The potential and pitfalls of de-extinction
Abstract
‘De-extinction’ is the nascent discipline that aims to one day literally revive now-extinct species from the dead. Although we have yet to see any successful attempts to truly resurrect an extinct species, several technologies are now in place that might one day provide a plausible solution. Thus, the area is receiving increased attention from both scientists and the general public. However, how far does present technology place us from the ultimate goal? We address the state of the art of several prominent de-extinction methods: back-breeding, cloning, synthetic genomics and genome editing, and discuss some of the major outstanding challenges for each. We also discuss some of the wider challenges facing de-extinction, including both what might constitute the definition of success and what might be needed to successfully take a recreated animal and confer on it the ability to establish itself back in the wild.
Introduction
The recreation of extinct species, otherwise known as ‘de-extinction’, has recently begun to receive a significant amount of attention at both the popular and scientific level. Media reports are increasingly presenting the topic in an optimistic framework, and conveying the impression that we face a real possibility of bringing mammoth or other charismatic beasts back from extinction within the near future (Harris 2000; Sherkow & Greely 2013; Zimmer 2013; Ogden 2014; Shapiro 2015a,b). Some of the other heavily discussed species include the auroch (Bos primigenius) – a large species of cattle previously numerous throughout Europe; the passenger pigeon (Ectopistes migratorius) – which used to number in billions in North America; and the famous dog-like marsupial of Australia – the thylacine (Thylacinus cynocephalus). It seems that people are thus most attracted by the peculiar and the gargantuan, although often citing reasons of ethical duty for de-extinction. The topic has also been the centre of somewhat heated debates in the scientific community regarding the correct allocation of scientific funds for ecosystem conservation and restoration.
Although a number of grounds have been presented to either encourage or justify de-extinction, including improving the ecosystems that humanity is dependent upon, ethical obligations to maintain nature and life forms, or simply to entertain an audience in a zoo or park (Zimov 2005; Sherkow & Greely 2013), an obvious drawback of any de-extinction project is the time, resources, equipment and expertise required to finalise a fully functioning self-sustaining species, and the financial cost of such an endeavour will be also substantial. One of the most highly resonating criticisms of de-extinction is the perspective that, at such a price, the goals do not justify the means, and urgent initiatives to save extant species or maintain ecosystems should be prioritised before the resurrection of the extinct (Yule 2002; Fletcher 2008; Sandler 2014). But opposing this view is the prospect that de-extinction may be a lucrative business (Cottrell et al. 2014; Whittle et al. 2015), perhaps only needing an initial subsidy. Charismatic animals are often the main attractions in wildlife tourism and zoos, and can evoke sustainability awareness in the public and promote nature conservation (Simberloff 1998; Williams et al. 2000). Conveniently, a common theme in many de-extinction candidates is ‘big’ – including big teeth, big claws and big horns. These animals could raise considerable sums through ticket sales, possibly sufficient to finance their own recreation as well as support further nature conservation.
A further argument that has been presented in support of de-extinction is that humans have caused the extinctions. Human hunting of megafauna and environmental impacts of human activity is considered to be one of the main drivers of global Holocene megafauna extinctions (Zimov et al. 1995; Miller et al. 1999; Alroy 2001; Fisher 2009; Gill et al. 2009). During this time frame, anatomically modern humans spread to most of their present range, and as they did, human presence generally correlated with extinctions (Burney & Flannery 2005; Sandom et al. 2014; Surovell et al. 2016). As Neolithic societies established and eventually became industrialised, global human populations modified increasing areas of the planet's surface, and hence virtually all ecosystems are today affected by human enterprise, causing wide ranging and accelerating extinction (Thomas et al. 2004; Harnik et al. 2012; Keith et al. 2015). The further back in time extinctions took place, the more difficult it is to determine the extent of the role humans played however, and early megafauna extinctions are especially hotly debated (Burney & Flannery 2005, 2006; Wroe et al. 2006; Lorenzen et al. 2011; Sandom et al. 2014; Willerslev et al. 2014). Nevertheless, there are many cases where it is absolutely certain that humans exterminated a species. These may be perceived by some to be ethically wrong acts which we as a species have a duty to right. Integrated in this line of thought is the idea that there is an order within nature which we are independent from, and that human-mediated extinctions are unnatural. Whether humans should be viewed as components of past or present ecosystems, and whether man-made de-extinction is more justified than man-made extinction is an ethical debate which is out of scope of this paper. However, setting ethics aside, and focusing on functionality, it has been reasoned that the introduction of extinct species could be useful to preserve and restore ecosystems. It is in humanity's best interest to maintain an environment that is functionally useful to society and aesthetically pleasing, supporting the range of ecosystem services required for us to thrive (DeFries et al. 2004; Zhang et al. 2007), and the introduction of keystone species may boost ecosystem function and stability. Species introductions in modern nature management are generally viewed as acceptable if the species was exterminated from the ecosystem, and its reintroduction is believed to be needed to complete the previous system (Duffy 2003; Svenning et al. 2016). Bringing back extinct species may also further our knowledge of evolutionary biology and our understanding of the breadth of possibilities for physiological functioning of animals that existed in different time periods and climates. Extinct genes could also be potentially used to improve livestock (FAO 2016).
