Transcriptome analysis of frontal fibrosis alopecia revealed involvement of immune cells and ferroptosis
Abstract
Background
Frontal fibrosis alopecia (FFA) is a primary cicatricial alopecia and has received increasing attention in recent years. However, the pathogenesis of FFA has not been fully elucidated.
Methods and results
Herein, we collected the transcriptome data of scalp lesions of seven patients with FFA and seven healthy controls. The differential expression analysis and weighted gene co-expression network analysis were conducted and we identified 458 differentially expressed genes (DEGs) in two key modules. Later, we performed functional enrichment analysis and functional modules identification, revealing the participation of immune response and fatty acid metabolism. Based on the results, we processed further studies. On the one hand, we analyzed the infiltrating immune cells of FFA through CIBERSORT algorithm, indicating the activation of M1 macrophage and CD8+ T cell. On the other hand, considering lipid metabolism of FFA and oxidative stress of hair follicle cells in alopecia, we explored the potential ferroptosis of FFA. By intersection of DEGs and ferroptosis-related genes from FerrDb database, 19 genes were identified and their expression was validated in an external dataset containing 36 FFA cases and 12 controls. Then, we used LASSO algorithms to construct a four-gene diagnostic model, which achieved an AUC of 0.924 in validation dataset. Additionally, the immune cells were found to be related to ferroptosis in FFA.
Conclusion
Taken together, this study contributed to reveal the molecular mechanisms of FFA and is expected to inspire future research on treatment.
1 INTRODUCTION
Frontal fibrosis alopecia (FFA) is a type of primary lymphocytic cicatricial alopecia, characterized by the progressive hair loss of eyebrow and frontotemporal.1 It mainly occurs in post-menopausal women and has a negative impact on the quality of patients’ life, and even might inducing psychological distress.2, 3 Some risk factors were found in recent studies, including sunscreen usage, tobacco consumption, thyroid disorders, and rosacea.4-6
More and more studies have showed increasing interests in cicatricial alopecia.7 In terms of the pathogenesis of FFA, it is still unclear and seldomly reported. E. Del Duca et al. preliminary investigated the biomarkers in scalp biopsies of patients with FFA.8 And they emphasized the robust immune activation with CD8+ cytotoxic T cells, increased regulatory T cells, increased interferon-γ aaZ production and JAK/STAT signaling. Similarly, Celina Dubin, BA et al .also studied the scalp and serum profiling of FFA patients and found significant gene marker related to Th1, T cell activation, fibrosis, T regulatory, and Janus kinase.9 In addition, major primary cicatricial alopecia variants, including FFA, were found to share dysregulated pathways involving mast cells.10 However, the mechanism of FFA has not been fully elucidated and needs to be further determined.
Ferroptosis is an iron-dependent and lipid peroxidation-involved programmed cell death type proposed in 2012.11 Distinguished from cell necrosis, apoptosis, and cell autophagy, it is characterized by mitochondrial atrophy, increased mitochondrial membrane density, participation of specific genes, and accumulation of iron and lipid reactive oxygen species.12 It has been found to have an impact on many diseases, including tumors, neurodegenerative diseases, inflammation, and infection.13 In recent years, the role of ferroptosis has been revealed in many skin diseases, such as melanoma, psoriasis, and vitiligo.14-16 Nevertheless, the relationship between ferroptosis and FFA, even alopecia, is unknown.
Herein, we aimed to analyze the gene expression profile of FFA from clinical scalp lesions through bioinformatics methods and identify key differentially expressed genes (DEGs) and functional modules. Furthermore, we explored the diagnostic performance of ferroptosis-related genes (FRGs) as the accurate biomarkers in FFA and analyzed the immune microenvironment of FFA. Considering that there are currently few studies on the mechanism of FFA, the present study is expected to provide new insight of it and contribute to future research on treatment and prevention strategies.
