Identification of VIM-2 metallo-β-lactamase-producing Pseudomonas aeruginosa isolated from dogs with pyoderma and otitis in Korea
Abstract
enBackground
Pseudomonas aeruginosa is a challenging pathogen cultured from cases of acute and chronic canine otitis and sometimes in cases of deep pyoderma. The spread of antimicrobial resistance, especially carbapenem resistance, is a serious therapeutic challenge worldwide.
Hypothesis/Objectives
To investigate the identification and characterization of resistant P. aeruginosa clinical canine isolates.
Materials
Clinical isolates (n = 80) were collected from dogs with pyoderma (n = 18) and otitis (n = 62) in Korea.
Methods
Antimicrobial susceptibility was determined using agar dilution and using Clinical and Laboratory Standards Institute guidelines for recording susceptibility for human Pseudomonas isolates; genetic relatedness of isolates was investigated by multilocus sequence typing (MLST) and SpeI macrorestriction analysis. The class 1 integrons were amplified and sequenced using primer walking.
Results
Most isolates were susceptible to colistin (97.5%), polymyxin B (96.3%), ciprofloxacin (81.3%) and meropenem (80.0%); whereas resistance to aztreonam (80%), piperacillin (52.5%), piperacillin/tazobactam (41.3%) and cefepime (37.5%) was high; 12 carbapenem-nonsusceptible isolates (15%) were detected. MLST revealed 45 different sequence types (STs) and macrorestriction analysis detected 55 distinct pulsotypes (PTs), which were divided into 25 clonal groups. Among carbapenem-nonsusceptible isolates, 10 (83.3%) were VIM-2-producing strains. Nine VIM-2-producing isolates were identified as ST1047 and harboured the same 2.8 kb class 1 integron. One remaining isolate was ST1203 with 2.1 kb class 1 integron.
Conclusions and clinical importance
This study demonstrated the diversity of the phenotype and genotype of clinical P. aeruginosa isolates from dogs with pyoderma and otitis. The identification of VIM-2-producing P. aeruginosa in dogs is alarming and warrants further surveillance.
Résumé
frContexte
Pseudomonas aeruginosa est un pathogène difficile, cultivé de cas d'otite aigue et chronique canine et parfois à partir de pyodermite profonde. La dissémination de résistance antimicrobienne, particulièrement la résistance aux carbapénèmes, est un défi thérapeutique mondial sérieux.
Hypothèses/Objectifs
Etudier l'identification et la caractérisation de souches cliniques canines résistantes de P. aeruginosa.
Matériels
Les isolats cliniques (n = 80) ont été collectés de chiens avec pyodermite (n = 18) et otite (n = 62) en Corée.
Méthodes
La sensibilité antimicrobienne a été déterminée par dilution d'agar et en suivant les recommandations du Laboratory Standards Institute pour l'enregistrement des sensibilité des souches de Pseudomonas chez l'homme; les correspondances génétiques des souches ont été étudiées par MLST (multilocus sequence typing) et analyse de macrorestriction SpeI. Les intégrons de classe 1 ont été amplifiés et séquencés par arpentage chromosomique.
Résultats
La plupart des souches étaient sensibles à la colistine (97.5%), à la polymyxine B (96.3%), à la ciprofloxacine (81.3%) et au méropénème (80.0%); tandis que la résistance à l'aztréoname (80%), à la pipéracilline (52.5%), la pipéracilline/tazobactame (41.3%) et à la céfépime (37.5%) était élevée; 12 souches non sensibles au carbapénème (15%) ont été détectés. MLST a révélé 45 différentes séquences types (STs) et les analyses de macrorestriction ont détectés 55 pulsotypes distincts (PTs) qui ont été divisés en 25 groupes de clones. Parmi les souches non sensibles à la carbapénème, 10 (83.3%) étaient des souches produisant VIM-2. Neuf souches produisant du VIM-2 ont été identifiées comme ST1047 et présentaient le même intégron de classe 1 de 2.8kb. Une souche restante était ST1203 avec intégron de classe 1 de 2.1kb.
Conclusions et importance clinique
Cette étude démontre la diversité du phénotype et du génotype des souches cliniques de P. aeruginosa des chiens avec pyodermite et otite. L'identification de P. aeruginosa produisant du VIM-2 est alarmant et doit entrainer une plus grande surveillance.
Resumen
esIntroducción
Pseudomonas aeruginosa es un patógeno de difícil control que se cultiva de casos de otitis canina aguda y crónica y, a veces, en casos de pioderma profunda. La propagación de la resistencia a los antimicrobianos, especialmente la resistencia a carbapenem, es un desafío terapéutico complejo en todo el mundo.
