Bovine Circadian Locomotor Output Cycles Kaput (CLOCK) and Clusterin (CLU) mRNA Quantitation in Ejaculated Crossbred Bull Spermatozoa
Contents
Mammalian circadian locomotor output cycles kaput (CLOCK) gene encodes a transcription factor that affects both the persistence and the period of circadian rhythms. Earlier reports suggested that CLOCK gene might be associated with male infertility in human. Present investigation, for the first time, reports that CLOCK gene expresses differentially between good and poor quality crossbred bull semen. The relative expression of CLOCK was significantly (p < 0.05) higher among good quality bull semen than motility-impaired ones. Clusterins (CLU) are series of genes associated with a variety of physiological activities including spermatogenesis, apoptosis and degenerative disease conditions. In the present context, we also investigated that the expression of CLU gene was significantly (p < 0.05) higher among motility-impaired crossbred bull semen compared to the good quality one.
Introduction
The current approach of bull selection is semen-based fertility parameters. As semen is available only in post-pubertal phase, sperm-/seminal plasma-based information cannot be used for the prediction of high-fertility males. This needs an hour hand to identify some candidate genes/biomarkers associated with bull fertility traits. Successful controlling of gene expression is a highly choreographed process and exhibited at each point of the pathway from transcription to translation.
There is growing evidence suggested that circadian rhythms have an important role on maintaining homoeostasis including reproductive capacity (Kennaway et al. 2012). This coordination is directed by numerous conserved transcriptional regulator genes among animals that may range from fruit flies to human beings (Beaver et al. 2002). This is highlighted most apparently among the mutant mouse models, and mutations in the Bmal1 gene significantly contract male fertility in murine model (Alvarez et al. 2008). Certain studies outlined that circadian system may alter the testosterone production in human beings (Cooke et al. 1993; Bribiescas and Hill 2010; Teo et al. 2011). Sex steroid levels in serum may also be influenced by genetic variants within circadian rhythm genes (Chu et al. 2008). Circadian locomotor output cycles kaput (CLOCK) gene encodes a basic helix–loop–helix–PAS transcription factor that may affect both the perseverance and epoch of circadian rhythms. Zhang et al. (2012) and Hodzic et al. (2013) reported that genetic variability within the CLOCK gene might be associated with semen quality parameters in humans. It was earlier studied that RNAi-based targeting of murine CLOCK gene may affect the sperm fertilization ability by regulating the sperm acrosin activity (Jiang et al. 2008).
Clusterin (CLU) family genes encode a heterodimeric protein (Kapron et al. 1997), which was the first protein to be identified as a major secretary glycoprotein of ram Sertoli cells (Blaschuk et al. 1983). The CLU gene was shown to have clustering effect among wide range of biological macromolecules including lipids, proteins and immunoglobulins among various types of mammalian cells (Fritz et al. 1983). CLU proteins are identified among numerous biological secretions such as milk, urine, CSF, synovial fluid, plasma and seminal fluids (Jenne and Tschopp 1992). The CLU gene thought to be concerned with a variety of physiological activities for illustration of apoptosis and degenerative disease conditions (Leger et al. 1987; Silkensen et al. 1994), spermatogenesis (Sylvester et al. 1991) and heat shock (Clark and Griswold 1997; Kimura et al. 1997) .
Perusal of literatures revealed that so far, no studies have been conducted for identifying the role of CLOCK and CLU genes on bull semen quality traits. Considering the fact, the aim of the present investigation was to analyse the differential expression profile of the CLOCK and CLU genes between motile and motility-impaired bull semen.
Materials and Methods
Fresh semen samples were taken from categorized crossbred Frieswal (Holestein-Friesan x Sahiwal) bulls according to their seminal quality traits, viz. semen volume (ml), progressive sperm motility in percentage (>50% considered as good and <50% as motility impaired) and post-thaw motility (PTM) in percentage. Semen was collected using artificial vagina (42–45°C) at early morning from each bull. Immediately after collection, the ejaculates were stored at 34°C in a water bath to evaluate the fresh semen quality traits. The fresh semen was then diluted with glycerol–egg yolk–citrate–Tris buffer, processed and cryopreserved. After storage in liquid nitrogen for 1–2 days, two straws were randomly obtained from each ejaculate and thawed at 37°C for 60 s and immediately evaluated for post-thaw motility (%) with phase contrast microscope. Ten extremely good and ten extremely poor bulls based on semen quality parameters were utilized for this study. The main criterion for classification was low sperm progressive motility supplemented with hypo-osmotic swelling response. The concentration of sperm was assessed using photometer readings (Accucell, IMV, France). Membrane integrity of spermatozoa was performed as per the procedure described earlier (Ganguly et al. 2013).