Today, although de-extinction leading to viable organisms has yet to be achieved, there are a number of technical options available that might eventually allow the possibility. The suitability of the different technologies themselves depends largely on the quality of genetic information that can be obtained from the extinct species, but includes using extant biological phenotypic and/or genomic variation to guide breeding so as to recreate extinct genomes/phenomes; cloning using well-preserved cells; in vitro fertilisation using preserved gametes or stem cells that would be transferred to a surrogate species; production of an entirely synthetic genome based on the genome sequence of the extinct species; and the editing of the genome of a species closely related to the extinct species. Each option possesses its own merits and challenges, which will be broadly addressed, together with some issues that span the range of options being considered using examples within the sections. One of the key considerations applicable to each option is whether true de-extinction is achievable for a species, with the result likely having some physiological or at least behavioural differences to the original species.
De-extinction through back-breeding
Before addressing the approaches that are most technologically demanding, it is worth discussing whether de-extinction warrants such methods, or if in at least some species, could be achieved through natural crosses between extant organisms – in particular where there are still sufficiently closely related extant species. Such pools of related genetic diversity can be in the form of either a sister species, a domesticate or hybrids, all of which can serve as a starting point to de-extinction by breeding ‘back’ an edition of the extinct species. To date, such efforts have largely focused on two iconic species, the quagga (Equus quagga; Quaggaproject 2016) and, as discussed in more detail here, the aurochs.
European cattle (Bos taurus) were first domesticated approximately 10 000 years ago (Larson et al. 2014) from the aurochs (Loftus et al. 1994; Bailey et al. 1996), and the original auroch species finally went extinct in Poland in 1627 following decline due to human pressure through the millennia (Rokosz 1995). The attempted de-extinction of the aurochs during the interwar period in Germany is the earliest and perhaps best known example of a back-breeding programme (Heck 1951; Van Vuure 2002). The ambition of the initiators – the two Heck brothers, was to mix European cattle breeds chosen based on key morphological factors, in an effort to resurrect at least the phenotype of wild aurochs (Heck 1951). Although Heck cattle do have certain resemblances to aurochs, they nevertheless maintain clear phenotypic differences (Van Vuure 2002, 2005; Fig. 1), and thus several subsequent initiatives have continued to cross Heck cattle with other cattle breeds in order to improve the phenotype (Stokstad 2015; TaurOs Project 2016).

While certainly an interesting approach, this example raises several questions that are fundamental to de-extinction. Firstly, how does one even define the phenotype to be pursued? In the case of the aurochs, the evidence available ranges from cave paintings, to archaeological remains, to historical texts and artwork, and thus which source(s) is/are the most appropriate? (Heck 1951; Van Vuure 2005). Secondly, even if we can define a phenotype, what level of reconstruction similarity might be acceptable? For example, although the Heck brothers claimed successful recreation of the aurochs 11–12 years after starting their breeding programme, as mentioned above others do not accept the Heck phenotype as sufficient (Van Vuure 2002, 2005). Ultimately, both questions are embedded within the broader question that all de-extinction studies will face – at what point can one (in this case) call cattle, aurochs? The answer to this question is by no means simple, as in answering this one must also face the challenges inherent in defining species themselves, and accounting for the geographic and temporal variation that the extinct form may have exhibited.
While to some degree relevant at the morphological level (as targeted by the Heck brothers), given its underlying role in defining phenotype, this variation is even more important at the genomic level. Thus, even if morphological similarity between back-bred aurochs and the original animal may be high, genomically the similarity may be no greater than that of conventional cow breeds. Given this, one future approach might be to draw on recent developments within the field of paleogenomics (i.e. the study of ancient samples at the full genomic level, e.g. Miller et al. 2008) and a growing understanding that due to past admixture between extant and now-extinct forms, the genomes of extant species may contain fossil remnants of their extinct relatives. First demonstrated in the case of Neanderthal and Denisovan relicts in human genomes (e.g. Green et al. 2010; Reich et al. 2010; Vernot et al. 2016), the comparison of a recently sequenced aurochs genome with those of modern cattle breeds has shown similar fossil remnants exist for aurochs (Park et al. 2015). Given this, plus the observation that different modern cattle breeds contain different genomic fossils, careful backcrossing could be used to progressively enrich the aurochs component of descendent genomes and in doing so gradually ‘restitch’ an aurochs genome together. While such an approach will still face the above-mentioned challenges of what exactly defines an aurochs, it may be less subjective than attempts guided purely by morphology.
Breeding back may be theoretically possible for the de-extinction of some species, but due to the high number of breeding generations that would be needed to alter most species, as well as the lack of control over the specific genetic make-up, this option is undesirable for obtaining a species that differs from its relative by more than a handful of traits. In such cases, there are other techniques that would in theory be more efficient, through relying on various forms of artificial genetic manipulation.