2 METHODS
2.1 Data source
The gene expression dataset of FFA was acquired from the Gene Expression Omnibus (GEO) database. The dataset numbered GSE125733 on platform GPL11154 was included in this analysis, which consists of scalp skins from seven patients with FFA and seven matched controls.17 Besides, we selected another dataset numbered GSE186075 as our validation data. It is based on platform GPL21290 and contains 36 FFA skin lesions and 12 healthy controls.10
2.2 Differential expression analysis and WGCNA
We used R package “limma” to conduct differential expression analysis. The criteria for DEGs were p value < 0.05 and |logFC| > 0.5.18 Subsequently, weighted gene co-expression network analysis (WGCNA) was processed to find the key modules.19 We used the top 50% of genes with the largest variance and hierarchical clustering analysis for outlier test to obtain a valid network. Then, under the best P power value, the correlation matrix was converted to the adjacency matrix (topological overlap matrix) through scale independence and mean connectivity. Later, we used the average-linkage hierarchical clustering method to cluster the genes and merged the similar clusters. Finally, we performed Pearson's correlation analysis to value the correlation between modules and FFA and confirmed the most related modules. We intersected genes in the key modules and DEGs and visualized it through the Venn diagram tool. The overlapping genes were regarded as key DEGs.
2.3 Functional analysis of key DEGs
Based on the expression of key DEGs, we conducted the gene set enrichment analysis (GSEA) of Gene Ontology (GO) annotations and the gene set enrichment analysis (GSEA) of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway to characterize biological and functional properties.20-22 Furthermore, we applied the MCODE app in Cytoscape to identified functional modules.23 And GO analysis was used to identify the characteristics of functional modules.
2.4 Identification and validation of DE-FRGs
The ferroptosis-related genes (FRGs) were collected from FerrDb database.24 After the intersection of FRGs and DEGs, the differentially expressed ferroptosis-related genes (DE-FRGs) were analyzed through a comprehensive functional enrichment analysis, including GO analysis on molecular function, cellular component, and biological process, and KEGG analysis for all pathways. Subsequently, we validated the DE-FRGs in GSE186075 dataset. The expression levels of DE-FRGs in FFA and control were compared by the t-test.
2.5 Ferroptosis-related biomarkers for diagnostic model of FFA
In order to reduce data dimension and identify ferroptosis-related biomarkers of FFA, the least absolute shrinkage and selection operator (LASSO) algorithm were employed under glmnet package.25 Then, based on the biomarkers obtained by LASSO algorithm, we developed a nomogram to quantify the probability of FFA.26 To evaluate the model's performance, we used calibration curve (CC) to show the difference between the observed and projected values, and receiver operating characteristic (ROC) curves in the validation dataset.27
2.6 Immune infiltration analysis
CIBERSORT, a deconvolution algorithm based on gene expression of leukocyte signature matrix reference, was adopted to explore the immune infiltration of FFA.28 We collected the CIBERSORT scores of immune cells and analyzed their correlation. In addition, we also analyzed the correlation between CIBERSORT scores of immune cells and expression of DE-FRGs.
3 RESULTS
3.1 DEGs and key modules in WGCNA
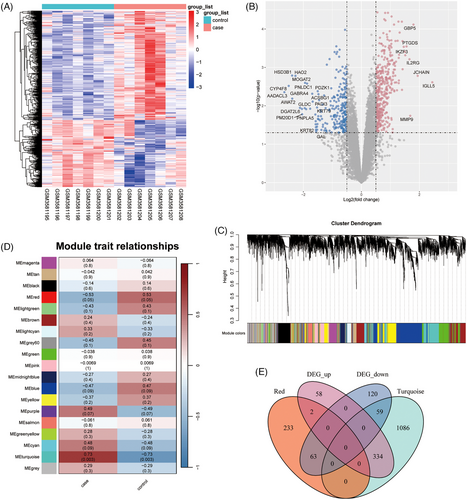
We processed the differential expression analysis in FFA, which was showed in the heatmap and volcano plots (Figure 1A and 1B). Finally, a total of 394 upregulated and 242 downregulated DEGs were found. The WGCNA analysis constructed the FFA network and clustered 19 modules (Figure 1C and 1D). And the turquoise and red modules were found significantly related to FFA and selected as the key modules. After the intersection of 636 DEGs with genes in the key modules, 458 key DEGs were identified (Figure 1E).
