Ubiquitin-specific protease 8 is a novel prognostic marker in early-stage lung adenocarcinoma
Abstract
Alterations of epidermal growth factor receptor (EGFR) expression frequently occur in early-stage lung adenocarcinoma. Ubiquitin-specific protease 8 (USP8) has been reported to stabilize EGFR protein at the plasma membrane through the recycling pathway. Here, we examined the correlation between USP8 expression and the expression or mutation status of EGFR, as well as the clinicopathological features of lung adenocarcinoma and patient outcome. Expression of EGFR and USP8 in surgically resected specimens of lung adenocarcinoma (82 cases) was examined by immunohistochemistry. Overexpression of EGFR was mutually correlated with that of USP8, and was also associated with clinicopathological features including pathological subtype, lymphatic permeation, and vascular invasion. Moreover, patients who had USP8-positive tumors had a significantly poorer outcome than those who were USP8-negative, not only overall but also patients who were EGFR-negative. Although EGFR was expressed in invasive adenocarcinoma but not in adenocarcinoma in situ (AIS), USP8 was overexpressed in not only invasive adenocarcinoma but also 38.1% of AIS cases. In vitro, USP8 regulated the expression and half-life of EGFR in immortalized AIS cells, and also cell proliferation. Our findings indicate that overexpression of USP8 in lung adenocarcinoma is an early event during the course of tumor progression, and is related to EGFR expression.
Mortality due to lung cancer has been increasing rapidly worldwide.1 Non-small cell lung cancer (NSCLC) accounts for about 80–85% of all lung cancers, the most common histological subtype being adenocarcinoma. The Noguchi classification of small lung adenocarcinomas (2 cm in diameter or less) is correlated with the postoperative 5-year survival rate.2 Types A and B in the Noguchi classification (adenocarcinoma in situ, AIS) have an extremely favorable outcome with a 5-year survival rate of 100%, and show stepwise progression to type C (early but invasive adenocarcinoma), which has a relatively poor outcome.2, 3 At the advanced stage, lung adenocarcinoma harbors multiple genetic abnormalities,4, 5 but interestingly, the mutation, amplification, and protein overexpression of epidermal growth factor receptor (EGFR) are often observed from the early stage. For complete cure, diagnosis and initiation of treatment at an early stage are essential. In this context, targeting of EGFR abnormality is thought to be a promising therapeutic strategy for lung adenocarcinoma.
Somatic mutation of EGFR is the most common driver mutation, and is particularly common in NSCLC patients. The most prominent mutations in EGFR occur in exons 18–21 of the tyrosine kinase domain, and patients harboring such mutations are responsive to treatment with tyrosine kinase inhibitors (TKIs) such as gifitinib and erlotinib.6 Although initially these treatments elicit a rapid antitumor effect, patients develop resistance to TKIs after a median of 10–16 months of drug administration.7, 8 Approximately 72–90% of non-Asian NSCLC patients who undergo mutation analysis have no detectable EGFR mutation, and show a lower response to TKIs. Recent studies have shown that as well as EGFR mutation status, a high copy number or expression of wild-type EGFR is also associated with tumor progression and patient survival.9, 10 However, no prognostic marker gene has yet emerged for lung adenocarcinoma patients with wild-type EGFR or low EGFR expression.
In addition to a high EGFR gene copy number and mutation, ligand-dependent activation as well as recycling back to the plasma membrane via the endocytosis-related pathway has been reported to play an important role in the early stage of lung cancer.11 Ubiquitin-specific protease 8 (USP8) is known to stabilize the EGFR protein at the plasma membrane through cleavage of poly-ubiquitin from EGFR, a process known as deubiquitination, which is reversible by ubiquitination and can lead to lysosomal degradation.
USP8 belongs to a ubiquitin-specific family of deubiquitination proteases (DUB) and is involved in endocytosis at endosomes.12 USP8 has an important physiological function in cell growth,13 and deletion of USP8 causes embryonic lethality in mice,14 similarly to deletion of EGFR.15 However, the relationship of USP8 to the expression or mutation status of EGFR in lung adenocarcinoma is still poorly understood.
