Circ_0068631 sponges miR-139-5p to promote the growth and metastasis of cutaneous squamous cell carcinoma by upregulating HOXB7
Abstract
Background
Circular RNAs (circRNAs) are often dysregulated in cancers and closely related to cancer progression, including cutaneous squamous cell carcinoma (CSCC). However, the role and mechanism of circ_0068631 in CSCC progression have not been reported.
Methods
The expression of circ_0068631, microRNA-139-5p (miR-139-5p), and homeobox B7 (HOXB7) was measured by real-time quantitative polymerase chain reaction (RT-qPCR). Cell counting kit-8 (CCK-8) assay, 5-ethynyl-2′-deoxyuridine (EdU) assay, and colony formation assay were used to measure cell proliferation. Cell apoptosis was assessed by flow cytometry. Cell migration was detected by transwell assay. The interaction between miR-139-5p and circ_0068631 or HOXB7 was confirmed by dual-luciferase reporter assay. A xenograft tumor model was established to confirm the function of circ_0068631 in vivo.
Results
Circ_0068631 was upregulated in CSCC tissues and cells, and its silencing could inhibit CSCC cell proliferation and metastasis while promoting apoptosis in vitro, as well as restrain CSCC tumor growth in vivo. Circ_0068631 acted as a sponge of miR-139-5p, and miR-139-5p inhibition reversed the repressive effect of circ_0068631 knockdown on CSCC cell progression. Furthermore, HOXB7 was a target of miR-139-5p, and miR-139-5p inhibited the malignant behaviors by downregulating HOXB7 expression in CSCC cells. Further, circ_0068631 sponged miR-139-5p to regulate HOXB7 expression.
Conclusion
Circ_0068631 functioned as a novel oncogene in CSCC progression by regulating miR-139-5p/HOXB7 axis, suggesting that circ_0068631 may be a potential target for CSCC treatment.
Highlights
- Circ_0068631 was overexpressed in CSCC tissues and cells.
- Circ_0068631 downregulation suppressed CSCC progression via miR-139-5p.
- Circ_0068631 regulated HOXB7 via sponging miR-139-5p.
1 INTRODUCTION
Cutaneous squamous cell carcinoma (CSCC) ranks as the second most frequent nonmelanoma skin malignancy, accounting for 20% of all cutaneous cancer types.1 Significant progress has been achieved in therapeutic, such as surgical excision, radiotherapy, chemotherapy, and topical drug, but the outcome is still not satisfactory owing to population aging and the focus on skin cancer screening.2 Therefore, identifying the molecular mechanisms that influence the progression of CSCC is essential to developing a novel theoretical basis for this cancer.
As a subset of noncoding RNAs (ncRNAs), circular RNAs (circRNAs) originate from the back-splicing events of pre-mRNAs and have covalently closed structures.3 Due to the lack of 5′ caps and 3′ poly (A) tails, circRNAs are less easily degraded by nucleases than well-known linear RNAs, which makes them more stable.4 There is increasing evidence that exhibited the important biological roles of circRNAs dysregulation in cellular functions and disease development.5, 6 More and more circRNAs have been reported as promoters or suppressors in the initiation of various human cancers, containing CSCC.7-9 CircRNA expression profile showed that circ_0068631 (chr3:195802029-195803993) derived from transferrin receptor gene was upregulated in CSCC. However, its function in CSCC is still unknown.
MicroRNAs (MiRNAs, another type of small ncRNAs) can interact with target mRNAs to inhibit their translation, thereby regulating gene expression at the posttranscriptional level.10 MiR-139-5p is a well-known suppressor in many cancers.11, 12 Moreover, homeobox B7 (HOXB7) has been demonstrated to promote CSCC development.13 Through bioinformatics tools, we found that HOXB7 3′UTR had the binding sequence with miR-139-5p. Based on the competitive endogenous RNA (ceRNA) networks hypothesis, circRNAs act as miRNA sponges to repress miRNA function, protecting the target genes from cleavage by specific miRNAs.14, 15 At present, this regulatory mechanism is also applicable to the research on CSCC progression.16 In this work, bioinformatics analysis exhibited that circ_0068631 possessed some binding sites with miR-139-5p. Hence, whether circ_0068631 regulated CSCC behaviors in a miR-139-5p-HOXB7-dependent manner was further investigated.