Hipótesis/Objetivos
investigar la identificación y caracterización de resistencia en aislamientos clínicos caninos de P. aeruginosa.
Materiales
Se obtuvieron aislados clínicos (n = 80) de perros con pioderma (n = 18) y otitis (n = 62) en Corea.
Métodos
La susceptibilidad a los antimicrobianos se determinó utilizando dilución de agar y utilizando las directrices del Instituto de Estándares Clínicos y de Laboratorio para registrar la susceptibilidad para aislamientos de Pseudomonas en humanos; la relación genética de los aislados se investigó mediante tipado de secuencia multilocus (MLST) y análisis de macrorrestricción SpeI. Los integrones de clase 1 se amplificaron y se secuenciaron usando iniciadores en camino (primer walking)
Resultados
la mayoría de los aislamientos fueron susceptibles a colistina (97,5%), polimixina B (96,3%), ciprofloxacina (81,3%) y meropenem (80,0%); mientras que hubo elevada resistencia a aztreonam (80%), piperacilina (52,5%), piperacilina/tazobactam (41.3%) y cefepima (37,.5%); se detectaron 12 aislamientos no susceptibles al carbapenem (15%). MLST reveló 45 tipos de secuencias diferentes (ST) y el análisis de macrorestricción detectó 55 pulsotipos distintos (PT), que se dividieron en 25 grupos clonales. Entre los aislados no susceptibles al carbapenem, 10 (83,3%) fueron cepas productoras de VIM-2. Nueve aislamientos productores de VIM-2 se identificaron como ST1047 y albergaban el mismo integrón de 2,8 kb de clase 1. Un aislado restante era ST1203 con 2,1 kb de integrón de clase.
Conclusiones e importancia clínica
este estudio demostró la diversidad del fenotipo y el genotipo de aislados clínicos de P. aeruginosa en perros con pioderma y otitis. La identificación de P. aeruginosa productora de VIM-2 en perros es alarmante y merece una mayor vigilancia.
Zusammenfassung
deHintergrund
Bei Pseudomonas aeruginosa handelt es sich um ein Pathogen mit Herausforderung, welches von Fällen akuter und chronischer caniner Otitis und manchmal bei Fällen tiefer Pyodermie kultiviert werden kann. Die Ausbreitung der antimikrobiellen Resistenz, vor allem eine Carbapenem Resistenz, ist weltweit eine wichtige therapeutische Herausforderung.
Hypthese/Ziele
Eine Untersuchung zur Identifizierung und Charakterisierung von klinischen Isolaten von Hunden mit resistenten P. aeruginosa.
Material
Klinische Isolate (n= 80) wurden von Hunden mit Pyodermie (n = 18) und Otitis (n = 62) in Korea genommen.
Methoden
Die antimikrobielle Empfindlichkeit wurde mittels Agarverdünnung und unter Zuhilfenahme der Guidelines des Clinical und Laboratory Standards Institut zur Festhaltung von empfindlichen humanen Pseudomonas Isolaten bestimmt; eine genetische Verwandtschaft der Isolate wurde mittels Multilokus Sequenzierung (MLST) und SpeI Makrorestriktionsanalyse untersucht. Die Klasse 1 Integrons wurden amplifiziert und mittels Primer Walking sequenziert.
Ergebnisse
Die meisten Isolate waren empfindlich auf Colistin (97,5%), Polymyxin B (96,3%), Ciprofloxacin (81,3%) und Meropenem (80,0%); während eine Resistenz auf Aztreonam (80%), Piperacillin (52,5%), Piperacillin/Tazobactam (41,3%) und Cefepime (37,5%) hoch war; 12 Carbapenem-nichtempfindliche Isolate (15%) wurden gefunden. Die MLST zeigte 45 verschiedene Sequenztypen (STs) und eine Makrorestriktionsanalyse zeigte 55 individuelle Pulsotypen (PTs), die in 25 klonale Gruppen einzuteilen waren. Unter den Carbapenem-nichtempfindlichen Isolaten waren 10 (83,3%) VIM-2 produzierende Stämme. Neun VIM-2-produzierende Isolate wurden als ST1047 identifiziert und beherbergten dasselbe 2,8 kb Klasse 1 Integron. Ein verbleibendes Isolat war ein ST1203 mit einem 2,1 kb Klasse 1 Integron.
Schlussfolgerungen und klinische Bedeutung
Diese Studie zeigte die Diversität der Phänotypen und der Genotypen von klinischen P. aeruginosa Isolaten von Hunden mit einer Pyodermie und einer Otitis. Die Identifizierung von VIM-2-produzierenden P. aeruginosa bei Hunden ist alarmierend und sollte weiter überwacht werden.