Here, we demonstrated that USP8 is correlated with the expression or mutation status of EGFR, as well as with the clinicopathological features of lung adenocarcinoma. USP8 showed overexpression in the early stage of lung adenocarcinoma and was significantly associated with shorter disease-free survival in patients overall, and also in those who were negative for EGFR expression. These findings suggest that USP8 might be a novel diagnostic and therapeutic target in early-stage lung adenocarcinoma.
MATERIALS AND METHODS
Sample collection
Specimens of lung adenocarcinomas that had been surgically resected at the University of Tsukuba Hospital (Ibaraki, Japan) between 1999 and 2014 were used for immunohistochemistry (IHC). We randomly collected 82 cases in which EGFR mutation had already been analyzed in order to validate chemotherapeutic options (LSI Medience Corporation, Tokyo, Japan). Follow-up information for all of the corresponding patients was obtainable from the medical records, and all of the patients provided informed consent for use of their materials. The study was approved by the Institutional Ethics Review Committee and the lung adenocarcinoma cases were classified according to the UICC TNM classification of malignant tumors (seventh edition) and the World Health Organization (WHO) classification of malignant tumors (fourth edition).16, 17
Immunohistochemistry (IHC)
Sections 4 µm thick were cut from formalin-fixed paraffin-embedded (FFPE) tissue blocks. The sections were deparaffinized and rehydrated, followed by blocking of endogenous peroxidase using 3% H2O2 for 30 min. Subsequently, antigen retrieval was performed using an autoclave with 10 mM Tris-EDTA buffer (pH 9.0) at 105°C for 10 min. Immunostaining was performed using a Dako Autostainer Link 48 (Agilent Technologies, Santa Clara, CA, USA) with the appropriate primary antibody and REAL Envision HRP rabbit/mouse (Agilent Technologies) as a secondary antibody. The immunoreactivity was detected with DAB (Dako REAL Envision Detection System; Agilent Technologies), and counterstaining was performed with hematoxylin for 1 min. Evaluation of USP8 and EGFR expression was based on the intensity of cytoplasmic staining. The staining was judged to be positive when the cytoplasm of the tumor cells was stained more strongly than that of the alveolar epithelium. Rabbit polyclonal anti-USP8 antibody (Bethyl Laboratories, Montgomery, TX, USA) and mouse monoclonal anti-EGFR antibody (Agilent Technologies, Clone DAK-H1-WT) were used as the primary antibodies. The evaluation of immunoreactivity was used two-tier grading as negative with non-stained and positive with diffusely positive.
Cell culture and conditions
The PL16T cell line was established in our laboratory from a surgically resected AIS of the lung.18 PL16T was maintained in MCDB153HAA (Wako, Osaka, Japan) supplemented with 2% FBS (Sigma-Aldrich, St. Louis, MO, USA), 0.5 ng/mL human EGF (Toyobo, Tokyo, Japan), 5 µg/mL human insulin (Wako), 72 ng/mL hydrocortisone (Wako), 40 µg/mL human transferrin (Sigma-Aldrich), and 20 ng/mL sodium selenate (Sigma-Aldrich). The cells were cultured in a 5% CO2 incubator at 37°C and passaged every 3–4 days.
Plasmid and siRNA transfection
Flag-USP8 plasmid was purchased from Addgene (Cambridge, MA, USA). The day before transfection, PL16T cells were plated to obtain 80% confluence on the day of transfection. Fugene HD (Promega, Madison, WI, USA) was used for plasmid transfection. USP8-specific siRNA (forward, GGACAACCAGAAAGUGGAAUUCUAA and reverse, UUAGAAUUCCACUUUCUGGUUGUCC) from Thermo Fisher Scientific (Waltham, MA, USA) and lipofectamine RNAiMAX (Thermo Fisher Scientific), were used for siRNA transfection. The final siRNA concentration used for PL16T cells was 5 nM. Transfections were performed in accordance with the manufacturer's protocol. The cells were incubated at 37°C in a 5% CO2 incubator for 24 or 48 h and then further analyzed.