To eliminate damaged spermatozoa and contaminating somatic cells, the semen samples were purified on a discontinuous gradient of Percoll 45 : 90 (Sigma-Aldrich Co., St. Louis, MO, USA). The Percoll solution was diluted in Tyrode's sperm medium (Sp-TALP; 100 mm NaCl, 3.1 mm KCl, 25 mm NaHCO3, 0.3 mm NaH2PO4, 21.6 mm Na lactate, 2.0 mm CaCl2, 0.4 mm MgCl2, 10 mm Hepes, 1 mm pyruvate and 50 mg/ml gentamycin). Briefly, samples were deposited on the Percoll gradient and centrifuged at 700 g for 30 min. The pellets were washed and centrifuged twice at 250 g for 5 min in Sp-TALP solution. The pellets containing the motile spermatozoa were kept at −80°C in RNA Later (Ambion, Austin, TX, USA) until RNA extraction. The motile spermatozoa were kept at −80°C in RNA Later (Ambion) until RNA extraction protocol. Extraction of total RNA from bovine spermatozoa was carried out using TRI Reagent (Sigma-Aldrich). The protocol was followed according to the manufacturer's recommendation with little modifications. For extraction, the TRI Reagent was heated at 60°C and the samples were incubated for half an hour to completely dissociate the membranes. Equal number of spermatozoa (100 million) was utilized for RNA isolation from each sample. The subsequent steps of the protocol were performed as recommended by the manufacturer. Total RNA sample (aqueous phase) was then passed through a RNA extraction column (GenElute Binding column, Sigma-Aldrich Co.) upon which an RNase-free, DNase I treatment (Ambion) was performed to eliminate contaminating genomic DNA. The quantity and purity of total RNA isolated from purified spermatozoa were measured using ND-1000 spectrophotometer (Nano-Drop; Fisher Thermo, Wilmington, DE, USA). Contamination of epithelial cells and germ cell in spermatozoal RNA was verified by RT-PCR using primers specific to molecular markers such as CDH1 and KIT, respectively (Table 1), where buffalo testicular cDNA was used as positive control. Again, primers targeting the CD4 antigen were used to evaluate the leucocyte contamination (Table 1). The positive control used here was the total RNA extracted from a blood sample using Histopaque 1077 and TRI Reagent (Sigma-Aldrich) as per the manufacture's instruction.
| Gene | Primer sequences | Annealing temperature |
|---|---|---|
| CLOCK |
F-5′GACCCAAAGGAACCATCTACTT-3′ R-5′CTGTGAGTGCGTTGTATTGTTC-3′ |
55°C |
| CLU |
F-5′GGAGAGTGGCTGGGCAATC-3′ R-5′TCACCTCCTTGAGGGCATTT-3′ |
55°C |
| PRM1 |
F-5′AGATACCGATGCTGCCTCAC3′ R-5′GTGGCATGTTCAAGATGTGG3′ |
55°C |
| Βeta-actin |
F-5′AGTTCGCCATGGATGATGA3′ R-5′TGCCGGAGCCGTTGT3′ |
55°C |
| PPIA |
F-5′ATGCTGGCCCCAACACAA3′ R-5′CCCTCTTTCACCTTGCCAAA3′ |
55°C |
| DAZL |
F -5′CAC CAG CCA AGG CTA TGT TT 3′ R 5′ CAC CAG TTC GAT CCG TGA TT 3′ |
54°C |
| CDH1 |
F- 5′CCG TGA GAG TTT TCC CAC AT3′ R -5′CAT TGG TGA CTG GGT CTG TG3′ |
55°C |
| v-kit |
F -5′GAC CTG GAG GAC TTG CTG AG3′ R- 5′AGG GGC TGC TTC CTA AAG AG3′ |
53°C |
| CD4 |
F -5′CAA TGG CAA AGT CCT GTT GG3′ R- 5′GAT CTG AGA CAT CCG TTC TGC3′ |
55°C |