要約
ja背景
Pseudomonas aeruginosaは、犬の急性および慢性耳炎、また時には深在性膿皮症の症例から分離され、臨床的問題となる病原体の一つである。抗菌剤耐性(特にカルバペネム耐性)の拡散は、世界中で治療上の深刻な問題となっている。
仮説/目的
本研究の目的は、犬からの臨床分離株における薬剤耐性P. aeruginosaの同定および特徴について解析することである。
材料
韓国において、膿皮症(n = 18)および耳炎(n = 62)を有する犬から臨床分離株(n = 80)を採取した。
方法
抗菌剤感受性の決定には寒天希釈法を用い、ヒト由来Pseudomonas株の薬剤感受性を記録するためのClinical and Laboratory Standards Instituteガイドラインを基準とした。分離株の遺伝的関連性については、multilocus sequence typing (MLST)およびSpeIを用いたマクロ制限酵素解析により評価した。またクラス1インテグロン遺伝子を増幅し、プライマーウォーキング法により塩基配列を決定した。
結果
ほとんどの分離株は、コリスチン(97.5%)、ポリミキシンB(96.3%)、シプロフロキサシン(81.3%)およびメロペネム(80.0%)に感受性であった。一方、アズトレオナム(80%)、ピペラシリン(52.5%)、ピペラシリン/タゾバクタム(41.3%)、セフェピム(37.5%)に対する耐性率は高く、カルバペネム非感受性株は12株(15%)検出された。 MLSTにより45の異なるsequence types (STs)が明らかとなり、マクロ制限酵素解析では55の異なるpulsotypes (PTs)が検出され、25の遺伝子群に分類された。カルバペネム非感受性分離株のうち、VIM-2産生株は10株(83.3%)であった。 VIM-2産生分離株のうち、9株はST1047として同定され、同じ2.8kbクラス1インテグロンを有していた。残りの1株は、2.1kbクラス1インテグロンを有するST1203であった。
結論と臨床的重要性
本研究により、膿皮症および耳炎を有する犬由来のP. aeruginosa臨床分離株の表現型および遺伝子型の多様性が証明された。犬からVIM-2産生P. aeruginosaが同定されたことは警戒されるべきであり、今後のさらなる調査が必要である。
摘要
zh背景
绿脓假单胞菌是一种具有挑战性的病原,可从犬急性和慢性中耳炎病例中培养出来,有时也存在于深层脓皮病病例中。抗生素耐药性的传播,尤其是碳青霉烯类耐药,是全世界面临的严峻治疗挑战。
假设/目的
探究犬临床上耐药绿脓假单胞菌的鉴别和表征。
材料
韩国境内收集的犬脓皮病(n = 18)和中耳炎(n = 62)病例中的临床菌株(n = 80)。
方法
依据临床实验室标准委员会评估人绿脓假单胞菌敏感性的参考标准,使用琼脂稀释法测定抗生素药物敏感性。采用多位点序列分型(MLST)和SpeI基因图谱分析,对菌株的遗传相关性进行研究。对1类整合子进行扩充,并设计引物测序。
结果
大多数菌株对黏菌素(97.5%)、多粘菌素B(96.3%)、环丙沙星(81.3%)和美罗培南(80.0%)敏感;对氨曲南(80%)、哌拉西林(52.5%)、哌拉西林-三唑巴坦(41.3%)和头孢吡肟(37.5%)高度耐药;并发现了12株(15%)耐碳青霉烯类假单胞菌。MLST显示了45种不同的序列类型(STs),基因图谱分析检测到55种独特的脉冲型,并分属于25个克隆组。耐碳青霉烯类的绿脓假单胞菌株中,有10 (83.3%)株是VIM-2型。9个VIM-2型菌株被鉴定为ST1047,并携带相同的2.8kbI型整合子。剩余的一株是携带2.1kbI型整合子的ST1203。
结论和临床意义
本研究针对犬脓皮病和中耳炎的病原绿脓假单胞菌,显示了其临床表型和基因型的多样性。犬VIM-2型绿脓假单胞菌的出现令人担忧,需要对其进一步监测。
Resumo
ptContexto
Pseudomonas aeruginosa é um patógeno desafiador isolado de casos de otite aguda crônica em cães e por vezes em casos de piodermite profunda. A disseminação da resistência a antimicrobianos, especialmente resistência ao carbapenem, é um importante desafio terapêutico ao redor do mundo.
Hipótese/Objetivos
Identificar e caracterizar os isolados caninos de Pseudomonas aeruginosa.
Materiais
Isolados clínicos (n = 80) foram coletados de cães com piodermite (n = 18) e otite (n = 62) na Coreia.
