The subcellular localisation of early photosystem II assembly in Synechocystis sp. PCC 6803
Abstract
The spatial organization of the de novo assembly of photosystem II (PSII) in cyanobacteria, particularly its subcellular starting point, is a matter of ongoing scientific debate. Here, we use fluorescence microscopy and biochemical experiments to demonstrate that the most likely marker of early PSII biogenesis, i.e., the precursor of the D1 protein of the PSII reaction centre, accumulates primarily at thylakoid convergence zones close to the periphery of the cell.
1 MAIN
In recent years, great strides have been made in elucidating the biogenesis of thylakoidal photosynthetic complexes, particularly photosystem II (PSII), at the molecular level (Komenda et al., 2024). The available evidence points to a conserved sequence of events in which various assembly factors enable the construction of the required sub-modules, whose structures are currently being resolved at close to atomic resolution (Xiao et al., 2021; Zabret et al., 2021; Zhao et al., 2023). However, much less is known about the spatial organization of this assembly process in photosynthetic cells and organelles. Basically, the initial steps of biogenesis involve the synthesis of photosynthetic subunits and their co-translational insertion into the thylakoid membrane bilayer. In the case of the cyanobacterial model organism Synechocystis sp. PCC 6803, two apparently contradictory models have been proposed for the initial steps in PSII de novo assembly. Based on the localization of the PSII-related assembly factor and manganese binding protein PratA, thylakoid convergence zones (TCZs), where thylakoid membranes fuse and converge upon the plasma membrane at the cell periphery, have been identified as potential sites for PSII initiation (Stengel et al., 2012). These regions may facilitate the efficient transfer of Mn ions to PSII, the primary Mn sink in the cell, a notion supported by the presence of the Mn-binding protein PratA within TCZs (Stengel et al., 2012) and the punctate localization of the Mn transporter Mnx at the Synechocystis cell periphery (Brandenburg et al., 2017). Furthermore, a dedicated biogenic sub-compartment for PSII assembly could accelerate the assembly process via substrate channelling while preventing the premature interaction of active and immature photosynthetic complexes, thereby reducing the risk of PSII damage. Additional evidence for this model comes from studies on morphological mutants affecting the thylakoid-shaping factors CurT and AncM, which show disrupted TCZs and defects specifically in PSII - but not PSI - assembly (Heinz et al., 2016; Ostermeier et al., 2022). On the other hand, recent work by Mahbub et al. (2020) has localised the psbA mRNA that encodes the D1 protein of PSII close to the inner layer of thylakoid sheets in the central cytoplasm of Synechocystis. These authors therefore suggest that the mRNA is anchored in this region by RNA-binding proteins and is subsequently translated (Mahbub et al., 2020). Both proposals are consistent with the accumulation of thylakoid-associated ribosomes in the respective regions, as revealed by in situ cryo-electron tomography (Rast et al., 2019).
To resolve the discrepancy between these two models, we have directly traced the early steps of PSII assembly in Synechocystis by specifically localizing the most probable and informative marker for early PSII assembly, i.e., the short-lived precursor of the D1 protein (pD1). In almost all phototrophic organisms, D1 proteins are synthesized as precursors with short C-terminal extensions, which are obligatorily removed by the endoprotease CtpA during the early phase of PSII assembly (Ivleva et al., 2000). It is worth mentioning that despite its evident protease activity, many aspects of how CtpA functions, its exact localization, as well as its interactions with other factors, remain unclear (Inagaki, 2022). In Synechocystis, the pD1 extension consists of 16 amino acids, which are cleaved off in a two-step process from all three homologous pD1 proteins encoded by the genes psbA1-3 (Jansson et al., 1987; Komenda et al., 2007). While psbA1 is exclusively expressed under low-oxygen conditions, psbA2 and psbA3 are responsible for D1 accumulation under normal physiological and most stress conditions, with pD1(2) accounting for 90% of these activities (Mohamed et al., 1993; Summerfield et al., 2008).