De-extinction through artificial reproductive technologies
Cloning
Cloning is a reproductive strategy in which new, genetically identical organisms are created from the cells of a ‘parent’. This principle can be applied to grow a fully viable multicellular diploid organism from a single intact cell nuclei without meiosis, and involves transplanting the nuclei from a cell of a target individual into a host egg cell that has been emptied of its own nuclei, then manipulating this cell into entering mitosis in such a way that it eventually develops into the full cloned individual (Colman 1999). Cloning has received great attention as a method for de-extinction, as a potential means to generated genotypically authentic living organisms from well-preserved cellular subsamples, either deliberately taken from species prior to extinction, or even recovered from naturally frozen extinct species (Benirschke 1984; Ryder & Benirschke 1997; Ryder et al. 2000; Kato et al. 2009). As such, cloning is currently envisaged as the quickest potential route to de-extinction, and furthermore, several near successful attempts have been made to recreate a recently extinct Pyrenean ibex (Capra pyrenaica pyrenaica), using frozen skin cells from the last surviving individual (Folch et al. 2009). Nevertheless, due to a number of technical reasons- not least because cloning suffers for a high failure rate even when using the most optimal conditions with perfectly fresh cell material and using the same species as surrogate- the use of cloning in de-extinction will be challenging, if not impossible, for most candidate species. We continue to focus on the de-extinction of mammals, but see Box 1 for a brief description of considerations for some other classes of animals.
Box 1. De-extinction prospects for non-mammalian systems
Here, the prospects of de-extinction of animals other than mammals are briefly discussed considering technologies other than back-breeding.
Birds
Due to the reproductive physiology of birds, whose oocytes are fertilised shortly after release before being covered in a hard shell, genetic engineering is much more difficult in general compared to mammals (Sang 2004). Cloning involves all sorts of additional barriers, with IVF and zygote transfer both fraught with problems, and possibly not achievable. Premature zygotes extracted by killing the mother can be brought to hatch using albumen as a culture, but with chickens at least it is not possible to see, and therefore manipulate, the nucleus of the oocytes (Sang 2004), so even if it is possible in theory to extract a viable oocyte before fertilisation, SCNT may not be achievable.
The use of genome editing via CRISPR/Cas9 would be more difficult and possibly not feasible due to the need to manipulate the cell line and discriminate between those which have and have not been altered in the desired way. The preservation of such a large cell for the number of years required to introduce all of the required changes may also be very difficult.
These barriers may significantly challenge some much discussed de-extinction candidates–at least in the present state of knowledge. These include the passenger pigeon, the 12-ft-tall giant moa of New Zealand, and the large, penguin-like species: the great auk (The Long Now Foundation 2016, http://longnow.org/).
De-extinction of birds may be possible by focussing on a method that does not rely on the manipulation of individual cells, however, via the production of chimeras. This relies on the availability of primordial germ cells (the embryonic procurers of germ cells; Naito et al. 1994; Tajima et al. 1993) or blastoderm cells (Petitte et al. 1990), and injection into the embryo or blastoderm, respectively. The aim would be the incorporation of the cells into the ovaries or testes, which results in chimeric animals that can produce some sperm or egg cells derived from the foreign cells. Producing two individuals by these methods and mating them could result in the birth of an animal containing only the incorporated DNA. Chimera production has involved the transfer of hundreds of the foreign cells (Petitte et al. 1990; Tajima et al. 1993; Naito et al. 1994), so de-extinction success via this method would most likely rely on the foresight of preservation of an abundance of these specific cells for currently endangered species.
Reptiles
Due to similarities in the reproductive physiology between reptiles and birds, the same hurdles would need to be overcome in order to achieve de-extinction.
Fish
Although there are no known cases of iSCNT using fish, intergenus embryo-derived nuclear transfer has been achieved, resulting in viable offspring (Sun & Zhu 2014). Thus, although it is unclear whether cloning using adult somatic cells is possible, de-extinction is certainly a possibility via the nuclear transfer of well-preserved stem cells, and genome editing may also be a possibility.
Amphibians
Cloning technology grew from initial experiments with amphibians, with the first embryonic nuclear transfer achieved in frogs in 1952 (Briggs and King 1952), a year before the double helical structure of DNA was first proposed by Watson and Crick (1953). The first cloning via somatic cell nuclear transfer (SCNT) was achieved in frogs a few years later (Gurdon et al. 1958). The nucleus used was from a tadpole cell, however, and to date there has not been an adult frog produced by cloning using somatic nucleus transfer from an adult frog cell (Wilmut et al. 2015).
This much slower rate of progress in the world of amphibian SCNT may make prospects slim of bringing back the gastric brooding frog (Rheobatrachus spp.) any time soon, which met their demise in the 1980s, due to the only available cells being somatic adult cells. These two frog species uniquely swallowed their eggs, and converted their stomachs into a womb for the development of their young before live frogs eventually hopped out of their mouths. If de-extinction were achieved, it may help to further our progress in the research of artificial uteri, as well as increase our knowledge of evolutionary biology.
Amphibians are going extinct more quickly than any other class of vertebrate (Clulow et al. 2014), so the collection and preservation of embryo or tadpole cells could be vital for species revival in the future. De-extinction may be possible through the same mechanisms described for fish.