Total RNA isolated from crossbred bull spermatozoa was reverse-transcribed to complementary DNA using random hexameric primers and M-MuLV reverse transcriptase (Protoscript, NEB). The cDNA product was stored at −20°C until use. Genomic DNA contamination was tested by polymerase chain reaction, using bovine PRM1 gene-specific intron-spanning primer. Primer sets used in this study are shown in Table 1. Genomic DNA isolated from blood by GenElute™ Blood Genomic DNA Kit (Sigma-Aldrich) was used as a positive control. cDNA samples diluted in the ratio of 1 : 10 were used to elucidate the differential expression of CLOCK and CLU genes among categorized bull semen. The expression profile of mRNAs was quantified by real-time PCR (Step One, Applied Biosystems, Foster City, CA, USA) using SYBR Green chemistry. Two housekeeping genes, peptidyl prolyl isomerase A (PPIA) and beta-actin, were used as an endogenous control. All the PCRs were accomplished in 48-well optical reaction plates in triplicate. The PCR was achieved using the SYBR Green Universal PCR Master Mix with ROX dye, which contains thermostable Taq DNA polymerase enzyme (Ampli-TaqGold) and all other combinations essential for carrying out the reaction. Following the manufacturer's instruction, a final volume of 10 μl reaction mixture contains 2 μl of template (50 ng/μl), 5 μl 2X SYBR Green Master Mix, 0.5 μl Primer (10 pMol) and 2 μl DNase-/RNase-free sterile water. All PCRs were executed in triplicate for accuracy, and the mean mRNA value was then estimated. For polymerase chain reaction, samples were first activated at 95°C for 10 min; amplification was performed for 40 cycles at 95°C for 15 s and 55°C for 60 s. Samples were then quantified by the ΔΔCt method (Livak and Schmittgen 2001). The expression values attained were normalized against the two housekeeping genes, viz. PPIA and beta-actin, permitting the comparison of samples self-sufficiently of the amount of total input cDNA. Data are presented as mean ± SEM and evaluated using SPSS statistical program (SPSS 10.0 for Windows; SPSS, Inc., Chicago, IL, USA). Gene expression pattern between good and poor quality semen producers was compared using Student's t-test.
Results
Bulls categorized into good and poor (motility-impaired) quality producer of semen were based upon the semen volume (ml), concentration (million/ml), initial progressive motility (%) and post-thaw motility (%) as presented in Table 2. Results demonstrated that all the parameters were significantly (p < 0.01) different between the groups.
| Classified bulls | Volume (ml) | Sperm concentration (million/ml) | Initial progressive motility (%) | Post-thaw motility (%) |
|---|---|---|---|---|
| Good (n = 10) | 4.93 ± 0.48a | 1136.28 ± 86.47c | 54.94 ± 1.19e | 37.06 ± 2.32g |
| Poor (n = 10) | 2.56 ± 0.32b | 686.64 ± 80.21d | 28.07 ± 2.57f | 16.80 ± 1.43h |
| Total (n = 20) | 3.69 ± 0.38 | 911.46 ± 77.17 | 41.51 ± 3.37 | 26.93 ± 2.67 |
- Mean bearing different superscripts (a, b, c, d, f, g, h) differs significantly at p < 0.01.
In the presence of genomic DNA, the intron-spanning primers produced an amplicon of 573 bp and 334 bp for DAZL and PRM1 genes, respectively (not shown). Sperm cDNA samples without genomic DNA contamination were used for further downstream works.
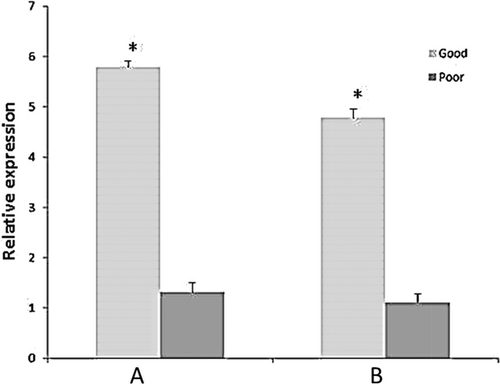
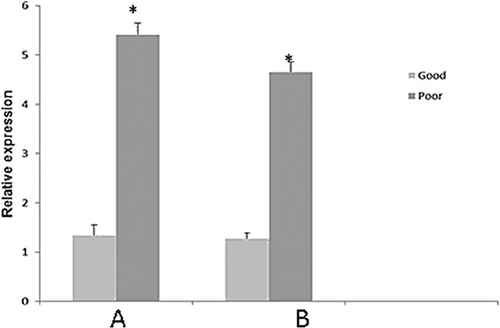
The relative expression of CLOCK was significantly (p < 0.05) higher among good quality bull semen than motility-impaired ones (Fig. 1). However, expression of CLU gene was significantly (p < 0.05) higher among motility-impaired bull semen than good quality motile spermatozoa (Fig. 2).
Discussion
Selection of high-fertility bulls is a prerequisite to cost-effective production of frozen semen. In the present investigation, we found that bovine CLOCK and CLU genes were differentially expressed among progressive and impaired motile bull spermatozoa, deserving the further confirmation as well as mechanistic examinations in other livestock.