The C-termini of either pD1(2) or pD1(3) were tagged with phiYFP by introducing its coding region into the 3´regions of either psbA2 or psbA3 genes, respectively. Non-tagged psbA copies were eliminated in the psbA2-YFP strain via insertional mutation or by introducing the psbA3-YFP gene into the psbA triple mutant TD41 (Figure S1a; Nixon et al., 1992). Expression of both pD1-YFP fusion proteins was verified by western blot analysis using a YFP antibody, and no significant break-down products were detected (Figure S2a and b). Strikingly, each of these strains exhibited a wild-type-like growth phenotype and accumulated wild-type-like levels of the mature D1 protein (Figure 1; Figure S2c). These data indicate that (1) each of these psbA genes can fully complement the loss of the other (Mohamed et al., 1993), and (2) the relatively bulky YFP extensions at the pD1 C-termini do not affect their correct C-terminal processing. Furthermore, no C-terminal processing product, i.e., “free” YFP, accumulates in these cells. When the subcellular distributions of pD1(2)-YFP and pD1(3)-YFP were monitored, punctate fluorescence signals were seen in regions at or near the periphery of the cells in both cases (Figure 2a). Earlier findings based on immunogold-treated ultrathin sections of Synechocystis had already detected clustering of pD1 close to the cellular periphery (Stengel et al., 2012). These patterns also strongly resemble the distribution of AncM, which marks attachment sites of thylakoids to the plasma membrane (thylapses) at TCZs (Ostermeier et al., 2022). Indeed, immunofluorescence experiments using an αAncM antibody revealed that both proteins co-localise with AncM in psbA2-YFP and psbA3-YFP strains (Figure 2b; Figure S3). In agreement with the localization in TCZs, pD1(2)-YFP was detected in lighter PDM (PratA-defined-membrane) fractions after membrane fractionation on sucrose gradients (Figure 2c and d; Schottkowski et al., 2009). PDMs have previously been shown to be enriched in the proteins PratA, YidC, YCF48, CurT, pD1 and AncM, suggesting that they represent a biogenic sub-fraction of thylakoids (Rengstl et al., 2011; Heinz et al., 2016; Ostermeier et al., 2022; Ostermeier et al., 2024). To test whether PDMs indeed harbor PSII de novo assembly intermediates 2D BN/SDS PAGE of each PDM and TM fraction from wild-type cells was performed. Subsequently, Western blot analyses revealed the distribution of D1-containing PSII complexes across the sucrose gradient (Figure 3). The data shows that PDM fractions accumulate some RC47 and RCC1 PSII intermediates but particularly the non-assembled pD1 precursor. In TM fractions, later assembly intermediates, as well as PSII monomers and dimers, predominate (Figure 3). Interestingly, in TM fraction 10/11 also a pD1 signal (roughly 15% of total pD1 signal) was detected parallel to RCC1 and RC47, consistent with pD1-YFP accumulation in the same fraction (Figures 2c and 3). These findings suggest that some PSII assembly but - due to the simultaneous appearance of later assembly factors and the absence of PratA (Eisenhut, 2019) - more likely repair takes place in TMs.
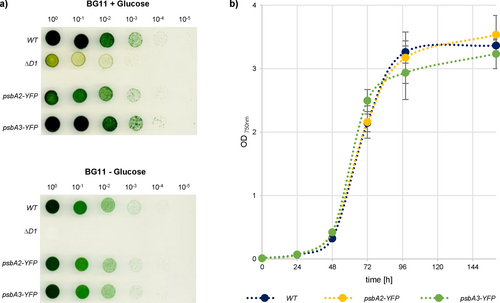
a) Dilution series of WT, ΔD1 (TD41, Nixon et al., 1992), psbA2-YFP and psbA3-YFP strains with (+) and without (−) 5% glucose. Logarithmically growing cells were set to OD750 = 1, diluted five times and plated.
b) Growth curves of WT, psbA2-YFP, and psbA3-YFP in liquid medium (n = three biological replicates).
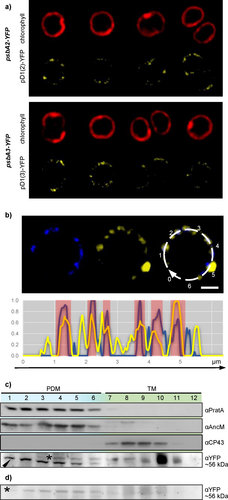
a) Live-cell fluorescence micrographs of psbA2-YFP and psbA3-YFP.
b) Immunofluorescence of psbA2-YFP with an AlexaFluor405-labeled αAncM antibody. AncM (blue) is on the left, pD1(2)-YFP (yellow) in the middle and the merged signals are on the right. Bar = 1 μm. The plot at the bottom shows an intensity blot of pD1(2) (yellow) and AncM (blue), which follows the dotted white line of the merged micrograph on the right of the upper panel. Strong overlaps are highlighted in red.
c) Membrane fractionations of psbA2-YFP according to Rengstl et al. (2011).The fractions were subjected to SDS-PAGE and immunoblotting, using antibodies against PratA, AncM, CP43 or YFP. Fractions 1–6 correspond to PDMs (light blue), whereas fractions 7–12 represent thylakoid membranes (green). The black arrowhead marks the αYFP signal, the asterisk marks a cross-reaction of the αYFP antibody (Suppl. Figure 2c).
d) Negative control in the WT background for membrane fractionation (c). Again cross-reactive material is marked by an asterisk.