Quantitative real-time PCR analysis
To confirm the transfection efficiency of the Flag-USP8 plasmid or siUSP8, PL16T cells were evaluated using quantitative real-time RT-PCR. Total RNA was extracted from Flag-USP8 plasmid- or siUSP8-transfected PL16T cells using an RNeasy Mini Plus Kit (QIAGEN, Hilden, Germany) and the quality was evaluated using an Agilent 2100 Bioanalyzer (Thermo Fisher Scientific). One microgram of total RNA per 20 µl of the reaction mixture was converted to cDNA using a High Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative real-time PCR was performed with SYBR Premix Ex TaqTM (Perfect Real Time; Takara Bio, Shiga, Japan) on a GeneAmp 7300 Sequence Detection System (Thermo Fisher Scientific) in accordance with the manufacturer's protocol.
Western blot analysis
Total protein from the cells was prepared on ice using Mammalian Protein Extraction Reagent (M-PER; Thermo Fisher Scientific) containing a Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). The total protein in the lysates was measured using a BCA protein assay kit (Thermo Fisher Scientific). Total protein aliquots (20 µg) were mixed with 5× sample loading buffer supplemented with DTT, denatured at 95°C for 5 min, and electrophoresed on 10% Mini-PROTEAN TGX Precast Gels (Bio-Rad Laboratories, Hercules, CA, USA). Proteins were transferred to polyvinylidene difluoride membranes using an iBlot gel transfer system (Thermo Fisher Scientific). The blots were then blocked and probed with various antibodies obtained from the following commercial sources: USP8 from Cell Signaling Technology (Danvers, MA, USA); EGFR from Medical & Biological Laboratories (Aichi, Japan); Flag and β-actin from Sigma-Aldrich. After extensive washing, immunoreactivity was detected with specific secondary antibodies conjugated to horseradish peroxidase (Thermo Fisher Scientific). Protein bands were visualized using SuperSignal West Femto Maximum sensitivity substrate (Thermo Fisher Scientific) and images were captured on a ChemiDoc Touch Imaging System (Bio-Rad Laboratories).
Immunofluorescence
PL16T cells were plated on collagen-coated cover slips (Iwaki Biosciences, Tokyo, Japan) and fixed with 10% neutral buffered formalin. They were then incubated with anti-EGFR conjugated with Alexa Fluor 488 antibody (Cell Signaling Technology) for 1 h at room temperature, and analyzed using a fluorescence microscope (Biorevo BZ-9000; Keyence, Osaka, Japan).
Pulse chase assay
Pulse-chase assay was performed followed by the protocol reported previously with some modification.19 After transfection with siUSP8 for 48 h, the cells were washed with PBS and incubated with prewarmed DMEM medium without Met/Cys for 30 min at 37°C in a 5% CO2 incubator. The cells were labeled with [35S]-Met/Cys (10 µCi/mL) as the pulse radioisotope in DMEM medium without Met/Cys for 30 min at 37°C in a 5% CO2 incubator. For chasing of the labeled protein, the isotope-labeled cells were washed 3 times with culture medium and incubated with the culture medium for 0, 2, 5, and 10 h. After chasing, total protein was extracted from the cells using IP Lysis Buffer (Thermo Fisher Scientific) containing a Halt protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). The labeled proteins were isolated from other cellular proteins by immunoprecipitation with EGFR antibody and subjected to Western blot analysis. For quantitative determination of the proteins, the membrane containing the metabolically labeled EGFR was subjected to β–ray scanning using a Typhoon FLA7000 (GE Healthcare, Chicago, IL, USA) image analysis system.
Proliferation assay
For analysis of cellular proliferation activity, a Cell Counting Kit-8 (WST-8) (Dojindo Laboratories, Kumamoto, Japan) was used in accordance with the manufacturer's protocol after plasmid or siRNA transfection.