Métodos
A suscetibilidade antimicrobianos foi determinada utilizando o método de diluição em ágar de acordo com as diretrizes do Clinical and Laboratory Standards Institute para suscetibilidade de isolados Pseudomonas aeruginosa humana. Investigou-se semelhança genética entre os isolados utilizando um método de tipificação multilocus (MLST) e análise de macro-restrição SpeI. Os integrons de classe 1 foram amplificados e sequenciados utilizando um primer móvel.
Resultados
A maioria dos isolados foi suscetível à colistina (97,5%), polimixina B (96,3%), ciprofloxacina (81,3%) e meropenem (80,0%); enquanto que a resistência a aztreonam (80%), piperacilin (52,5%), piperacillin/tazobactam (41,3%) e cefepime (37,5%); foram detectados 12 isolados (15%) não-suscetíveis ao carbapenem. A MSLT revelou 45 tipos de sequências diferentes e a análise de macro-restrição detectou 55 pulsotipos distintos, que foram divididos em 25 grupos clonais. Entre os isolados não-suscetíveis, 10 (83,3%) eram cepas produtoras de VIM-2. Nove cepas produtoras de VIM-2 isoladas foram identificadas como ST1047 e eram portadoras do mesmo integron classe 1 de 2,8 kb. Um isolado remanescente foi ST1203 com integron classe 1 de 2,1 kb.
Conclusões e importância clínica
Este estudo demonstrou a diversidade do fenótipo e genótipo de isolados clínicos de P. aeruginosa oriundas de cães com piodermite e otite. A identificação de P. aeruginosa produtora de VIM-2 em cães é alarmante em e demanda maior investigação.
Abbreviations
-
- AMR
-
- antimicrobial resistance
-
- MBLs
-
- metallo-β-lactamases
-
- MLST
-
- multilocus sequence typing
-
- STs
-
- sequence types
Introduction
Pseudomonas aeruginosa is a major opportunistic human pathogen that causes nosocomial infections, especially in immunocompromised patients.1 In dogs, it is a problematic pathogen often cultured from cases of acute and chronic canine otitis, and occasionally found in cases of deep pyoderma.2, 3 Dogs, given close physical contact with their owners, may be reservoirs of antimicrobial-resistant bacteria which could lead to zoonotic infections.4
The emergence and worldwide spread of carbapenemase-producing P. aeruginosa have increased recently, and these bacteria have become a serious clinical threat in human medicine.5-7 Diverse kinds of carbapenemases have been identified in P. aeruginosa, which are classified as class A penicillinases, class B metallo-β-lactamases and class D oxacillinases.8 Metallo-β-lactamases (MBLs) are the most common carbapenemases in P. aeruginosa worldwide and, among them, VIM-2 and IMP-6 have been commonly reported in Asia and Korea, respectively.7, 9
Antimicrobial resistance (AMR) is an emerging problem in veterinary medicine,10, 11 and carbapenemase-producing bacterial strains have been reported in companion animals, in food-producing animals and the environment.12-14 Although the carbapenem class of antibiotics, including imipenem and meropenem, has not been approved by the FDA for use in animals, veterinarians have been prescribing these antimicrobials for animals with chronic treatment-resistant infection as extra-label drugs.15, 16 Owing to the chronic and treatment-resistant nature of P. aeruginosa infections in dogs, long-term antimicrobial treatment often is required.2 However, few studies have assessed antimicrobial resistance in clinical P. aeruginosa isolates from dogs and previous reports have found that the incidence of carbapenem-nonsusceptible P. aeruginosa isolated from dogs was low.17, 18 There is a report on carbapenemase-producing P. aeruginosa from an anal swab of a healthy dog in China, which was highly similar to that from humans in the same area of China.19 However, to our knowledge, no studies have been reported on carbapenemase-producing P. aeruginosa from lesional areas of animals, including companion animals and food-producing animals.
The primary objective of this study was to assess the pathogenic phenotype, including antimicrobial susceptibility and genotype of clinical P. aeruginosa isolates from canine pyoderma and otitis. For genotypic comparison, we used nucleotide sequence-based typing method [multilocus sequence typing (MLST)] and band-based typing method (macrorestriction analysis). The present study also investigated the spread and molecular characteristics of carbapenem-resistant P. aeruginosa from clinical isolates of dogs.