3.2 Functional enrichment of key DEGs
The GSEA-GO analysis on 458 key DEGs mainly suggested the role of immune response, stimulus response, and lipid metabolic process in FFA (Figure 2A). And in GSEA-KEGG results, these genes were mostly enriched in some autoimmune diseases, antigen processing and presentation, and immune response (Figure 2B). Later, five functional modules were identified by MCODE of Cytoscape (Figure 2C-G). The results of enrichment analysis of functional modules showed that module 1, 2, and 3 were all related to immune response and immune cells; module 4 played a role in fatty acid metabolism; and module 5 was associated with membrane potential and synapse (Figure S1).

3.3 DE-FRGs and validation
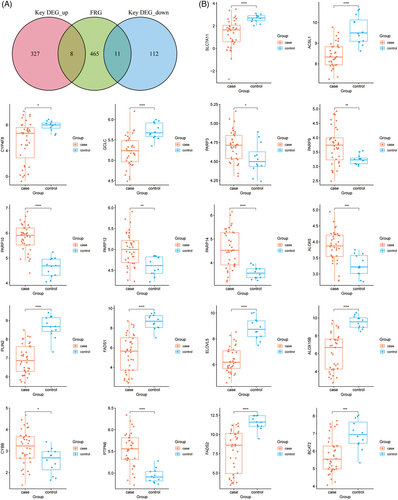
We further investigated the ferroptosis-related genes in FFA. We acquired 484 FRGs from FerrDb database and took the intersection of FRGs and key DEGs, indicating 19 DE-FRGs, including SLC7A11, ACSL1, CYP4F8, GCLC, PARP3, PARP9, PARP10, PARP12, PARP14, ALOX5, PLIN2, FADS1, ELOVL5, ALOX15B, CYBB, PTPN6, FADS2, BCAT2, and HILPDA (Figure 3A). Furthermore, we explored the functional enrichment of 19 DE-FRGs. The results of GO analysis on molecular function, cellular component, and biological process and KEGG analysis were showed in Figure 4. The KEGG results pointed out the involving pathways including ferroptosis, fatty acid metabolism, biosynthesis of unsaturated fatty acids, arachidonic acid metabolism, metabolic pathways and PPAR signaling pathway. Subsequently, we verified the gene expression of 19 DE-FRGs in validation dataset GSE186075. And all DE-FRGs were found to be significantly differentially expressed (p < 0.05), except HILPDA was not detected for the different platforms (Figure 3B).

3.4 Diagnostic model and biomarkers
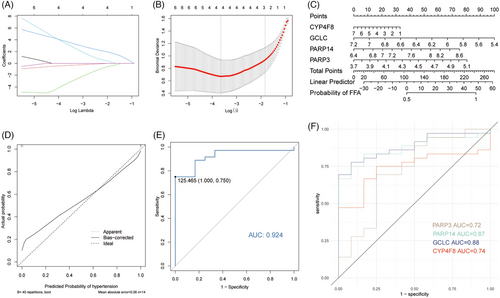
The LASSO algorithm obtained four DE-FRDs as biomarkers, including CYP4F8, GCLC, PARP14, and PARP3 (Figure 5A and 5B). We established a diagnostic model for FFA based on these four DE-FRDs and the CC shows a small difference between the observed and projected probability of FFA (Figure 5C and 5D). Then, the performance of the model in validation dataset GSE186075 were explored in ROC analysis with a good AUC of 0.924 (Figure 5E). Additionally, the ROC analyses of every biomarker in validation dataset were processed and the results were AUC of 0.72 for PARP3, 0.87 for PARP14, 0.88 for GCLC, and 0.74 for CYP4F8 (Figure 5F).