Statistical analysis
Group results are expressed as mean ± SD. Data were compared between groups using the t test for 2-tailed distributions and the paired t test. Differences at P *<0.05, **<0.01, and ***<0.001 were considered significant. SPSS 22 statistical software (SPSS, Chicago, IL, USA) was used for IHC data analysis as follows. Correlations of clinicopathological features with the expression and mutation status of EGFR or expression of USP8 were analyzed using the chi-squared test. Disease-free survival was examined using the Kaplan-Meier method, and the significance of differences between survival curves was evaluated using log-rank test. Univariate and multivariate analysis was conducted using the Cox proportional hazards model.
RESULTS
Overexpression of EGFR and correlation with clinicopathological features
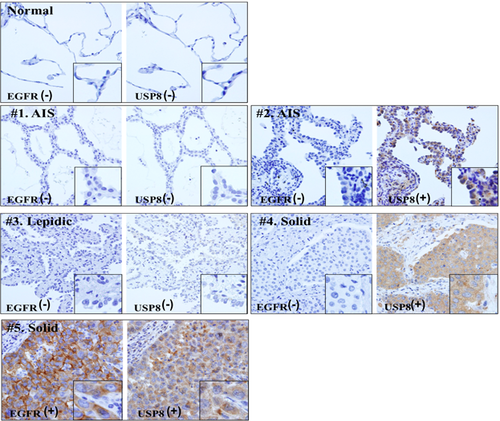
We examined EGFR expression in both normal lung tissue and tumor tissue (Fig. S1a, b). EGFR expression in tumor tissue was higher than that in normal tissue, and staining was strong in the cytoplasm and on the cell membrane of tumor cells. EGFR expression was detected in 26.8% (22/82) of the cases and was significantly correlated with pathological subtype, pathological stage, lymphatic permeation, and vascular invasion (Table 1, left).
| EGFR Expression | EGFR mutation status | |||||||
|---|---|---|---|---|---|---|---|---|
| Clinicopathological features | Negative | Positive | Total patients | P-value | Wild-type | Mutant | Total patients | P-value |
| Age (years) | 0.285 | 0.684 | ||||||
| ≤60 | 18 | 4 | 22 | 15 | 7 | 22 | ||
| >60 | 42 | 18 | 60 | 38 | 22 | 60 | ||
| Gender | 0.465 | 0.002 | ||||||
| Female | 30 | 13 | 43 | 21 | 22 | 43 | ** | |
| Male | 30 | 9 | 39 | 32 | 7 | 39 | ||
| Noguchi classification | 0.066 | <0.001 | ||||||
| Type A | 8 | 0 | 8 | 8 | 0 | 8 | *** | |
| Type B | 12 | 1 | 13 | 13 | 0 | 13 | ||
| Type C' | 2 | 0 | 2 | 2 | 0 | 2 | ||
| Type C | 4 | 3 | 7 | 2 | 5 | 7 | ||
| Type D | 1 | 0 | 1 | 1 | 0 | 1 | ||
| Total | 27 | 4 | 31 | 26 | 5 | 31 | ||
| Pathological subtype | 0.021 | 0.001 | ||||||
| AIS | 20 | 1 | 21 | * | 21 | 0 | 21 | |
| MIA | 2 | 0 | 2 | 2 | 0 | 2 | ** | |
| Invasive adenocarcinoma | ||||||||
| Lepidic | 10 | 2 | 12 | 5 | 7 | 12 | ||
| Acinar | 9 | 9 | 18 | 6 | 12 | 18 | ||
| Papillary | 9 | 3 | 12 | 5 | 7 | 12 | ||
| Micropapillary | 1 | 0 | 1 | 1 | 0 | 1 | ||
| Solid | 7 | 7 | 14 | 11 | 3 | 14 | ||
| IMA | 2 | 0 | 2 | 2 | 0 | 2 | ||
| Pathological stage† | 0.001 | 0.001 | ||||||
| Stage I | 37 | 5 | 42 | 33 | 9 | 42 | ||
| Stage II | 11 | 5 | 16 | ** | 9 | 7 | 16 | ** |
| Stage III | 8 | 12 | 20 | 10 | 10 | 20 | ||
| Stage IV | 4 | 0 | 4 | 1 | 3 | 4 | ||
| Lymphatic permeation | 0.035 | 0.012 | ||||||
| Negative | 40 | 9 | 49 | * | 37 | 12 | 49 | * |
| Positive | 20 | 13 | 33 | 16 | 17 | 33 | ||
| Vascular invasion | 0.002 | 0.006 | ||||||
| Negative | 39 | 6 | 45 | ** | 35 | 10 | 45 | ** |
| Positive | 21 | 16 | 37 | 18 | 19 | 37 | ||
- † Stage I includes IA and IB, stage II includes IIA and IIB, stage III includes IIIA and IIIB. Correlation between expression of EGFR or mutation status and clinicopathological features was analyzed using chi-squared test.