Materials and Methods
Bacterial strains
Pseudomonas aeruginosa clinical isolates were collected from the Veterinary Medical Teaching Hospital of Seoul National University (n = 40) and the diagnostic laboratory of the Neodin Medical Institute (n = 40) in Korea between 2013 and 2017. The isolates were obtained from 80 dogs with pyoderma (18 isolates, 22.5%) and otitis (62 isolates, 77.5%). Species identification was performed using the Vitek 2 automated ID system (Biomérieux; Lyon, France). Isolates confirmed to be P. aeruginosa were cryopreserved at −80°C in 20% (v/v) glycerol until further examination.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing and interpretation were performed using disk diffusion according to the Clinical and Laboratory Standards Institute (CLSI).20 The antimicrobial agents tested were piperacillin, piperacillin/tazobactam, ceftazidime, cefepime, aztreonam, imipenem, meropenem, colistin, polymyxin B, gentamicin, tobramycin, amikacin, ciprofloxacin and ofloxacin. The minimum inhibitory concentrations (MICs) of imipenem and meropenem were determined using agar dilution according to the CLSI guidelines.20
It is important to note that the disk-diffusion method and MIC determination were performed to assess susceptibility and interpretation was based on guidelines for human P. aeruginosa isolates due to the lack of information about interpretative criteria of animal isolates and the comparison with the results of human isolates.
Identification of β-lactamase genes
Genes encoding β-lactamase were detected using PCR with primers listed in Table 1, as previously described.21-23 Bacterial DNA from 12 isolates with reduced susceptibility to imipenem and meropenem was prepared using the QIAamp® DNA Mini Kit (Qiagen; Hilden, Germany) according to the manufacturer's instructions. The DNA was used as a template for PCR amplification and amplified PCR products were purified using the AccuPrep™ PCR purification kit (Bioneer; Daejeon, Korea) and sequenced using the ABI PRISM 3730xl DNA analyzer (Applied Biosystems; Foster City, CA, USA). Further PCR and full sequencing of class 1 integrons using the primer walking was performed to analyse the genetic structure of class 1 integrons, as described previously.24, 25 The nucleotide and protein sequences were analysed with BLAST (http://blast.ncbi.nlm.nih.gov/Blast).
| Primer name | Primer sequence (5′→3′) | Target gene | Enzyme class | Product size (bp) | References |
|---|---|---|---|---|---|
| GES-F | ATGCGCTTCATTCACGCAC | blaGES | Class A penicillinases | 844 | 22 |
| GES-R | CTATTTGTCCGTGCTCAGG | ||||
| KPC-F | CGTCTAGTTCTGCTGTCTTG | blaKPC | 798 and 232 | 22 | |
| KPC-R | CTTGTCATCCTTGTTAGGCG | ||||
| IMP-F | GGAATAGAGTGGCTTAAYTCTC | blaIMP | Class B metallo-β-lactamases | 232 | 21 |
| IMP-R | GGTTTAAYAAAACAACCACC | ||||
| VIM-F | GATGGTGTTTGGTCGCATA | blaVIM | 390 | 21 | |
| VIM-R | CGAATGCGCAGCACCAG | ||||
| NDM-F | GGTTTGGCGATCTGGTTTTC | blaNDM | 621 | 21 | |
| NDM-R | CGGAATGGCTCATCACGATC | ||||
| OXAI-F | TCAACAAATCGCCAGAGAAG | blaOXA-Igroup | Class D oxacillinases | 276 | 23 |
| OXAI-R | TCCCACACCAGAAAAACCAG | ||||
| OXAII-F | AAGAAACGCTACTCGCCTGC | blaOXA-IIgroup | 478 | 23 | |
| OXAII-R | CCACTCAACCCATCCTACCC | ||||
| OXAIII-F | TTTTCTGTTGTTTGGGTTTT | blaOXA-IIIgroup | 427 | 23 | |
| OXAIII-R | TTTCTTGGCTTTTATGCTTG | ||||
| INT1-F | GGCATCCAAGCAGCAAG | 5′CS | - | 24 | |
| INT1-R | AAGCAGACTTGACCTGA | 3′CS | - | ||
| tniCF | CGATCTCTGCGAAGAACTCG | tniC | - | 25 |
- F forward, R reverse; the INT1-F, INT-R and tniCF were combined with others and the product size depends on the combination.
MLST
MLST was performed by sequencing the internal fragments of seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA and trpE).26 The allele numbers and sequence types (STs) were assigned in the P. aeruginosa MLST database (http://pubmlst.org/paeruginosa/). For cluster analysis, the minimum spanning tree based on allelic profiles was constructed using Bionumerics software (Applied Maths; Sint-Martens-Latem, Belgium).