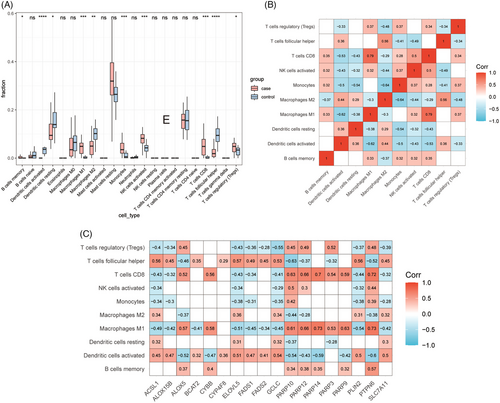
3.5 Immune infiltration of FFA
The immune microenvironment in FFA was explored utilizing the CIBERSORT algorithm. Memory B cells, activated dendritic cells, resting dendritic cells, M1 macrophages, M2 macrophages, monocytes, activated NK cells, CD8+ T cells, follicular helper T cells, and regulatory T cells were found to be significantly different between FFA lesions and controls (Figure 6A). And the correlation analysis of immune cells yielded out a high correlation of CD8+ T cells and M1 macrophages (Figure 6B). Besides, the relationshio analysis of immune cells and DE-FRDs showed a strong correlation between DE-FRDs and helper T cells, CD8+ T cells, M1 macrophages, and activated dendritic cells (p < 0.05) (Figure 6C).
4 DISCUSSION
This study analyzed transcriptome data of 14 clinical skin samples and collected 48 skin samples as validation dataset. After differential expression analysis and WGCNA, a total of 458 key DEGs were identified. They yielded five functional modules, including three immune-related modules, a fatty acid metabolism-related module, and a membrane potential and synapse-related module. Considering the robust immune infiltration of FFA suggested by the results of functional enrichment analysis and the functional modules, we further processed the immune infiltration analysis through CIBERSORT. And the results stressed the role of M1 macrophages and CD8+ T cells in FFA and ferroptosis. This was partly consistent with prior studies. Previous immunohistochemical studies on inflammatory microenvironment of hair follicles in FFA showed a CD8-biased T-cell infiltration.29 And the study of E. Del Duca et al. also suggested a strong CD8+ cytotoxic T cells activation, juxtaposed to follicular stem cells, both in FFA lesions and non-lesional scalp.8 Besides, the role of M1 macrophages also was emphasized in the pathogenesis of FFA. FFA is commonly considered a variant of lichen planopilaris (LPP) because of their similar histological features. However, the macrophages polarization seemed to play a vital role in the determination of these two diseases. The M1 macrophage marker CD86 in LPP were found to be downregulated compared with FFA, while M2 marker CD163 were upregulated in LPP.30
In present studies, the key DEGs also showed the involvement in fatty acid metabolism in mechanism of FFA. Genes on cholesterol biosynthesis, fatty acid biosynthesis and metabolism were found to be downregulated in FFA lesions in previous study.10 The lipid metabolism is highly correlated with ferroptosis and lipid peroxidation.31 Besides, oxidative stress has been found in multiple hair loss disorders, including alopecia areata and androgenetic alopecia.32-34 The free radical-mediated lipid peroxidation could induce the apoptosis of hair follicle cells.35 Notably, reactive oxygen species generation has been considered tightly linked to ferroptosis, as the result of the activation of ferroptosis by excessive reactive oxygen species production.36 Additionally, ferroptosis were found to be associated with the development of many fibrotic diseases, such as renal fibrosis, liver cirrhosis, idiopathic pulmonary fibrosis, and cardiomyopathy.37 Whereas FFA was described as a type of scarring alopecia clinically and histologically and many fibrosis markers were founded to be significantly elevated in FFA.9, 38 Based on these clues of the association between FFA and ferroptosis, we further identified potential FRGs in FFA. And finally, 19 DE-FRGs, named SLC7A11, ACSL1, CYP4F8, GCLC, PARP3, PARP9, PARP10, PARP12, PARP14, ALOX5, PLIN2, FADS1, ELOVL5, ALOX15B, CYBB, PTPN6, FADS2, BCAT2, and HILPDA, were selected and validated for differential expression in an external dataset.