2 METHODS
2.1 Specimen collection
Authorization to perform this project was acquired from the Ethics Committee of The First College of Clinical Medical Science before it was carried out. Twenty-four cases of CSCC tissue samples and normal samples were provided by The First College of Clinical Medical Science. The detailed clinical feature of sufferers is described in Table 1. None of these patients received any treatment prior to surgical resection, and they signed informed consents. Tissue specimens were preserved at −80°C.
| HOXB7 expression | ||||
|---|---|---|---|---|
Characteristics (n = 24) |
Low (n = 12) | High (n = 12) | p-Value | |
| Age (years) | 0.4136 | |||
| ≤50 | 11 | 4 | 7 | |
| >50 | 13 | 8 | 5 | |
| Gender | >0.9999 | |||
| Male | 13 | 7 | 6 | |
| Female | 11 | 5 | 6 | |
| Lymph node metastasis | 0.0361* | |||
| Positive | 14 | 4 | 10 | |
| Negative | 10 | 8 | 2 | |
| Tumor size | 0.0123* | |||
| ≤4 cm | 13 | 10 | 3 | |
| >4 cm | 11 | 2 | 9 | |
- * p < 0.05.
2.2 Cell culture and transfection
The cell culture medium was Dulbecco's Modified Eagle Medium (DMEM; Solarbio, Beijing, China) including 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA). Meanwhile, all cell lines in this project were provided by BeNa Culture Collection (Beijing, China) under standard conditions (37°C, 5% CO2).
The oligonucleotides (RiboBio, Guangzhou, China) contain short hairpin (sh)-circ_0068631, miR-139-5p mimic/inhibitor (miR-139-5p/anti-miR-139-5p), controls (sh-NC, miR-NC/anti-miR-NC), and plasmids (RiboBio) include HOXB7-overexpressing plasmid (HOXB7) and pcDNA were introduced into tumor cells by Lipofectamine 3000 Reagent (Invitrogen).
2.3 Real-time quantitative polymerase chain reaction (RT-qPCR)
After TRIzol Reagent (Invitrogen) treatment, total RNA from cells or clinical samples was prepared, followed by reverse transcription with miRNA First-Strand Synthesis Kit (for miRNA; Clontech, Mountain View, CA, USA) or a Primescript RT Reagent (for mRNA/circRNA; TaKaRa, Kusatsu, Japan). Polymerase chain reaction (PCR) detection was prepared by SYBR Master Mix (Roche, Basel, Switzerland). After being normalized with U6 (for miRNA) or Glyceraldehyde-3-phosphate Dehydrogenase (GAPDH) (for mRNA/circRNA), the collected data were calculated by the 2−ΔΔCt method. RT-qPCR assay was performed at least three times. Primer information was listed in Table 2.
| Names | Sequences (5′−3′) |
|---|---|
| circ_0068631: Forward | AAGGCCAATGTCACAAAACC |
| circ_0068631: Reverse | GCCGAGTCTACAGTGCACGTGTC |
| miR-139-5p: Forward | GCCGAGTCTACAGTGCACGTGTC |
| miR-139-5p: Reverse | CAGTGCGTGTCGTGGAGT |
| HOXB7: Forward | ATCTACCCCTGGATGCGAAGCT |
| HOXB7: Reverse | GCGTCAGGTAGCGATTGTAGTG |
| U6: Forward | CTCGCTTCGGCAGCACATATACT |
| U6: Reverse | ACGCTTCACGAATTTGCGTGTC |
| GAPDH: Forward | GTCTCCTCTGACTTCAACAGCG |
| GAPDH: Reverse | ACCACCCTGTTGCTGTAGCCAA |
2.4 CircRNA identification
To check its stability, total RNA from SCL-1 and A431 cells at room temperature was mixed with RNase R (Seebio, Shanghai, China) for 0.5 h, followed by RT-qPCR analysis. The assay was triplicated.
2.5 Cell proliferation assays
Cell counting kit-8 (CCK-8) assay was utilized for examining cell viability. Shortly, transfected tumor cells (2000 cells per well) were mixed with CCK-8 reagent (10 μl, Beyotime, Jiangsu, China) at indicated time points. After being incubated for 3 h, absorbance was detected according to a microplate reader. The experiment was conducted thrice.