SpeI macrorestriction analysis
SpeI macrorestriction analysis of clinical P. aeruginosa isolates was carried out as described previously27 with some modifications. Each 1% (w/v) SeaKem Gold Agarose (Lonza) plug containing DNA was digested with 20 U SpeI (Takara; Tokyo, Japan) for 4 h at 37°C in 1× reaction buffer in a shaking incubator. After digestion, each plug slice was transferred to the well of 1% Seakem agarose gel and DNA fragments were separated using a CHEF Mapper pulsed-field electrophoresis system (Bio-Rad Laboratories; Hercules, CA, USA) according to the manufacturer's instructions. Gels were stained with 2 μg/mL of ethidium bromide for 60 min, destained in distilled water and imaged on a GelDoc UV transilluminator (Bio-Rad). The resulting band profiles were analysed using Bionumerics software (Applied Maths), with Dice bFFand-based comparison at a band position tolerance of 1.7%.28
Results
Bacterial strains and susceptibility testing
Eighty P. aeruginosa clinical isolates from dogs in Korea were investigated. Among all the isolates, most showed high susceptibility to colistin (97.5%), polymyxin B (96.3%), ciprofloxacin (81.0%) and meropenem (80.0%); with high resistance rates to aztreonam, piperacillin, piperacillin/tazobactam and cefepime, of 80%, 52.5%, 41.3% and 37.5%, respectively. Twelve P. aeruginosa isolates (15%) were carbapenem-nonsusceptible strains, which were intermediately susceptible or resistant to imipenem and meropenem. Antimicrobial susceptibility rates are presented in Table 2.
| Carbapenem-susceptible isolates (n = 68) | Carbapem-nonsusceptible isolates (n = 12) | Total Isolates (n = 80) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial agents | Disk content | %S | %I | %R | %S | %I | %R | %S | %I | %R |
| Piperacillin | 100 μg | 55.9 | 41.2 | 2.9 | 0.0 | 58.3 | 41.7 | 47.5 | 43.8 | 8.8 |
| Piperacillin/Tazobactam | 100/10 μg | 67.6 | 32.4 | 0.0 | 8.3 | 58.3 | 33.3 | 58.8 | 36.3 | 5.0 |
| Ceftazidime | 30 μg | 86.8 | 10.3 | 2.9 | 25.0 | 0.0 | 75.0 | 77.5 | 8.8 | 13.8 |
| Cefepime | 30 μg | 72.1 | 16.2 | 11.8 | 8.3 | 0.0 | 91.7 | 62.5 | 13.8 | 23.8 |
| Aztreonam | 30 μg | 17.6 | 70.6 | 11.8 | 33.3 | 58.3 | 8.3 | 20.0 | 68.8 | 11.3 |
| Imipenem | 10 μg | 86.8 | 13.2 | 0.0 | 0.0 | 16.7 | 83.3 | 73.8 | 13.8 | 12.5 |
| Meropenem | 10 μg | 94.1 | 1.5 | 4.4 | 0.0 | 16.7 | 83.3 | 80.0 | 3.8 | 16.3 |
| Colistin | 10 μg | 97.1 | 0.0 | 2.9 | 100.0 | 0.0 | 0.0 | 97.5 | 0.0 | 2.5 |
| Polymyxin B | 300 units | 95.6 | 0.0 | 4.4 | 100.0 | 0.0 | 0.0 | 96.3 | 0.0 | 3.8 |
| Gentamicin | 10 μg | 73.5 | 10.3 | 16.2 | 16.7 | 0.0 | 83.3 | 65.0 | 8.8 | 26.3 |
| Tobramycin | 10 μg | 89.7 | 2.9 | 7.4 | 16.7 | 0.0 | 83.3 | 78.8 | 2.5 | 18.8 |
| Amikacin | 30 μg | 77.9 | 11.8 | 10.3 | 8.3 | 0.0 | 91.7 | 67.5 | 10.0 | 22.5 |
| Ciprofloxacin | 5 μg | 80.9 | 0.0 | 19.1 | 83.3 | 8.3 | 8.3 | 81.3 | 1.3 | 17.5 |
| Ofloxacin | 5 μg | 64.7 | 7.4 | 27.9 | 66.7 | 16.7 | 16.7 | 65.0 | 8.8 | 26.3 |
| Piperacillin | 100 μg | 55.9 | 41.2 | 2.9 | 0.0 | 58.3 | 41.7 | 47.5 | 43.8 | 8.8 |
- %S percentage of susceptibility, %I percentage of intermediate, %R percentage of resistant.
- Susceptibility assessed according to CLSI guidelines,20 so caution should be exercised when extrapolating the interpretation of the results for veterinary isolates.