- AIS, adenocarcinoma in situ; MIA, minimally invasive adenocarcinoma; IMA, invasive mucinous adenocarcinoma.
- P-Value* < 0.05, **< 0.01, and ***< 0.0001.
EGFR mutation status and correlation with clinicopathological features
Next, we investigated the mutation status of EGFR in the same cases. Similarly to previous reports, mutant EGFR containing the E746-A750 deletion in exon 19 and L858R in exon 21 was detected in 35.4% (29/82) of the cases and was significantly correlated with patient gender, the Noguchi classification, pathological subtype, pathological stage, lymphatic permeation, and vascular invasion (Table 1, right). The frequency of EGFR mutation was significantly higher in women (75.9%, 22/29) than in men. Acinar adenocarcinoma was the most common dominant histological subtype with mutant EGFR (12/29; 41.4% of all mutant cases, 12/18; 66.7% of cases with an acinar pattern). Moreover, EGFR mutation status was correlated with EGFR expression; mutation was detected in 63.6% (14/22) of cases that were EGFR-positive (Table S1).
Overexpression of USP8 and correlation with clinicopathological features
USP8 showed higher expression in tumor tissue than in normal lung tissue (Fig. S1c, d) and was stained mainly in the cytoplasm. USP8 expression was observed in 65.9% (54/82) of the cases and was correlated with the Noguchi classification, pathological subtype, lymphatic permeation, and vascular invasion (Table 2). Overexpression of USP8 was detected in not only invasive adenocarcinoma (44/57, 77.2%) but also AIS (8/21, 38.1%).
| USP8 Expression | ||||
|---|---|---|---|---|
| Clinicopathological features | Negative | Positive | Total patients | P-value |
| Age (yr) | 0.434 | |||
| ≤60 | 9 | 13 | 22 | |
| >60 | 19 | 41 | 60 | |
| Gender | 0.750 | |||
| Female | 14 | 29 | 43 | |
| Male | 14 | 25 | 39 | |
| Noguchi classification | 0.018 | |||
| Type A | 7 | 1 | 8 | * |
| Type B | 6 | 7 | 13 | |
| Type C' | 2 | 0 | 2 | |
| Type C | 1 | 6 | 7 | |
| Type D | 0 | 1 | 1 | |
| Total | 16 | 15 | 31 | |
| Pathological subtype | 0.021 | |||
| AIS | 13 | 8 | 21 | * |
| MIA | 2 | 0 | 2 | |
| Invasive adenocarcinoma | ||||
| Lepidic | 3 | 9 | 12 | |
| Acinar | 3 | 15 | 18 | |
| Papillary | 4 | 8 | 12 | |
| Micropapillary | 0 | 1 | 1 | |
| Solid | 3 | 11 | 14 | |
| IMA | 0 | 2 | 2 | |
| Pathological stage† | 0.060 | |||
| Stage I | 20 | 22 | 42 | |
| Stage II | 4 | 12 | 16 | |
| Stage III | 3 | 17 | 20 | |
| Stage IV | 1 | 3 | 4 | |
| Lymphatic permeation | <0.001 | |||
| Negative | 25 | 24 | 49 | *** |
| Positive | 3 | 30 | 33 | |
| Vascular invasion | 0.002 | |||
| Negative | 22 | 23 | 45 | ** |
| Positive | 6 | 31 | 38 | |
- † Stage I includes IA and IB, stage II includes IIA and IIB, stage III includes IIIA and IIIB. Correlation between expression of USP8 and clinicopathological feature was analyzed using chi-squared test.