The ideal cell source for de-extinction success would be relatively undifferentiated cells such as those taken from a foetus (Wilmut et al. 1997), or other early developmental stage in animals that undergo metamorphosis (Gurdon et al. 1958). However, due to the natural limits of available suitable starting material, cloning for de-extinction would likely require the use of somatic cell nuclear transfer (SCNT) using cells taken from a fully developed animal. At face value, such attempts seem promising – this approach has been successfully applied to extant mammals (Verma et al. 2015). Although most notably first used in 1996 to give rise to ‘Dolly’ the sheep, who was produced from a mammary cell nucleus inserted into an enucleated sheep oocyte (Wilmut et al. 1997), to date successes span a wide taxonomic diversity, including five orders and 13 genera (Verma et al. 2015). Furthermore, only a few years after the birth of Dolly we saw the first live mammalian offspring produced by interspecies somatic cell nuclear transfer (iSCNT), in the form of an endangered gaur (Bos gaurus) birthed from a Bos taurus mother (Lanza et al. 2000). Although the original animal died only 2 days after birth due to a bacterial infection, this is considered unrelated to the cloning process itself (Vogel 2001), and a subsequent attempt has resulted in the birth and subsequent survival of a healthy gaur (Srirattana et al. 2012). Cloning by iSCNT involves the transfer of a somatic cell nucleus to the enucleated oocyte of a different species or subspecies, and would be the method used for de-extinction due to the obvious impossibility of using the same species as a surrogate. This technology has triumphed only when using very closely related animals, with other cases including cloning of the coyote (Canis latrans) using domestic dog (Canis lupus familiaris) cells and surrogates (Hwang et al. 2013), wild cat (Felis silvestris lybica) and sand cat (Felis margarita) using the domestic cat (Felis catus; Gómez et al. 2004, 2008), and a mouflon (Ovis orientalis musimon), an endangered subspecies of sheep, using domestic sheep (Ovis aries) oocyte and surrogate following nucleus retrieval from a cell of a deceased mouflon (Loi et al. 2001). It is clear, therefore, that de-extinction by cloning using this technique is almost certainly achievable in theory. In practice, however, it has not been so successful, with the most successful attempt so far represented by the above-mentioned Pyrenean ibex. While this species was brought back into the world using a domestic goat (Capra aegagrus hircus) oocyte and surrogate, and nuclear transfer from a specially preserved Pyrenean ibex cell line prepared from the last extant ibex prior to its extinction, the recreated ibex died only minutes after birth due to lung defects (Folch et al. 2009). Death of cloned offspring shortly after birth is not, in fact, uncommon in SCNT on extant species (Watanabe 2013). Thus, this ‘failure’ should not necessarily be held in repudiation of prospects de-extinction success. The survival of the ibex would have been more of a symbolic, than technological, breakthrough. Perhaps more problematic for larger scale application in de-extinction is that due to various incompatibilities, this method is (most likely) limited to animals that can (or could in the past) naturally interbreed. Furthermore, even if such incompatibilities can be overcome, success will require excellent understanding of the reproductive biology of the surrogate (something we lack for most vertebrate species, Comizzoli 2015) and will only be applicable to extinct species from which exceptionally well-preserved cells can be found.
The challenge of incompatibility is apparent in the problems facing even conventional SCNT approaches – the number of viable offspring that develop as a proportion of total embryos transferred to surrogates is low (estimated at 1–5% in 2007 [Oback 2008], but due to technical improvements ca. 5–10% today [Watanabe 2013; Long et al. 2014]). iSCNT is harder still to achieve. Due to incompatibility between cell and nucleus, and the cell and the nucleus and surrogate, reported success rates are 1–6% (Loi et al. 2011). More promising, however, is that although a further possible conflict is between mitochondria and nucleus (Hiendleder et al. 2004), working cells have been created using depletion of mitochondria in the oocyte and addition of mitochondria from the same species as the nuclear DNA (Jiang et al. 2011). Mitochondria can also be added by using the cell fusion method for importation of the nucleus (Moro et al. 2015).
The viability of cloning as an option for de-extinction also depends on whether the animals created are capable of living healthy lives during both adolescence and maturity. The incomplete reprogramming of the somatic nuclei after insertion into the oocyte is a common problem and is strongly linked to developmental issues, causing both abortion and birth defects (Long et al. 2014). In cattle, up to 50% of those born die within the first 200 days, but those that survive after this point appear normal (Watanabe 2013). It is possible, however, that inefficiency through incomplete reprogramming can be overcome. For example, the insertion of a mammalian nucleus into a frog oocyte can result in more efficient reprogramming via de-methylation (Byrne et al. 2003), and treatment of nuclei with frog oocyte extract can increase the efficiency of SCNT (Yang et al. 2012). Another potential problem is shortened telomeres in the clone. The first successfully SCNT cloned animal (Dolly the sheep), died prematurely of cancer (at 7 years old), possibly relating to telomere length (Wilmut et al. 2015). However, telomere length has not been demonstrated to influence survival time in cloned cattle (Watanabe 2013).
We must then consider, in case of successful birth of a fully healthy animal, whether the process may cause differences in physiology or behaviour compared to pre-extinction populations. One of the potential problems would be phenotypic plasticity. It has been found that subtle differences during development, such as temperature and chemical cues, can greatly influence the phenotype of individuals (Gilbert 2001). Perhaps the differences between the species would thus result in some abnormal phenotypes. Cytoplasmic factors have also been found to alter development, resulting in abnormal numbers of vertebrae in offspring of interspecies embryonic nuclear transfer experiments in fish (Sun & Zhu 2014).
