that Card, and (i1, …, in) as an hierarchical order on .
We are going to sum out following the inverse order of the given hierarchical order.
Let us assume that the result is right till step ℓ (in other words marginalizing out in−ℓ+1) and let′s proof the result for step ℓ + 1 (in other words marginalizing out in−ℓ).
What justify the proof by induction is that once the marginalizing out in, …, in−ℓ+1 these last elements will not interfere in the next steps of marginalizing out in−ℓ, …, i1.
The result is right till step ℓ means that we dispose on A = S ∪ {i1, …, in−ℓ}, of a partition that contains the subsets {T(C)∩S+} for , and the nodes {k} such that , (i.e., ), where is the S-hypermoral partition associated to A.
Showing the result for ℓ + 1 means to find a partition of S ∪ {i1, …, in−ℓ−1} constituted of subsets of type 1 et 2 mentioned above.
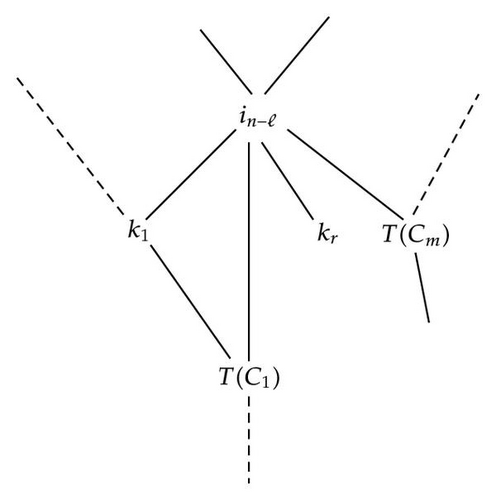
On one hand, let′s first try to find the partition obtained by application of the SRA. We know that, the marginalization of in−ℓ creates a new node, , which contains the close descendants of in−ℓ in the BN2 on A (), shown by Figure 9.
So if we write J = {k1, …, kr}, the set of K such that for all j ∈ {1, …, r}, and L = {C1, …, Cm}, the set of such that for all s ∈ {1, …, m}, T(Cs)∩cd(in−ℓ) ≠ ∅.
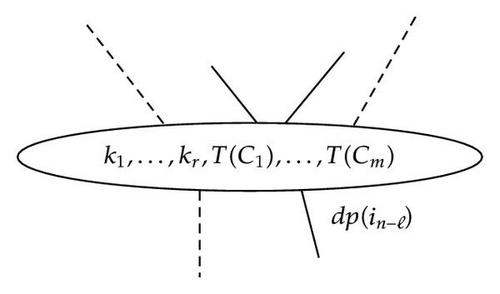
In this case, the partition of B = A − {in−ℓ} = S ∪ {i1, …, in−ℓ−1} is constituted of the new node , and all the other nodes that are left isolated, in other words all the nodes k ∈ (K − J) and all the {T(C)∩A+} where .
If we write so the partition of B by application of the SRA is composed of the following two types of sets:
- (1)
,
- (2)
all other isolated nodes, that is, the nodes of K′.
On the other hand, let′s now try to determine the partition of B = A − {in−ℓ} using the D-Separation properties.
We have on A a partition composed of nodes {T(C)∩A+} where , and all nodes {k} such that .
Since B = A − {in−ℓ}, and no descendant of in−ℓ is in i1, …, in−ℓ−1, (by definition of the hierarchical order), so only one equivalent subset is associated, , it results by definition, the partition of B = S ∪ {i1, …, in−ℓ−1} is composed of the following types of subsets:
- (1)
,
- (2)
all other isolated nodes R such that
(5.2)
So we have .
This shows that this partition is same partition obtained by application of the SRA.