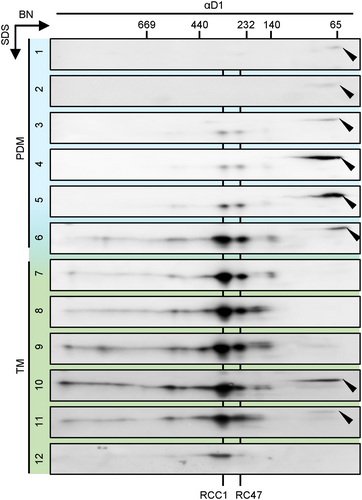
Each membrane fraction (1 to 12) of WT was separately subjected to BN/SDS–PAGE analysis and probed with a D1 antibody. Black arrowheads indicate pD1. Fractions 1–6 correspond to PDMs (light blue), whereas fractions 7–12 represent thylakoid membranes (green). RCCI, PSII monomer; RC47, reaction center complex lacking CP43. Identical volumes of each fraction corresponding to protein in fraction 7 were loaded. The exposure time was equivalent to 120 s.
Based on these findings, a refined model for PSII biogenesis is proposed in Figure 4. After transcription, mRNAs that code for PSII subunits are targeted to the surface of the innermost thylakoid sheet by RNA-binding proteins (Mahbub et al., 2020). Polypeptides are co-translationally inserted into the membrane and subsequently form mobile assembly modules, as recently outlined in Eckardt et al. (2024). pD1, in particular, is synthesized at, and/or is transferred to TCZs, where uptake of manganese ions is likely to occur, and very early steps of PSII de novo assembly, especially C-terminal pD1 processing, primarily takes place (Stengel et al., 2012; Eisenhut, 2019).
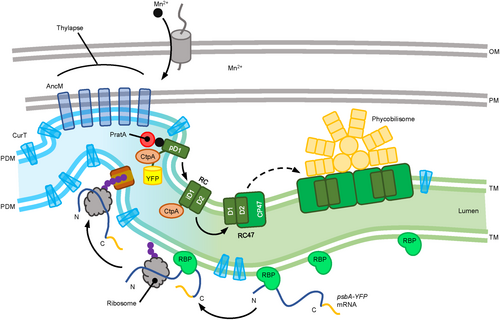
The thylakoid membrane is divided into two regions. The light-blue region represents biogenic PDMs, and the green region represents photosynthetically active TMs. Outer and plasma membranes (OM and PM) are depicted in grey. CurT is distributed throughout the membranes, whereas AncM forms distinct spots anchoring the thylapse to the PM (Ostermeier et al. 2022). The psbA mRNA is captured and transported by RBPs, as described by Mahbub et al. (2020). In the thylapse region, the mRNA is translated into pD1, followed by C-terminal cleavage by CtpA (Based on the predicted orientation of the D1 C-terminus within the lumen, however, direct experimental confirmation of this topology is still lacking.). This is followed by the assembly of RC, RC47, and ultimately the PSII dimer with attached phycobilisomes proceeds.
2 METHODS
Synechocystis 6803 strains were grown autotrophically on solid or liquid BG11 medium (Rippka et al., 1979) at 30°C under continuous illumination at an irradiance of 30 μmol photons m−2 s−1 of white light. For heterotrophic growth conditions, the media were supplemented with 5 mM glucose. Cell density was monitored photometrically at 750 nm.
Generation of strains, protein analysis, membrane fractionation, live-cell fluorescence microscopy and immunofluorescence were performed as described previously (Ostermeier et al., 2022). For BN/SDS-PAGE (Ostermeier et al., 2022), each membrane fraction (1 to 12) was first separated on a native gel, excised, and subsequently run on individual SDS gels. All 12 gels were then blotted, and signals were detected using a D1 antibody.
Gene fragments were ordered as gBlocks® gene fragments (Integrated DNA Technologies), cloned into the pJET1.2 vector (Thermo Fisher Scientific) and transformed into WT Synechocystis or TD41, respectively (Figure S1; Table S1).
AUTHOR CONTRIBUTIONS
M.O., S.H., and J.N. designed the research; M.O., I.B. and S.H. performed research; M.O., S.H. and J.N. analyzed data; M.O. and J.N. wrote the paper.
ACKNOWLEDGEMENTS
We thank Paul Hardy for critical reading of the manuscript. This work was funded by the Deutsche Forschungsgemeinschaft (Ni390/13-1).