For animals that either have no sufficiently closely related species to act as a surrogate, or in which practical issues such as size difference make the match unsuitable, a potential future solution would be the use of an artificial uterus and placenta. This area of research is still in its infancy, so it is difficult to speculate on whether and when the complete in vitro development of offspring would be achievable. Placenta has also been found to be able to continue to function following abortion, so the possible implantation of placenta in to an artificial uterus could make partial in vitro development possible (Bulletti et al. 2011). This would negate the need to construct an artificial placenta, and also means that if an embryo could be kept alive within a surrogate for only a period of the gestation, it could be sufficient to allow development to complete in vitro. Implantation of this kind has been attempted, but not achieved (Bulletti et al. 2011).
Although the incompatibilities discussed above represent major challenges, it is conceivable that they will be solved through future technological breakthroughs. There is, however, a second major challenge to the direct cloning of almost all extinct organisms that might be much harder to overcome – specifically whether we will actually be able find intact cell nuclei to clone. Intact nuclei preservation requires rapid freezing of cells, and thus with the exception of very recently extinct species that had their tissues harvested and directly frozen prior to their demise, specimens from permafrozen environments (such as the extinct megafauna from Beringia) are perhaps the most likely candidates for this. Indeed, identifiable nuclei have even been reported from such specimens (Kato et al. 2009). Nevertheless, it should be stressed that even if preserved cells are found, the DNA within such nuclei may well be degraded beyond the point at which they can be used in cloning (Shapiro 2015a). For example, Kato et al. (2009) attempted somatic cell nuclear transfer of ~15 000-year-old nuclei found in mammoth muscles and bone marrow cells, into mouse oocytes, but found that they did not begin to develop. Although it is unclear whether this is due to the condition of the DNA or nucleus, or the incompatibility between the mouse oocyte and the mammoth nucleus, both are likely (Kato et al. 2009). Limited choice of cell type for cloning can also affect the probability of de-extinction, with nuclei derived from different cell types found to distinctly impact the prospect of successful development (Saini et al. 2015), so the retrieval of a healthy, intact nucleus is not the only important consideration.
Given these challenges, there is a growing interest in alternative de-extinction methods that do not require frozen cells, but rather involve first sequencing the full genome of extinct organisms, and then using these sequences as template for either (i) resynthesising the extinct genome – a method referred to as synthetic genomes, or (ii) the direct editing of genomes in extant relatives – a method referred to as genome editing. Obtaining an ancient genome as template, generating synthetic genomes or editing of an existing genome is not trivial, but as discussed in the following in more detail, it is at least hypothetically possible.
Synthetic genomes
Synthetic genomes are those that are functional, yet literally created de novo in a laboratory, through the synthesis and joining of smaller oligonucleotide strings (Baker 2011). Since the first early steps when Khorana et al. (1972) synthesised a single gene, the size of synthetic genomes has continued to increase, up to sizes of ca. 1 million base pairs in bacteria, synthesised by in vitro recombination (Gibson et al. 2008, 2010). The synthesis of functional eukaryotic genomes is much more complicated due to their much larger sizes and additional complexities related to eukaryotic cells. Nevertheless, a relatively short eukaryotic chromosome of around 0.27 million bases from a yeast has been recently synthesised in vivo, using recombination to gradually replace sections of the original chromosome with synthetic strands (Annaluru et al. 2014). While this represents less than 0.01% of the length of most animal genomes, and thus using current techniques the synthesis of a full-length vertebrate genome represents a considerable economic and labour challenge, it is likely that things will not remain this way as genome synthesis techniques improve. Greater challenges will be to ensure that (a) the reconstructed genome is of sufficient completeness to be functional – something particularly challenging given the incompleteness of palaeogenomes, and (b) the architecture of the DNA and its surroundings is correct for it to fulfil its functions within a cell.
The challenge of completeness relates to the fact that even if it were possible to synthesise the full genome sequence recovered through palaeogenomic analysis of an extinct sample, there is a high probability that it will not be the complete genome sequence. It has long been known that DNA rapidly degrades following death (Lindahl 1993), with a maximum survival time in optimal conditions (cold and dry) of possibly 1–2 million years (Smith et al. 2001; Willerslev et al. 2007). Thus, for most of the species that once lived on the planet but are now gone, and that have left biological remains to the present, there will simply be no DNA surviving, and thus their de-extinction is a non-starter. As for those that are represented by biological material containing traces of their DNA, such tissues are not the flash frozen tissues preferred by biobanks, and the DNA in such materials is generally heavily fragmented, with molecules often in the range of less than 100 bp in length (e.g. Poinar et al. 2006). This short size precludes the de novo sequencing of such genomes; instead, their sequences must be recreated through mapping against the reference genomes of their closest extant relatives (e.g. Prüfer et al. 2010; Shapiro & Hofreiter 2014). For example, the available woolly mammoth (Mammuthus primigenius) draft genomes (Miller et al. 2008; Lynch et al. 2015; Palkopoulou et al. 2015) were constructed by mapping to the Broad Institute African elephant (Loxodonta africana) genome LoxAfr. An almost guaranteed limitation of such approaches is that in addition to presenting difficulties relating to detecting genomic rearrangements, because mapping requires sequence homology, regions of low complexity and/or rapid evolution will be harder to map and thus may be absent from the draft (Prüfer et al. 2010; Shapiro & Hofreiter 2014). This problem will be exasperated as evolutionary divergence increases, and ultimately one can expect that any recreated genome that is based upon a palaeogenomic blue print will be only partially complete.
