Genetic and signalling pathways of dry fruit size: targets for genome editing-based crop improvement
Summary
Fruit is seed-bearing structures specific to angiosperm that form from the gynoecium after flowering. Fruit size is an important fitness character for plant evolution and an agronomical trait for crop domestication/improvement. Despite the functional and economic importance of fruit size, the underlying genes and mechanisms are poorly understood, especially for dry fruit types. Improving our understanding of the genomic basis for fruit size opens the potential to apply gene-editing technology such as CRISPR/Cas to modulate fruit size in a range of species. This review examines the genes involved in the regulation of fruit size and identifies their genetic/signalling pathways, including the phytohormones, transcription and elongation factors, ubiquitin-proteasome and microRNA pathways, G-protein and receptor kinases signalling, arabinogalactan and RNA-binding proteins. Interestingly, different plant taxa have conserved functions for various fruit size regulators, suggesting that common genome edits across species may have similar outcomes. Many fruit size regulators identified to date are pleiotropic and affect other organs such as seeds, flowers and leaves, indicating a coordinated regulation. The relationships between fruit size and fruit number/seed number per fruit/seed size, as well as future research questions, are also discussed.
Introduction
The term ‘fruit’ normally refers to the fleshy seed-containing structure of a plant that is edible in the crude state, such as apple, banana, grape, lemon, orange, strawberry and tomato (Schlegel, 2003). It also includes the structures that are not commonly called ‘fruit’, such as pod, silique, kernel and grain. Fruit, therefore, account for a substantial part of the world’s agricultural output for human and livestock diet (Giovannoni, 2004; Tanksley, 2004), and they are a major target for crop improvement. Genome editing offers the potential to accelerate fruit size breeding gains and facilitate the introduction of novel mutations that are unavailable in current germplasm (Scheben and Edwards, 2017; Scheben et al., 2017).
Botanically, the fruit is a feature of angiosperms that develop a gynoecium derived from carpels after flowering. As such, fruit represents the reproductive organ for seed development and a structure that offers protection from insects and pests as well as a mechanism for seed dispersal (Bennett et al., 2011; Giovannoni, 2004; Pesaresi et al., 2014; Seymour et al., 2013). Fruit types can be classified using several characteristics: dry or fleshy, dehiscent or indehiscent and apocarpous or syncarpous carpels (Karlova et al., 2014; Pesaresi et al., 2014). Capsules and siliques (as seen in Arabidopsis and relatives) are dry, dehiscent and syncarpous (Figure S1); achenes and nuts are dry, indehiscent and unicarpellate; berries are fleshy, indehiscent and syncarpous; and drupes (e.g. stone fruit) are fleshy and indehiscent with the single seed enclosed in a hard endocarp (Seymour et al., 2013).
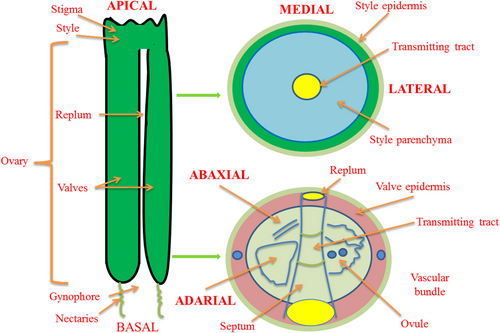
From a biological point of view, fruit size is a vital fitness character for plant evolution. From a human perspective, fruit size is an important agronomic trait for crop improvement, and is, therefore, a target for artificial selection (Pesaresi et al., 2014; Seymour et al., 2013; Tanksley, 2004). A classic example is the fruit of fleshy, tomato, which is nearly 1,000 times larger than its ancestor, and where size is modulated by the additive contribution of tens of quantitative trait loci (QTL), some of which have been cloned (Lin et al., 2014; Tanksley, 2004). Also, studies in Arabidopsis have identified several critical regulators for fruit development (Giovannoni, 2004; Seymour et al., 2008, 2013). As a model plant and member of the Brassicaceae family, Arabidopsis has contributed significantly to our understanding of fruit size regulation, with identification and functional characterization of many genes. From an ontogenetic standpoint, final fruit size is determined by successive processes of gynoecium formation, fertilization, fruit growth involving cell proliferation, differentiation and expansion, with partial overlaps in time (Tanksley, 2004; Wang et al., 2016). Before fertilization, the first patterning event in Arabidopsis gynoecium is the construction of the apical-basal, mediolateral and abaxial–adaxial axes (Figure 1) that determine fruit length, width and thickness, respectively (Seymour et al., 2013). Fruit size in Arabidopsis is mainly determined by fruit length that is governed by elongation of the apical-basal axis. The developmental switch that turns gynoecium into growing fruit is dependent on the fertilization of ovule, which otherwise senesces and dies (Seymour et al., 2013). After the fertilization of fruit, it enters a stage where ovary growth and maturation are tightly cooperated with seed development.