For the 5-ethynyl-2′-deoxyuridine (EdU) assay, transfected SCL-1 and A431 cells in 24-well plates were reacted with 20 μM of EdU solution (Beyotime) for 2 h, followed by a 4% paraformaldehyde fixture. After being added with Click Additive Solution, cells were subjected to staining nucleic acids using 4′,6-diamidino-2-phenylindole. Under a fluorescence microscope (×100; Leica, Wetzlar, Germany), EdU-positive cells were analyzed. This assay was carried out at least three times.
In addition, tumor cells in six-well plates were cultured 14 days later. The generated colonies (cell mass containing > 50 cells) were fixed using methanol (Sangon Biotech, Shanghai, China) and stained with Giemsa (Beyotime), followed by calculating using a microscope. Colony formation assay was conducted at least three times.
2.6 Flow cytometry analysis
Shortly, transfected cells in six-well plates were labeled with Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (Beyotime) and assessed by means of FACScan flow cytometer. This assay was performed at least three times.
2.7 Transwell assay
In short, the bottom chambers were full of 10% FBS (as chemoattractant) medium, and CSCC cells in FBS-free medium were introduced into the top counterpart with Matrigel (BD Biosciences, San Jose, CA, USA). After 24 h, cells invaded cells were fixed and stained, followed by capturing images using a microscope (Leica). This assay was executed at least three times.
2.8 Western blot assay
According to RIPA lysis buffer (Solarbio), total proteins were prepared. Next, 30-μg protein samples were size-fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. After transferring onto PVDF membranes (2 h, 100 V), primary antibodies (4°C, overnight) were incubated. After incubation with secondary antibody, protein bands were observed according to the enhanced chemiluminescence solution (Vazyme, Nanjing, China). Antibodies (Abcam, Cambridge, UK) contains E-cadherin (ab231303; 1:1500), N-cadherin (ab98952; 1:1000), HOXB7 (ab152454; 1:1500), β-actin (ab8227; 1:2000), and secondary antibodies (ab205718/ ab205719; 1:4000). Western blot assay was conducted at least three times.
2.9 Dual-luciferase reporter assay
Fragments of circ_0068631 or HOXB7 3′-untranslated regions (3′UTR) with or without miR-139-5p binding sequences were generated, followed by incubation into the pmirGLO vector (YouBia, Changsha, China), generating wild-type/mutant-type (WT/MUT)-circ_0068631 or WT/MUT-HOXB7 3′UTR luciferase vectors. After that these above vectors were introduced into SCL-1 and A431 cells with t miR-139-5p/miR-NC for 48 h, followed by dual-luciferase reporter assay system (Promega, Madison, WI, USA) detection. Luciferase reporter assay was performed at least three times.
2.10 In vivo assay
After being approved by the Animal Care and Use Committee of The First College of Clinical Medical Science, BALB/c nude mice (female; Vital River, Beijing, China) were introduced into this research. Following segmented into two groups (n = 6 per group), 5–6 weeks old mice (n = 12) were injected with A431 cells (1 × 107, circ_0068631 silencing or control). Eight days later, the tumors volume was recorded and calculated according to the equation: volume = 1/2 (length × width2). At 23 days postinoculation, the mice were killed, and harvested tissues were further tested.
2.11 Immunohistochemistry analysis
Tumor tissue samples from nude mice were fixed in formaldehyde (10%) and embedded in paraffin. Subsequently, these tissues were then cut into the 4-μm-thick section, and incubated with Ki67 (ab15580; 1:500; Abcam) HOXB7 (D262980; 1:200; Sangon Biotech), E-cadherin (ab231303; 1:500; Abcam), N-cadherin (ab98952; 1:500; Abcam), Matrix metalloproteinase9 (MMP9; ab76003; 1:1000), B-cell lymphoma-2 (Bcl-2; ab182858; 1:500), and Bcl-2-related X protein (Bax; ab32503; 1:250), followed by incubation using corresponding secondary antibody (ab205718/ab205719; 1:2000). After diaminobenzidine staining and haematoxylin counterstaining, the observation of positive cells was performed under a microscopy. CSCC tumor tissues and normal tissues were subjected to immunohistochemistry (IHC) staining analysis, according to the above steps.
2.12 Statistical analysis
All data in this research were represented as mean ± standard deviation. Each assay was repeated independently at least three times. Significance differences were determined using student's t-test or a one-way analysis of variance with GraphPad Prism 6.0. Expression association was assessed according to Pearson correlation analysis. p < 0.05 was defined as statistically significant.