- AIS, adenocarcinoma in situ); MIA, minimally invasive adenocarcinoma; IMA, invasive mucinous adenocarcinoma.
- P-value*< 0.05, **< 0.01, and ***< 0.001.
Correlation between expressions of USP8 and the expression and mutation status of EGFR
Next, we analyzed the correlation between expressions of USP8 and mutation status of EGFR. We found that all cases showing EGFR overexpression also had USP8 overexpression, the two being significantly correlated with each other (Table 3, upper). Fig. 1 shows representative cases in which expression of EGFR was consistent with that of USP8. Moreover, we confirmed that USP8 expression was in correlation with EGFR mutation status (Table 3, lower). Similarly to EGFR expression, USP8 expression and EGFR mutation status were significantly correlated, and 86.2% (25/29) of cases with EGFR mutation showed USP8 overexpression.
| USP8 expression | ||||
|---|---|---|---|---|
| Negative | Positive | Total patients | P-value | |
| EGFR expression | <0.001 | |||
| Negative | 28 (46.7%) | 32 (53.3%) | 60 | *** |
| Positive | 0 | 22 (100%) | 22 | |
| EGFR mutation status | 0.004 | |||
| Wild-type | 24 (45.3%) | 29 (54.7%) | 53 | ** |
| Mutant | 4 (13.7%) | 25 (86.2%) | 29 | |
| Exon 19 (E746-A750 del) | 2/4 | 9/25 | 11/29 | |
| Exon 21 (L858R) | 2/4 | 16/25 | 18/29 | |
- P-value*< 0.05, **< 0.01, and ***< 0.001.
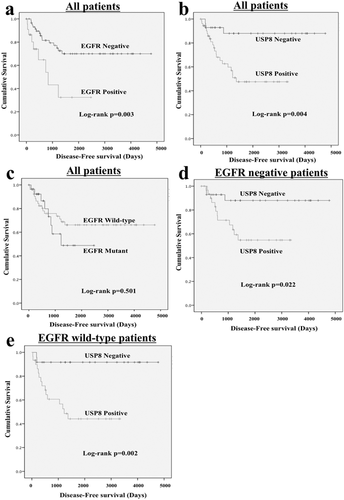
Analysis of EGFR and USP8 expression in relation to survival
To examine the prognostic implications of EGFR mutation status and expression of EGFR or USP8, we analyzed the disease-free survival of the patients. The Kaplan-Meier curves indicated that patients with positive expression of EGFR or USP8 had a significantly poorer outcome than those lacking such expression (Fig. 2a, b). However, the mutation status of EGFR did not show any association with patient outcome (Fig. 2c).
Additionally, multivariate analysis of the variables shown to be significant by univariate analysis revealed that vascular invasion, lymphatic permeation, and pathological stage were independently associated with disease-free survival, whereas EGFR or USP8 expression was not (Table S2).
Since our IHC results showed that USP8 overexpression was present even in AIS, we speculated that USP8 overexpression might be an earlier event than the appearance of EGFR abnormalities and possibly related to prognosis, even in patients who had no EGFR abnormalities including overexpression or mutation. To explore this possibility, we selected EGFR-negative or EGFR wild-type cases and analyzed patient outcome using the Kaplan-Meier curves obtained. Interestingly, in the EGFR-negative or EGFR wild-type population, patients with USP8 overexpression had significantly poorer outcome than those without it (Fig. 2d, e), indicating that USP8 might be a useful prognostic marker for patients with no EGFR abnormalities.