Quality assessment of the isolated sperm RNA samples was tested by RT-PCR. The DAZL and PRM1 genes were targeted to perceive the presence of genomic DNA (gDNA) contamination. Spermatozoa contain on average of 0.015 pg of mRNA per cell (Miller et al. 2005); thus, contamination from any somatic cell type was checked prior to the experiment. The absence of impurity by somatic and germ cells in extracted sperm RNA was always confirmed by visual inspection of the semen fractions. Besides, the absence of contamination of germ cells, epithelial cells and leucocytes in all the RNA extractions was confirmed by RT-PCR by targeting specific molecular markers such as KIT, CDH1 and CD4, respectively. Contamination-free RNA sample was chosen for further processing. The similar strategy was also followed by Ganguly et al. (2013).
The circadian clock is an internal rhythmic system that offers adaptation during environmental deviations (El-Helaly et al. 2010; Ortiz et al. 2010). In addition to the direct effect of circadian system, it also indirectly impacts on many physiological processes including the functional attribution of male reproductive process. Genetic variability in the CLOCK gene was reported to be connected with obesity (Sookoian et al. 2008) and male infertility (Hodzic et al. 2013) traits. In the present study, for the first time we investigated the transcript abundance of CLOCK gene between good quality and motility-impaired bull semen. Our results demonstrated that CLOCK gene expression was significantly (p < 0.05) higher among good quality semen than the motility-impaired ones. Earlier reports suggested that CLOCK genes of moth and flies were associated with sperm release (Gvakharia et al. 2000). Giebultowicz (2001) reported that CLOCK genes are also expressed in the testes of mouse, rat, zebra fish and humans, suggesting a likelihood that circadian clocks may be intricate in fecundity across animal phyla. The expression of CLOCK gene among good quality bull semen may be due to the effect of sperm acrosin activity, which is earlier known to be regulated by CLOCK gene (Jiang et al. 2008). As circadian system influences many biological processes including release of testosterone (Alvarez et al. 2008), further studies need to be conducted in bulls to correlate the expressivity of CLOCK gene with hormonal profiles of the animals, particularly testosterone and LH that influence the process of spermatogenesis.
Clusterins are one of the seminal proteins that have been implicated in many activities such as apoptosis inhibition, cell cycle regulation, DNA repair mechanism and sperm maturation (Jones and Jomary 2002). In Salehi et al. (2013) study, the relationship between human secretory clusterin (sCLU) in seminal plasma with sperm parameters, protamine deficiency and DNA fragmentation was investigated. Their findings revealed that sCLU negatively correlated with protamine deficiency, sperm DNA fragmentation and abnormal spermatozoal morphology. Jonathan et al. (2007) reported that CLU gene expressions in the bovine testes are essential for the functional activity of sertoli cells. The present findings revealed that expression of CLU was gene significantly (p < 0.05) higher among motility-impaired bull semen than good quality motile spermatozoa. Han et al. (2012) have raised the possibility that the CLU protein found in spermatozoa could be in a different form from the sCLU found in seminal plasma and could have different functions. They identified a form of clusterin in normal human spermatozoa, which exhibited molecular weight, heterodimeric structure, as well as the hydrophobic binding activity similar to that of sCLU in semen and on the surfaces of abnormal spermatozoa. Clusterin in the mammalian reproductive tract is not always involved in decreased sperm motility, sperm aggregation or infertility as it has been stated that clusterin can defend bull spermatozoa against oxidative damage and then enhance sperm motility (Reyes-Moreno et al. 2002). Therefore, the exact function of clusterin in bovine spermatozoa has yet to be established. Further study on clusterin in normal bovine spermatozoa may aid to expose the innovative purposes of clusterin during spermatogenesis as well as the maturation of spermatozoa.
Although our preliminary reports may provide an insight for understanding the relations between investigated genes and their influence on bull sperm quality, further validation of this experiment at functional level would be required before drawing a final conclusion. Whether the differences in expression of investigated genes were connected with genotype of bulls or related to their physiological status needs to be address?
Acknowledgement
We acknowledge the World Bank-funded National Agricultural Innovation Project, ICAR, New Delhi, for providing financial support. We are also thankful to the Director, CIRC, Meerut, for providing necessary facilities.
Conflict of interest
None of the authors have any conflict of interest to declare.
Author contributions
Sushil Kumar, Rajib Deb, Umesh Singh and Indrajit Ganguly designed and drafted the manuscript. D.K. Mandal, Shrikant Tyagi and Mahesh Kumar analysed the data. Gyanendra Sengar, Sheetal Sharma, Rani Singh and Rupali Singh involved in wet laboratory work.