3 RESULTS
3.1 Circ_0068631 was upregulated in CSCC tissues and cells
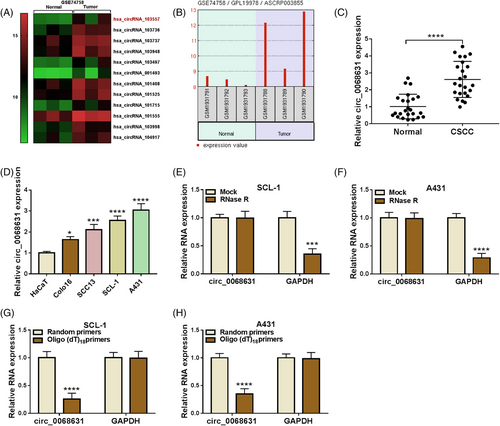
To identify circRNAs involved in CSCC tumorigenesis, we searched the GEO database. According to the previous report,16 GEO database (accession number GSE74758) showed that hsa_circRNA_103557 (also known as circ_0068631) was upregulated in punch biopsies from CSCC (Figure 1A), and its expression was presented in Figure 1B. Apart from that, circ_0068631 content was increased in CSCC sufferers and CSCC cell lines (SCL-1 and A431) (Figure 1C,D). Next, we analyzed the characteristics of circ_0068631 in CSCC cells. As displayed in Figure 1E,F, circ_0068631, rather than GAPDH, was resistant to RNase R. Next, circ_0068631 expression was lower when oligo (dT)18 primers were used (Figure 1G,H). Collectively, these results mentioned above demonstrated that circ_0068631 was a circRNA and upregulated in CSCC.
3.2 Circ_0068631 absence constrained CSCC cell malignant behavior
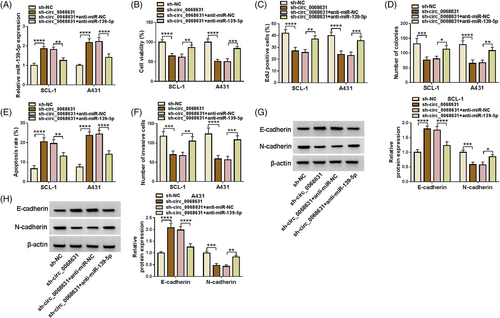
Subsequently, the effect of circ_0068631 on cell behavior was investigated. The knockdown efficiency of sh-circ_0068631 was determined by RT-qPCR (Figure 2A). Circ_0068631 downregulation suppressed SCL-1 and A431 cell viability (Figure 2B). Meanwhile, the percentage of EdU-positive cells was obviously decreased via the interference of circ_0068631 in tumor cells (Figure 2C), indicating repression of circ_0068631 silencing on DNA synthesis. Besides, circ_0068631 absence reduced SCL-1 and A431 cell colony-forming ability (Figure 2D). Furthermore, circ_0068631 downregulation increased SCL-1 and A431 cell apoptosis (Figure 2E). Also, circ_0068631 silencing repressed SCL-1 and A431 cell invasion (Figure 2F). In addition, E-cadherin (an epithelial marker) was increased, and N-cadherin (a mesenchymal marker) was downregulated via circ_0068631 knockdown (Figure 2G,H). To sum up, circ_0068631 absence inhibited CSCC cell malignant behaviors.

3.3 Circ_0068631 could sponge miR-139-5p
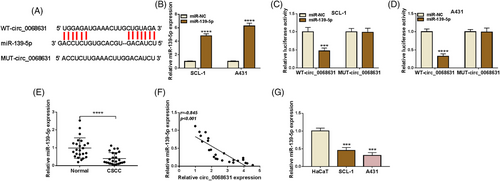
Accumulating evidence suggests that circRNAs are able to sponge miRNAs to exert their functions.17 As presented in Figure 3A, circinteractome analysis displayed that circ_0068631 had binding sites with miR-139-5p. RT-qPCR analysis of the transfection efficiency of miR-139-5p mimic (Figure 3B). Next, miR-139-5p overexpression obviously reduced the luciferase activity of WT-circ_0068631, rather than the MUT-circ_0068631 group (Figure 3C,D). Moreover, we found that miR-139-5p was downregulated in CSCC tissues and negatively correlated with circ_0068631 expression in CSCC tissues (Figure 3E,F). Likewise, an apparent reduction of miR-139-5p was viewed in tumor cells (Figure 3G). The above-mentioned results suggest circ_0068631 targeted miR-139-5p.