Identification of β-lactamase-producing clinical isolates
PCR amplification and subsequent sequence analyses identified 10 isolates (83.3%) among the 12 carbapenem-resistant strains that harboured blaVIM-2 (Figure 1). None of the isolates showed genes encoding other β-lactamases except blaVIM-2. All blaVIM-2-producing isolates presented a multidrug-resistant phenotype and high MICs for imipenem and meropenem (64–256 μg/mL and 32–64 μg/mL, respectively). However, the remaining two isolates that were carbapenem-resistant but β-lactamases-nonproducing showed relatively low resistance, with the MIC ranging from 4 to 32 μg/mL for imipenem and 2–8 μg/mL for meropenem.

Two types of class 1 integron were identified by primer walking (Table 3). Nine blaVIM-2-producing isolates shared an unusual class 1 integron (InVIM-2A), in which the 3′ conserved sequence (3′CS) was substituted by the tniC gene (GenBank accession no. KT768111.1). One remaining blaVIM-2-harbouring isolate had a gene cassette array (InVIM-2B) between the 5′CS and 3′CS, which did not contain the blaVIM-2 gene inside (GenBank accession no. KJ546444.1).
| Type | Structure of the class 1 integron | Size (kb) | No. of isolates | GeneBank accession no. |
|---|---|---|---|---|
| InVIM-2A | 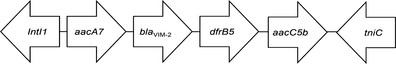 |
2.8 | 9 | KT768111.1 |
| InVIM-2B | 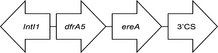 |
2.1 | 1 | KJ546444.1 |
- The arrows indicate the genes with the direction of transcription. IntI1 gene for the class 1 integrase, aacA7 AAC(6′)-II aminoglycoside acetyltransferase gene, blaVIM-2 metallo-beta-lactamase VIM-2 gene, dfrB5 and dfrA5 dihydrofolate reductase genes, aacC5b aminoglycoside 3-N-acetyltransferase gene, tniC resolvase, ereA erythromycin esterase gene.
MLST analysis
Eighty P. aeruginosa clinical isolates from dogs were assigned to 45 different STs (Figure S1). Among them, 41 STs were already registered in the P. aeruginosa MLST database, whereas four were novel STs (ST2596, ST2601, ST2741 and ST2742). Of the 45 different STs, there were no distinctly dominant STs; however, in descending order, ST1047, ST244, ST319 and ST2601 were identified in nine, six, six and four isolates, respectively. All nine isolates sharing an identical blaVIM-2 gene-carrying class 1 integron were identified as ST1047. One isolate with a different type of class 1 integron belonged to ST1203. The two remaining β-lactamases-nonproducing isolates of carbapenem-resistant strains were ST1428 and ST871, respectively.
SpeI macrorestriction analysis
Genetic diversity among the 80 P. aeruginosa isolates was analysed by constructing a dendrogram of macrorestriction patterns based on Dice's coefficient. Genotyping of all 80 isolates detected 55 distinct pulsotypes (PTs) (Figure S2). Based on a Dice similarity of more than 85% for SpeI-restricted DNA fragments,29 the isolates were divided into 25 clonal groups. All blaVIM-2-producing P. aeruginosa ST1047 isolates were closely related (90.7% similarity), whereas the similarity between ST1047 and ST1203 isolates was low (71.6% similarity), indicating that they displayed different patterns.
Discussion
A total of 80 P. aeruginosa clinical isolates from 80 dogs with pyoderma and otitis were analysed in this study. The antimicrobial susceptibility results revealed that resistance to most antibiotics, except gentamicin and ciprofloxacin, were higher compared to those identified in previous studies,18, 30, 31 but most organisms remained susceptible to colistin (97.5%), polymyxin B (96.3%) and ciprofloxacin (81.3%). Compared with carbapenem-susceptible isolates, carbapenem-resistant isolates showed higher resistance rates to other antibiotics, such as piperacillin (2.9% versus 41.7%), ceftazidime (2.9% versus 75.0%), cefepime (11.8% versus 91.7%), gentamicin (16.2% versus 83.3%), tobramycin (7.4% versus 83.3%) and amikacin (10.3% versus 91.7%). All carbapenem-nonsusceptible strains were susceptible to colistin and polymyxin B (Table 2). It must be emphasized that the assessment of susceptibility of the isolates was based on data used for human isolates and antimicrobial therapies; extrapolation to isolates from dogs must be performed with care because there is no certainty that there is a direct relationship.