The challenge of genomic architecture relates to a number of considerations, not least the enclosing of DNA within a nuclear membrane. Although we have some knowledge of how nuclear envelopes form around DNA from studying cells during telophase – the final stage in cell replication when the nucleus re-forms, we do not fully understand the mechanisms that govern the process (Larijani & Poccia 2009), and the ability to artificially manipulate nuclear envelope formation would constitute another great milestone in cellular biology. Nucleus-like structures can form spontaneously around DNA inserted into a frog cell (Blow & Laskey 1986), but it is unclear whether one nucleus could form around multiple chromosomes, and whether the structure is fully functional. Nuclear envelope formation appears to rely on the presence of chromatin (Grant & Wilson 1997), which has also been found to form around ‘naked’ DNA inserted into frog eggs (Hirano 1991). DNA within a nucleus is usually assembled in these protein complexes which affect transcription and cell replication (Hayes & Lee 1997), so incorrect assembly could have significant consequences on development. Centromeres are also vital components of chromosomes for replication, but even the sequencing of these regions is difficult (Hayden & Willard 2012) and the assembly of the proteins that comprise this region constitutes another barrier (Aldrup-Macdonald & Sullivan 2014). The nucleolus is another vital consideration, with the presence of a structure called the nucleolus precursor body (which has structural differences from the somatic cell nucleolus) being a necessary component of the oocyte in order for successful embryonic development (Kyogoku et al. 2014).
Genome editing
An alternate option for recreating extinct species, which is possibly more immediately viable, would be to edit the genome of a closely related living species using a draft genome of the extinct species as a guide. The woolly mammoth based on editing of an Asian elephant (Elephas maximus) genome is used as an example here, but the principles are applicable to other possible cases of de-extinction using ancient DNA, and given the obvious challenges in working with a large mammal such as the elephant as a surrogate, it may be that smaller systems represent a more attractive model for exploring such methods further (Box 2).
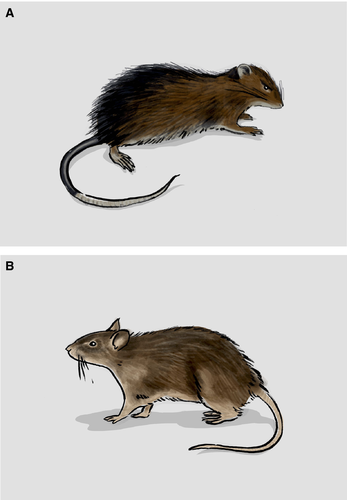
Box 2. The Christmas Island rat as a model for de-extinction?
While initial de-extinction attempts based around genome editing are largely being discussed in the context of mammoths using elephants (Shapiro 2015a,b), to progress faster it may be useful to employ a model system that is less charismatic, yet ultimately more tractable. In this regard, we propose the little known Christmas Island rat (Rattus macleari; Fig. 2A) – a species that is not (as far as we know) a major candidate being explored by current de-extinction projects. We argue, however, it is a potentially valuable model organism to work on due to its close relationship to the brown rat (Rattus norvegicus; Fig. 2B) that would be an ideal host. In comparison with Asian elephants, the brown rat has (i) fast gestation times (23 days [King 1913] vs the 22 months of elephants [Meyer et al. 2004]) and large litters (ca 14), (ii) a well-characterised genome and physiology, (iii) likely considerably fewer ethical challenges given its status as a standard laboratory model, and perhaps equally importantly (iv) it will simply be a much cheaper system to explore. Furthermore, the evolutionary divergence between the two rat species is within a few million years (Robins et al. 2008), the extinction happened relatively recently (ca 1900, MacPhee & Flemming 1999) and museum collections of skin samples with documented DNA survival exist (Wyatt et al. 2008). Thus, although it would likely be argued that more rats are not something that our planet is lacking, attempts to apply genome editing to this pair of species will certainly be a faster route to exploring both the potential and challenges of the methods, both during the genome engineering itself, but also for survival after birth. We furthermore believe that media attention on such a project would more likely be focused on the small gains of successful manipulation of few genes in a model system (something that has additional relevance to general research into gene function and evolution), as opposed to large losses such as captivation and/or death of elephants, and this in turn would help to generate research funds to support the project.