3.4 Circ_0068631 regulated CSCC cell behaviors by binding with miR-139-5p
As shown in Figure 4A, circ_0068631 absence increased the miR-139-5p level, which was reversed via miR-139-5p inhibition in SCL-1 and A431 cells. Moreover, miR-139-5p downregulation counteracted circ_0068631 knockdown-triggered proliferation inhibition and apoptosis promotion (Figure 4B–E). Furthermore, circ_0068631 silencing-mediated invasion and epithelial mesenchymal transformation (EMT) suppression in SCL-1 and A431 cells also were abolished by the miR-139-5p inhibitor (Figure 4F–H). That was, circ_0068631 sponged miR-139-5p to regulate CSCC development.
3.5 miR-139-5p targeted HOXB7
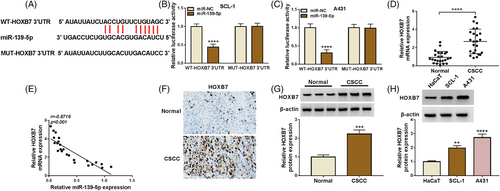
According to the application of Starbase software, HOXB7 3′UTR possessed binding sites with miR-139-5p (Figure 5A). Simultaneously, increased miR-139-5p inhibited the luciferase activity of WT-HOXB7 3′UTR, instead of the MUT-HOXB7 3′UTR group (Figure 5B,C). Moreover, HOXB7 content was enhanced and negatively correlated with miR-139-5p in CSCC tissues (Figure 5D,E). To probe the correlation of HOXB7 content with clinicopathologic features, the 24 patients with CSCC were then classified in Table 1. Data presented that HOXB7 content had an association with Lymph node metastasis and Tumor size. Besides, IHC staining displayed that HOXB7 content was enhanced in CSCC tissues (Figure 5F). Apart from that, an upregulated HOXB7 was viewed in CSCC tissues and cells (Figure 5G,H). Our data suggested that miR-139-5p could target HOXB7.
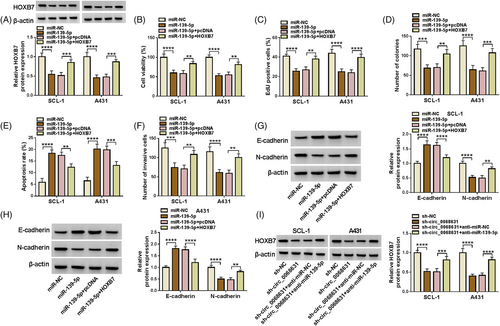
3.6 MiR-139-5p targeted HOXB7 to hamper CSCC cell malignant behaviors
Western blot discovered repression of miR-139-5p mimic on HOXB7 content was partly abolished via HOXB7 introduction (Figure 6A). Functionally, increased miR-139-5p-caused CSCC cell proliferation and invasion suppression, and apoptosis acceleration was reversed via HOXB7 (Figure 6B–F). Beyond that the increase of E-cadherin protein expression and the decrease of N-cadherin protein expression caused by miR-139-5p upregulation were inverted by HOXB7 overexpression (Figure 6G,H). Next, we observed that circ_0068631 knockdown decreased the protein level of HOXB7 in tumor cells, while this effect was overturned via miR-139-5p inhibition (Figure 6I). All results confirmed that miR-139-5p exerted an anticancer role via regulating HOXB7.
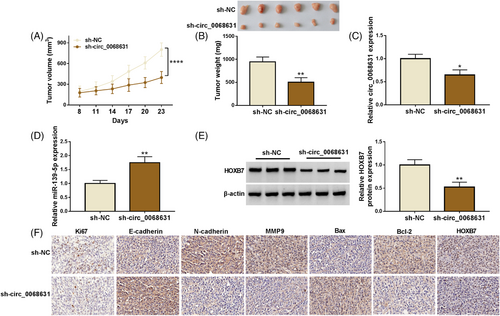
3.7 Circ_0068631 knockdown inhibited CSCC tumor growth in vivo
To clarify the biological function of circ_0068631, a xenograft model was established. As expected, circ_0068631 knockdown might constrain CSCC tumor growth (Figure 7A,B). Beyond that, circ_0068631 expression and HOXB7 protein expression were reduced in the sh-circ_0068631 group, while miR-139-5p expression was enhanced (Figure 7C–E, Figure S1). IHC analysis identified that Ki67 (a proliferation marker), N-cadherin, MMP9, Bcl-2, and HOXB7 expression were decreased, while E-cadherin and Bax expression was elevated via circ_0068631 downregulation (Figure 7F). All in all, circ_0068631 knockdown inhibited CSCC progression in vivo.