Regulation of EGFR expression by USP8 in immortalized AIS cells
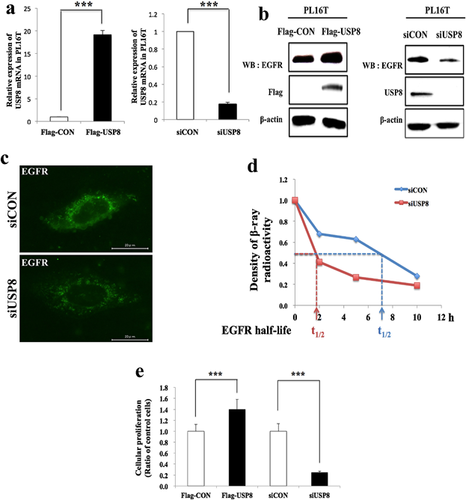
Our IHC results indicated that USP8 was overexpressed in lung adenocarcinoma from an early stage, such as AIS or minimally invasive adenocarcinoma (MIA). Therefore, we employed an immortalized AIS cell line, PL16T, for analysis of USP8 function in relation to EGFR expression. To examine the effects of USP8 overexpression or knockdown on EGFR expression in PL16T, we transfected the cells with Flag-USP8 or siUSP8. To confirm the transfection efficiency, we examined the mRNA and protein of USP8 (Fig. 3a, b). Overexpression of USP8 led to up-regulation of EGFR expression, whereas knockdown of USP8 led to down-regulation of total EGFR, not only on the cell surface but also in the cytoplasm (Fig. 3b, c). In addition, knockdown of USP8 shortened the half-life of EGFR relative to the control, indicating that USP8 helps to stabilize EGFR by inhibiting its degradation (Fig. 3d). Furthermore, cellular proliferation was reduced after USP8 knockdown, and accelerated after USP8 overexpression, relative to the control (Fig. 3e). These changes in cellular proliferation are thought to result from regulation of EGFR expression by USP8. Thus, our in vitro results suggested that USP8 controls the expression of EGFR, thus possibly affecting the clinical outcome.
DISCUSSION
In this study, we demonstrated that expression of EGFR and USP8 in lung adenocarcinoma was higher in tumor tissue than in normal lung tissue, and was associated with clinicopathological features such as the pathological subtype, lymphatic permeation, and vascular invasion (Tables 1, 2). Moreover, the expression and mutation status of EGFR were mutually correlated.20 Since EGFR mutation accelerates tumor cell proliferation and results in gene amplification,7, 10, 21 EGFR abnormalities such as mutation, amplification, and overexpression might occur sequentially in tandem with the stepwise progression of lung adenocarcinoma, particularly at the early stage such as AIS.3, 7 Additionally, consistent with a previous report,22 the frequency of EGFR mutation was found to be associated with histological phenotype.
Although many researchers have investigated the association between EGFR expression and amplification, the results have not been consistent; Lee et al. and Sasaki et al. found a significant correlation between them,10, 23 whereas Tang et al. did not.11 This discrepancy suggests that not only genetic alteration but also various regulatory mechanisms occurring at the protein level might influence EGFR expression. USP8 is one of the EGFR-regulating factors that induce EGFR protein recycling through deubiquitination.24 In this study, we showed that the expression of USP8 was significantly associated with that of EGFR. Overexpression of USP8 showed 38.1% of AIS cases (Table 2), suggesting that alteration of USP8 might be an early event similar to overexpression of EGFR. Based on these findings, we suggest that these alterations occur sequentially and are closely related to the stepwise progression of lung adenocarcinoma.