The majority of the mammoth's genes are functionally identical to the Asian elephant (Lynch et al. 2015), so as opposed to attempting to build a genome from scratch, the sections that are different could be overwritten with mammoth variants. This would both be simpler than creating a synthetic genome, and much faster than back-breeding. This can potentially be achieved by taking advantage of recent advances in genome editing technology: using CRISPR/Cas9, which cuts DNA at specific nucleotide sequences, guided by an adjoined RNA sequence. Genomes can be edited by cutting the DNA at the required sites using custom built guide RNAs, followed by insertion of a DNA sequence of choice carried with the complex (Pu et al. 2015). There are some issues with the accuracy and efficiency of the system though. Cuts are sometimes made in unwanted areas (O'Geen et al. 2015) and CRISPR/Cas9 is not able to work at every location of a genome due to requirements of the DNA sequences that the complex is able to interact with (Seruggia & Montoliu 2014). Furthermore, currently the insertion of a new strand of DNA cannot be completely controlled, and occurs at a lower frequency than the simple repair of the break which the Cas9 produces, with reports in the range of 0.5–20% of cuts resulting in successful insertions (Wang et al. 2015). This is due to non-homologous end joining being the primary mechanism used by cells to repair double-strand breaks: a process which often results in random deletions and insertions during the repair process. To achieve the desired insertion, homology directed repair must instead be employed, which occurs at a much lower frequency. Although efficiency can be improved to some extent by influencing the repair mechanism via inhibitors (Song et al. 2016), these molecules may interfere with embryo development (Wang et al. 2015), and regardless, the efficiency is still fairly low, which may pose a problem for large scale genome editing. After each attempted deletion and insertion, checks would be required to see whether the insertion was actually made. If unwanted mutations are instead made, they may inhibit the CRISPR/cas9 complex from locating the same region in the same cell on subsequent attempts (Seruggia & Montoliu 2014). Each locus that needs to be changed would most likely need to be edited in this somewhat haphazard way, one at a time, until the desired outcome is achieved. This could be problematic when wanting to introduce a large number of specific modifications into one genome. Multiple knock-in targeting has been achieved, but it can be error prone so is most likely not currently a viable option (Seruggia & Montoliu 2014).
The number of potential sites that would need to be changed would mainly depend on substitutions that alter the amino acid composition of genes, as well as those that affect promoter and enhancer regions. There may also be some elephant genes which would need to be switched off by mutations that render the resulting proteins useless, or by inserting a stop codon into the gene, or by altering the promoter region. Lynch et al. (2015) sequenced the mammoth genome and found over two thousand amino acid differences between the genes of the mammoth and the Asian elephant. The total number of gene insertions required might be fewer than this if there are multiple substitutions in close proximity, or if some of the amino acid substitutions are expected to not functionally affect the protein. While making this quantity of alterations to a genome in order to create a woolly mammoth seems quite reasonable, it is still an enormous undertaking, and to successfully generate this number of knock-ins into a genome would be a very lengthy process. Most studies regarding CRISPR knock-ins focus on only achieving one insertion, which alone is not an easy task.
George Church, whose team are attempting to recreate mammoth traits from an Asian elephant using this technology (Shapiro 2015a), has speculated that only a few dozen genes may need to be altered in order to create an animal that fulfils the ecological functions of a mammoth (Church 2013). This number of edits is probably much more achievable within the near future, although of course whether the restoration of only a handful of genes represents species de-extinction is questionable. The result would instead be the de-extinction of a selection of genes rather than a whole species. The challenge of obtaining the complete genome of an extinct organism is, however, relevant to having the full knowledge of what genes to edit in the organism subject to modification.
What other challenges are there?
Ultimately, while the above-mentioned techniques may not be sufficiently developed today to ensure successful de-extinction, given the rapid progress of modern molecular biology, it is reasonable to expect that within the not too distant future we will see the recreation of organisms with at least partial genomic similarity to their extinct relatives. At this point, a number of other challenges arise that cannot be ignored, in particular relating to the interactions between the recreated ‘species’ and the natural world as it is today. The species may have adverse effects on ecosystems and the current environment may not be suitable for it.
One important consideration is the microbiome of an organism, which has been referred to as the second genome. Microbial interactions with an animal can have a major influence on behaviour, health and nutrition obtained from food (Turnbaugh et al. 2006; Zhu et al. 2010; Grice & Segre 2012), and microbiomes differ greatly across species or even populations (Muegge et al. 2011; Yatsunenko et al. 2012). Little is known about how to equip a de-extinct individual with a functional microbiome – bridging symbiotic capabilities with the needs of the practical world. Even if we could recreate ancient microbiomes, doing so does not guarantee that it will function in the climate or environment the de-extinct individual will face today.
Secondly, we must consider the disease angle. This relates to both the susceptibility of the de-extinct form to modern diseases and parasites, with which it stopped co-evolving at time of extinction (Seddon et al. 2014), and whether the de-extinction form could represent a vector of disease to other species (including livestock; Cohen 2014; Seddon et al. 2014). An extreme example of susceptibility is the Christmas Island rat (Rattus macleari, Box 2), whose extinction appears to have been caused by an infectious disease (Wyatt et al. 2008). Therefore, bringing this species back (with an aim of rewilding) would require equipping it with immunity to the trypanosome that likely led to its demise. As the de-extinction method of choice for the rat would most likely be via genome editing of the brown rat (Rattus norvegicus), it is possible that this could be achieved by maintaining some of the immune genes of this species rather than replacing with those of the Christmas Island rat. The brown rat has established itself on Christmas Island (Armstrong 1992) so it appears that it is equipped with the required immunity for survival here. If the brown rat is used as a surrogate, the individual should also obtain antibodies for resistance during gestation. The more we alter the species, the less its creation will be viewed as a de-extinction, however. A trade-off exists between maintaining the integrity of the original species, and creating a species that is able to survive and function well in today's ecosystems. We must therefore decide how important the label of de-extinction is, and carefully consider what the real objectives of the process are, in order to decide whether these compromises are acceptable.