Among the 19 DE-FRGs, PARP3, PARP9, PARP10, PARP12, PARP14 are the members of poly ADP-ribose polymerase (PARP) family, which are involved in DNA repair, DNA methylation, transcriptional regulation, and metabolic regulation.39 Previous study has showed the role of PARP in promoting ferroptosis via regulating the expression of SLC7A11 in some diseases.40, 41 And many members of PARP family were found to be upregulated in FFA cases compared with healthy controls in this study. This might be a direction for FFA treatment that deserves further investigation. Moreover, we constructed a four-gene-model for FFA diagnosis based on four DE-FRGs through LASSO algorithm. The model achieved the AUC of 0.924 in validation dataset, suggesting its accuration and specificity in distinguishing FFA samples from healthy controls. This also explains to some extent that ferroptosis signaling is a potential pathway in the development of FFA.
The expression of Solute Carrier Family 7 Member 11, SLC7A11, in FFA was downregulated in our study. SLC7A11 is a key gene of ferroptosis, as its downregulation could indirectly inhibit the activity of GPX4 which can lead to the accumulation of lipid peroxides and finally induce ferroptosis in cells.42 The role of SLC7A11-GPX4 axis in ferroptosis may function in some inflammatory and immune dermatosis. For instance, in vitiligo, an autoimmune skin disease charactered by the disappearance of melanocytes due to the overactivity of CD8+ T cells, ferroptosis might be triggered by oxidative-related production, including the reduced activity and expression of GPX4.43, 44 It was tightly related to oxidative stress and might initiate autoimmunity of vitiligo.45 Interestingly, vitiligo seems to be associated with FFA as some studies have reported cases of coexistence of FFA and vitiligo.46-48 These two diseases are both immune-mediated disorders and the infiltration of most lymphocyte are of CD8+ cytotoxic origin. Besides, they both emphasized the interferon-γ-induced immune collapse and the potential role of oxidative stress. Mechanistically, CD8+ T cells can secrete interferon-γ to downregulate the expression of SLC7A11, thereby promoting ferroptosis induction secondary to GSH depletion.49 Therefore, similar pathogenic pathways might explain the coexistence and ferroptosis might also take place in the development of FFA. Besides, although the role of macrophage in ferroptosis is unclear, a previous study indicated that M1 macrophage could enhance the functions of CD8+ T cell via stimulating the secretion of interferon-γ of CD8+ T cell.50 This implies that M1 macrophage may play a role in ferroptosis by increasing interferon-γ from CD8+ T cell.
Despite the fact that we have gone as far as possible into the pathogenesis of FFA and preliminarily explored the ferroptosis in the development of FFA, there are still some limitations in this study. First, although the DE-FRGs have been validated in another dataset, further experimental verification of gene expression and functions in FFA is absent due to a lack of clinical cases. Secondly, the ferroptosis sensitivity might differ in multiple cells of the epidermis.50 Thus, the specific situation of ferroptosis in different pathological locations of FFA is worthy of further study.
In conclusion, we identified 458 key DEGs and five functional modules through transcriptome data of FFA scalp samples, stressing the involvement of immune and fatty acid metabolism in FFA. And further study suggested that ferroptosis and immune cells, mainly M1 macrophage and CD8+ T cell, might contribute to the development of FFA. This study is expected to improve the understanding of the pathogenesis of FFA and provide potential therapeutic targets for the future.
ACKNOWLEDGMENTS
The authors thank the authors of the databases mentioned in the manuscript.
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
CONFLICT OF INTEREST STATEMENT
None of the authors have any potential financial conflict of interest related to this manuscript.
ETHICS STATEMENT
No ethical approval is required.
Open Research
DATA AVAILABILITY STATEMENT
Data will be made available on request.