Overexpression of USP8 was detected in more than half of the cases of lung adenocarcinoma (Table 2). Chiara et al. screened alteration of DUBs in human cancers including those of the breast, colon-rectum, lung, stomach, kidney, prostate, non-Hodgkin's lymphoma, and melanoma, and found that USP9X, USP10, USP11, USP22, and USP24, but not USP8, were overexpressed in lung cancer.25 The observed discrepancy of USP8 positivity might be attributable to differences in the antibody or methodology used for IHC, and the freshness of the specimens employed.
Moreover, in IHC, the number of cases positive for USP8 was higher than that of cases positive for EGFR. We selected 60 cases that lacked EGFR expression and examined the association between USP8 expression and patient outcome. Interestingly, patients whose cancers were positive for USP8 had a significantly poorer outcome than those whose cancers were USP8-negative (Fig. 1, 2c), suggesting that USP8 might be a novel prognostic marker even in patients with EGFR-negative cancers.
Because we collected the samples in which EGFR mutation had already been analyzed, it can be easily envisaged that they might include high number of recurrence cases. Indeed, recurrence rate of our tested sample (36/82 cases, 43.9%) was higher than overall lung adenocarcinoma cases (156/652 cases, 23.9%) between 1999 and 2014 at university of Tsukuba Hospital. Therefore, in order to understand our result more correctly, we are planning additional large scale examination for expression of USP8 and EGFR as well as mutation status of EGFR.
Additionally, our in vitro experiments using immortalized AIS cells revealed that USP8 regulates EGFR expression at the cell membrane and in the cytoplasm, as well as its half-life, and cellular proliferation (Fig. 3). Therefore, our results imply that overexpression of USP8 might stabilize EGFR expression by inducing deubiquitination of EGFR from the early stage of lung adenocarcinoma such as AIS which does not show invasiveness.
USP8 activity is tightly controlled by scaffold proteins such as 14-3-3 proteins26 or post-translational modification such as phosphorylation.27 Most DUBs undergo phosphorylation by protein kinases that can switch their activity into on or off.27 In case of USP8, its stability and phosphorylation are regulated by AKT28 and Src,29 which are representative oncogenic signaling factors located in the downstream EGFR. In addition, USP19 was reported to have auto-deubiquitination function, removing ubiquitin moieties from USP19 protein itself.30 USP8 might also have similar function to control its own stability. Based on these facts, we expect that oncogenic signaling such as AKT and Src and the auto deubiquitination activity of USP8 may contribute overexpression of USP8 in lung adenocarcinoma.
Similarly to USP8, heat shock protein 90 (HSP90) acts as a chaperone protein that is known to stabilize not only wild-type but also mutant EGFR by regulation of its degradation after chemotherapy and radiotherapy.31, 32 Moreover, HSP90 inhibitor such as AUY922, potential agents for cancer treatment, effectively decreased cellular proliferation in lung adenocarcinoma cells harboring mutant EGFR by downregulation of EGFR and MET expression, which subsequently led to reduction of AKT-pathway33 likewise USP8 inhibitor effect on RTKs.34 However, recent clinical study of AUY922 in EGFR mutated patients of lung adenocarcinoma observed partial responses of this treatment but the dose and duration of the combination treatment with AUY922 and erlotinib to avoid rapid tumor development was limited by toxicities.35 Similarly to HSP90, USP8 might be also worth verifying its diagnostic or therapeutic usefulness.
Unlike the current treatment strategy for advanced adenocarcinoma, no therapeutic approach for early-stage lung adenocarcinomas such as AIS has yet been established, except for surgical resection.36 Based on our findings, we believe that USP8 could be an attractive therapeutic target for early-stage lung adenocarcinoma. Additionally, small-molecule inhibitors targeting USP8 have been developed, and are very selective. Therefore, our finding would seem to justify the development of a USP8 inhibitor for treatment of lung adenocarcinoma.
In conclusion, based on our findings, we believe that USP8 appears to be a suitable protein for use as a prognostic marker in early-stage lung adenocarcinoma, and might also be a promising therapeutic target.
DISCLOSURE STATEMENT
None declared.