A third challenge (again at least for rewilding) is the move from producing healthy and developmentally correct individuals, to producing a stable, self-sustaining, population of the species. This could potentially require the de-extinction of a very large number of genetically diverse individuals to ensure adequate genetic diversity of a resurrected population. This in turn raises the concern as to whether enough reference samples exist from which to generate appropriate diversity? One major challenge facing small populations with low genetic diversity is the accumulation of deleterious mutations through inbreeding (Charlesworth & Charlesworth 1987), and in this regard, an interesting solution that would likely be possible (should we have the technology to recreate the extinct species in the first place), could be the use of genome editing technology to remove deleterious mutations from the population.
A fourth group of challenges relate to the wider environment. This includes providing a suitable habitat for a de-extinct individual (Cottrell et al. 2014; Seddon et al. 2014), whose original niche might no longer be available in the form or extent required, thus necessitating land management action. Altering whole ecosystems to accommodate the de-extinct form might well defy one of the most attractive and logical arguments for de-extinction: ecosystem restoration via introduction of extinct keystone species. If the original ecosystem does exist, it is likely under pressure by human activity, and the de-extinct species may compete with, or predate on, potentially already threatened species (Seddon et al. 2014). Outside of the problems a de-extinct form might pose for other species, it might also not be welcomed by humans, due to the ecological changes that it causes, even if the result is a reversion to a previous system. Consider as an example the Christmas Island rat. Since its extinction a little over 100 years ago (Wyatt et al. 2008), the red crab has established a large population of around 40 million (Misso & West 2014), possibly affected by ecological changes resulting from a niche left open following the rat's extinction (Harper & Bunbury 2015). The migration of this crab is considered by some to be one of the most magnificent natural spectacles on earth (Misso & West 2014; Collins 2016; The Telegraph 2016), and the reintroduction of the Christmas Island rat may drastically reduce red crab numbers, thus resulting in cultural conflict and potential economic loss in the form of tourism. Species introduction within an ecosystem can also have indirect effects on species which can be difficult to predict, such as a case of rat introduction reducing crab numbers, which results in increased vegetation, which in turn decreased the population of a bird species that requires clear ground for take-off (Harper & Bunbury 2015). This highlights the delicacy of such an introduction, as it could alter a whole ecosystem. The focus on an introduction of a de-extinct species from the public, media and scientific community would likely be immense, with much pressure to obtain the desired outcome, and any adverse effects likely to be highly criticised. It has been suggested that, as opposed to viewing the introduction of de-extinct animals as the restoration of an ecosystem to its correct state, it should instead be considered in the same light as that of bringing extant species to new ecosystems, with the IUCN Red List criteria for suitable introductions applied to any de-extinction candidate (Seddon et al. 2014).
Finally, it is important to remember that an animal is not just a physical entity, but also potentially an accumulation of behavioural information that is passed down from one generation to the next. For every species that walks the earth, the journey to their current state began billions of years ago and has continued, unfaltered, until now. Behavioural evolution and transfer possibly ranges as far back as the evolution of the ability to learn within a species’ lineage. In social animals, along with extinction was the loss of social information which might not be easily restored, or may even be completely irretrievable, such as hunting techniques, migration routes and possibly even communication methods that aid bonding or mating.
Conclusions
In summary, we have access today to a growing number of methods that might one day render the de-extinction of species possible – at least under a very loose understanding of what exactly constitutes a recreated species. We hope that in our text, we have made it clear that while each of the methods holds promise, they also hold particular limitations, and thus ultimately the choice of which is most suitable will depend both on the individual species under consideration and on the desired target for success. Furthermore, one aspect that all those discussed share is the requirement for a close living relative to use in the process, whether for back-breeding with, or as a viable surrogate for gestation, or in order to provide a reference and/or editable genome at close enough evolutionary divergence. For those that do have a potential surrogate, the procedure may still be fraught with both ethical and practical hurdles, both during the pregnancy, and any subsequently attempted acclimatisation and introduction of the species to the desired environment.
Ultimately, despite the considerable progress in de-extinction technology, due to the challenges discussed, it may be that we will never be able to recreate most extinct species in their purest form. This poses the key question that all future attempts will have to tackle: What will teams undertaking the work define as successful de-extinction? Is trying to create something that meets phenotypic goals enough, while ignoring non-immediately visually apparent genes such as those affecting behaviour or internal physiology? Or is creation of something that has 90%, or 99%, or 100% of the same genomic content as the original (as well as epigenetic profiles and even microbiomes) enough? Or is creating an animal that largely fulfils the same ecological function as the original species, while possibly forgoing some of the aesthetic specifics, enough? Clearly, it will be important to both consider, and openly discuss, such issues at the starting point of any de-extinction project, and to have a clearly identifiable set of planned attributes from the outset that could allow for determination of at what point the process can be declared to have been a success.
Acknowledgements
We thank Mia Mottelson and Gustav Pontoppidan for the illustrations and ERC Consolidator Grant (681396 – Extinction Genomics) for funding.




