Characterization of the novel HLA-B*08:302 allele by sequencing-based typing
Abstract
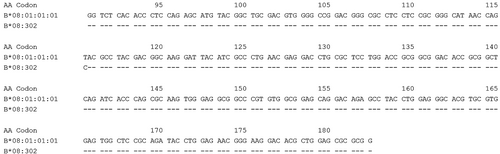
HLA-B*08:302 differs from HLA-B*08:01:01:01 by one nucleotide substitution in codon 116 in exon 3.
We report here a novel HLA-B*08 allele, now named B*08:302 that carries one nucleotide substitution in exon 3 when compared to the B*08:01:01:01 allele, identified in a volunteer bone marrow donor. The HLA typing was performed using Next-Generation Sequencing (AllType NGS, One Lambda, Canoga Park, CA) on the Ion S5 system platform (ThermoFisher Scientific, Waltham, MA),1 from exons 1 to 7. The reads were analyzed using the TypeStream Visual Software version 2.1 (One Lambda). This donor was found to have a new B*08 allele and was consequently typed A*01:01, 11:01; B*08:302, 35:08; C*03:03, 07:01; DRB1*03:01, 13:03; DRB3*01:01, 01:01; DQA1*05:01, 05:05; DQB1*02:01, 03:01P; DPA1*01:03, 02:01; DPB1*02:01, 11:01. Using the IPD-IMGT/HLA Database,2 nucleotide sequence alignment with HLA-B alleles shows that this new allele has one nucleotide change from B*08:01:01:01 in codon 116 in exon 3 where T → C, resulting in a coding change (TAC → CAC, Tyrosine → Histidine, Figure 1). This nucleotide change was confirmed using other NGS reagents provided by Immucor (Mia Fora NGS Flex, Norcross, GA) run on the Illumina MiSeq system (San Diego, CA) and analyzed with the Mia Fora Flex software (Immucor, version 5.1). We were very confident in the phasing as the sample displayed a mean read length of 324 base pairs over all the loci, the mismatched C base was attributed 179 times to the new HLA-B*08 allele and can be only attributed to this allele because it was possible to discriminate from the associated HLA-B*35:08:01:01 allele by virtue of 11 variant positions each distant by less than 100 base pairs. HLA typing by Luminex reverse sequence-specific oligonucleotide (SSO) was performed (One Lambda Labtype XR, Canoga Park, CA).3 With this assay (lot 006, catalog RSSOX1B_007_02), the pattern of positive beads did not result in an acceptable HLA typing. We had to modify the bead #600 and bead #706 from negative to positive to obtain a HLA-B*08:01 result. Indeed beads #600 and #706 displayed oligonucleotides targeting the sequence surrounding codon 116. The analysis of the localization of this amino-acid and its antibody accessibility with the pHLA3D database4 indicated that this amino-acid is located into the peptide binding groove. As such this could have an importance in both allogeneic hematopoietic stem cell transplantation and solid organ transplantation. The coding nucleotide sequence of the new allele has been submitted to the GenBank database (Accession No. ON933816) and to the IPD-IMGT/HLA Database (Submission No. HWS10062188). The name B*08:302 has been officially assigned by the WHO Nomenclature Committee for Factors of the HLA System in August 2022. This follows the agreed policy that, subject to the conditions stated in the most recent Nomenclature Report,5 names will be assigned to new sequences as they are identified. Lists of such new names will be published in the following WHO Nomenclature Report.
AUTHOR CONTRIBUTIONS
Marine Cargou and Jonathan Visentin contributed to the design of the study. Marine Cargou and Jonathan Visentin participated in the writing of the paper. Marine Cargou, Vincent Elsermans, Isabelle Top, Elodie Wojciechowski and Jonathan Visentin participated in the performance of the research. Marine Cargou, Vincent Elsermans, Isabelle Top, Elodie Wojciechowski and Jonathan Visentin participated in data analysis. Vincent Elsermans, Isabelle Top and Elodie Wojciechowski were involved in critical revision of the manuscript.
ACKNOWLEDGMENTS
The authors thank the technicians of the Bordeaux and Lille Immunology laboratories for their technical expertise.
CONFLICT OF INTEREST
The authors confirm that there are no conflicts of interest.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.