To the best of the authors’ knowledge, this study presents the first observation of VIM-2 metallo-β-lactamase from P. aeruginosa originating from skin and ear lesions in dogs. The screening of carbapenemase genes by PCR amplification and subsequent sequencing identified blaVIM-2 in 83.3% (n = 10) of carbapenem-resistant P. aeruginosa isolates. Although blaVIM-2 is known to be the most commonly detected MBL gene in Asia, blaIMP-6 is the most commonly identified carbapenemase gene from human patients in Korea.7, 9 The absence of carbapenemase in the remaining two carbapenem-nonsusceptible P. aeruginosa isolates suggested that carbapenem resistance might have been induced by other factors, such as decreased expression of outer membrane proteins.32 The MICs of imipenem and meropenem were higher in VIM-2-producing isolates than in β-lactamases-nonproducing, carbapenem-nonsusceptible P. aeruginosa isolates (Figure 1).
Two types of class 1 integrons carrying the blaVIM-2 were identified. Nine VIM-2-producing isolates shared an identical 2.8 kb class 1 integron carrying blaVIM-2 between 5′CS and tniC. This integron also harboured other resistance gene cassettes, including one with a gene for a dihydrofolate reductase (dfrB5) and two types of gene for an aminoglycoside acetyltransferase (aacA7 and aacC5b). Integrons carrying MBL genes often have a variety of resistance genes, such as aminoglycoside resistance genes, thus harbouring integron contributes to acquire multidrug resistance.33 Several types of blaVIM-2-harbouring integrons with tniC instead of 3′CS at the end have been reported in the United States, Russia and India (GenBank accession nos DQ522233 and AM296017) and have been considered as ancestral integrons, which are progenitors of 3′CS-carrying integrons.25 The remaining VIM-2-producing isolate in the present study harboured a 2.1 kb class 1 integron containing gene cassettes coding for a dihydrofolate reductase (dfrA5) and for an erythromycin esterase (ereA) between 5′CS and 3′CS, but blaVIM-2 was not detected inside the class 1 integron. Previous studies have described that the VIM-2 gene was most commonly located in the class 1 integron of VIM-2-producing P. aeruginosa,34, 35 but the blaVIM-2 could not be found in the class 1 integron in this study.
For genetic typing of P. aeruginosa isolates, we performed MLST and SpeI macrorestriction analysis. The “gold standard” method of P. aeruginosa typing is considered to be macrorestriction analysis, which is a more discriminatory technique than MLST,29, 36 but for assessment of strain lineage we also performed MLST and compared the results of the two methods. Among the 80 isolates, MLST revealed 45 different STs and macrorestriction analysis revealed 55 distinct PTs, which were classified into 25 clonal groups according to Dice's similarity above 85%. These results of MLST and macrorestriction analysis suggested that P. aeruginosa isolated from dogs with pyoderma and otitis showed high genetic diversity, which is in contrast to the findings in a previous study.37
Among 10 VIM-2-producing stains, nine were ST1047 and one was ST1203. The nine ST1047 P. aeruginosa strains showed highly homologous macrorestriction analysis patterns with 90.7% Dice similarity and they differed significantly from the one ST1203 strain (71.6% similarity). ST1047 was first reported as a VIM-producing isolate associated with import from India in Norway (P. aeruginosa PubMLST id: 746) and it has been reported in Europe and Asia.38, 39 ST1203 was reported as a GES- and OXA-producing isolate from a patient with an eye infection in India.40 Ten VIM-2-producing strains were isolated at different periods from different individuals, and all dogs except two dogs never had contact with each other. These two dogs were hosts of PA25_SNU and PA26_SNU, respectively (Figure 1), and lived in the same household. The possibility of P. aeruginosa carriage related to the household contact is proposed when considering the similarity of phenotype and genotype of isolates; however, further studies are needed to elucidate this possibility.
The major MBL-producing P. aeruginosa from human isolates in Korea was ST235, which is an international high risk clone.41 One ST235 isolate was identified in the present study; however, the isolate showed sensitivity to most antimicrobials, except piperacillin and aztreonam. Considering the different types of MBL genes and STs of carbapenemase-producing P. aeruginosa between dogs and humans in Korea, the occurrence of carbapenemase-producing P. aeruginosa infection in dogs in this study appeared to be associated with individual infection rather than human-to-dog transfer. However, the emergence of carbapenemase-producing P. aeruginosa that causes the active lesions in dogs suggests that continued surveillance of interspecies transmission is needed.
In conclusion, our data demonstrated the diversity of the phenotype and genotype of P. aeruginosa from dogs with pyoderma and otitis, and the emergence of VIM-2 MBL-producing P. aeruginosa. The antimicrobial resistance of all P. aeruginosa isolates in this study was higher than the resistance reported previously, albeit based on guidelines for interpretation of human isolates. It is necessary to build a surveillance system to monitor antimicrobial susceptibility and assess the presence of carbapenemase in P. aeruginosa on a large scale in veterinary medicine.