4 DISCUSSION
During the past decades, the morbidity and mortality of CSCC sufferers have been constantly increasing.18 Notably, it has been confirmed that the dysregulation of ncRNAs, particularly circRNAs, enrolled in the regulation of CSCC progression and tumorigenesis.16, 19 Herein, we found that circ_0068631 is upregulated in CSCC, and its downregulation repressed CSCC progression by regulating miR-139-5p/HOXB7 axis.
Due to their stability, conservation, and abundance, circRNAs have been thought of as potential biomarkers in human diseases.20, 21 Advances in sequencing technology have directly improved the knowledge of many human circRNAs.22 Moreover, the majority of circRNAs are being found correlated with disease processes.23, 24.Furthermore, some circRNAs were also reported to play pivotal roles in CSCC development in some kinds of literature. It has been confirmed that circ_0067772 was upregulated in CSCC and aggravated the malignant progression of CSCC.23 Additionally, circSEC24A promoted CCSC progression by increasing cell migration, proliferation, glycolysis, and invasion.24 However, there are still many circRNAs to be discovered. Here, we found a dramatic enhancement of circ_0068631 in CSCC, implying that it has a tumor-promoting role in CSCC. Importantly, circ_0068631 silencing constrained cell proliferation and invasion and promote apoptosis. At the same time, in vivo experiments validated the tumor-promoting of circ_0068631 in CSCC. The earlier study corroborated that downregulation of circ_0068631 repressed breast cancer cell growth and migration.25 Collectively, these observations reasonably proposed that circ_0068631 performs a carcinogenic factor in some tumors.
Mechanistically, circRNAs serve as miRNA sponges to release their repression on 3′UTR of downstream target mRNAs, thereby regulating cancer progression.26 Whether circ_0068631 served as a miRNA sponge was further explored. The current work validated that circ_0068631 could act as a miR-139-5p sponge. MiR-139-5p was dysregulated in multiple tumors and partake in cancer development, including endometrial cancer,27 prostate cancer,28 gastric cancer,29 cervical cancer,30 breast cancer,31 and lung adenocarcinoma.32 More importantly, miR-139-5p upregulation inhibited oral squamous cell carcinoma cell proliferation and mobility.33 However, its role in CSCC has not been explored. In this research, low expression of miR-139-5p was detected in CSCC cell lines and ESCC tissues. Rescue assays disclosed that miR-139-5p inhibitor partly counteracted circ_0068631 deficiency-mediated CSCC cell development, implying the oncogenic function of circ_0068631 exerted was mediated via binding to miR-139-5p.
Using the bioinformatics tool (starBase), we searched for potential targets of miR-139-5p. HOXB7 belongs to a family of transcription factors that can maintain adult tissue homeostasis and regulate growth and differentiation during embryonic development.34 HOXB7 is of10 abnormally expressed and acts as a tumor promoter via modulating cell behaviors in many cancers, such as gastric,35 prostate,36 colorectal,36 and breast37 cancers. Previous research showed that HOXB7 upregulation might facilitate CSCC tumor progression.38 Another study indicated that HOXB7 deficiency might repress CSCC cell malignant biological behaviors via the Wnt/β-catenin signaling pathway.39 In agreement with these results, we also detected an upregulation of HOXB7 expression in CSCC cells and tissues. Further rescue assays suggested miR-139-5p restoration restrained CSCC cell proliferation and metastasis via suppressing HOXB7 expression. Additionally, circ_0068631 sponged miR-139-5p to modulate the HOXB7 level.
In conclusion, the present study revealed that circ_0068631 facilitated CSCC progression via regulation of the miR-139-5p /HOXB7 axis, providing a novel theoretical basis for CCSS.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
FUNDING INFORMATION
The authors received no specific funding for this work.
ETHICS STATEMENT
The present study was approved by the ethical review committee of The First College of Clinical Medical Science. Written informed consent was obtained from all enrolled patients. Patients agree to participate in this work.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