Although dry fruit types (such as cereal and oilseed crops) account for the majority of plants, fruit size studies have focused primarily on fleshy-fruit species because of their importance in the human diet (Giovannoni, 2004). Despite the importance of fruit size to grain production, there are little-published reviews in dry fruit types.
Whilst tomato is an ideal system for studying fruit development, including size regulation (Karlova et al., 2014; Pesaresi et al., 2014; Seymour et al., 2008, 2013), significant advances have been made in Arabidopsis since it was sequenced in 2000 (Kaul et al., 2000). Information from Arabidopsis is often directly applicable to the polyploid crop relatives of the Brassicaceae, such as rapeseed, as well as other taxa including legumes, and lesser extent cereals. As genome editing technology and plant transformation protocols make knockout and knockin of most fruit size genes and regulatory elements in crops feasible (Scheben et al., 2017), the challenge becomes selecting appropriate editing targets. Identifying and reviewing the broad range of genes and regulatory elements controlling fruit size, therefore, provides a foundation for crop improvement through genome editing. This review summarizes the genes and regulatory networks affecting fruit size and classifies the individual genetic/signalling pathways. We aim to provide new insights into the molecular mechanisms of fruit size regulation, which may help identify novel targets for genome editing and facilitate crop genetic improvement, especially for dry fruit types, including many important crops such as soya bean, peanut and rapeseed.
Fruit size regulators
Phytohormones
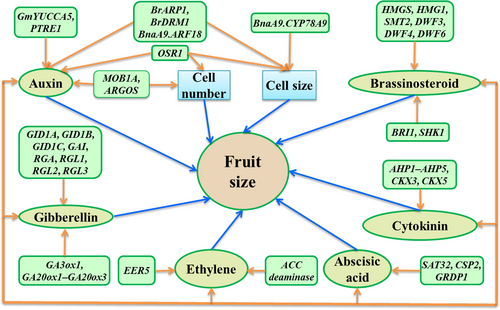
Plant hormones (also known as phytohormones) are a group of low-abundance chemical substances (signal molecules) produced within plants, which can act either locally or more remotely by long-distance transport through the vascular system (Lacombe and Achard, 2016). Phytohormones can regulate or influence almost all aspects of plant growth and development (including fruit size) in response to environmental and endogenous signals (https://en.wikipedia.org/wiki/Plant_hormone). At the cellular and molecular levels, phytohormones can affect gene expression and transcription, cell division and growth. According to their chemical structures and physiological effects, phytohormones are divided into ten classes: abscisic acid (ABA), auxins (AUX), brassinosteroids (BR), cytokinins (CTK), ethylene (ETH), gibberellins (GA), jasmonates (JA), salicylic acid (SA), strigolactones (SL) and others (Lin and Tan, 2011); their roles in regulating fruit size are summarized below (Figure 2; Table 1).
| Pathways | Gene name | Species | Accession number | Biological function | Reference(s) |
|---|---|---|---|---|---|
| Gibberellin (GA) | GA3ox1 | Arabidopsis | AT1G15550 | GA biosynthetic process | Hu et al. (2003) |
| GA3ox4 | Arabidopsis | AT1G80330 | GA biosynthetic process | Hu et al. (2003) | |
| GID1A | Arabidopsis | AT3G05120 | Cellular response to hypoxia | Griffiths et al. (2006) | |
| GID1B | Arabidopsis | AT3G63010 | Catabolic process | Griffiths et al. (2006) | |
| GID1C | Arabidopsis | AT5G27320 | Catabolic process | Griffiths et al. (2006) | |
| GA20ox2 | Arabidopsis | AT5G51810 | Oxidation–reduction process | Plackett et al. (2012) | |
| GAI | Arabidopsis | AT1G14920 | Gibberellic acid homeostasis | Peng et al. (1997) | |
| RGA | Arabidopsis | AT2G01570 | Multicellular organism development | Silverstone et al. (2016) | |
| RGL1 | Arabidopsis | AT1G66350 | DELLA proteins response to GA | Wen and Chang, (2002) | |
| RGL2 | Arabidopsis | AT3G03450 | Defence response | Lee et al. (2006) | |
| RGL3 | Arabidopsis | AT5G17490 | Multicellular organism development | Fuentes et al. (2012) | |
| Auxin (IAA) | PTRE1 | Arabidopsis | AT3G53970 | Regulates auxin signalling | Yang et al. (2011) |
| MOB1A | Arabidopsis | AT5G45550 | Promotes auxin signalling | Cui et al. (2016) | |
| GmYUCCA5 | Arabidopsis, soybean | AT5G43890 | Auxin biosynthetic process | Wang et al. (2017c) | |
| BnaA9. ARF18 | Rapeseed | BnaA09g55580D | Auxin response factor | Liu et al. (2008a) | |
| BrARP1 | Brassica rapa | AT1G43170 | Auxin-repressed protein 1 | Lee et al. (2016) | |
| BrDRM1 | Brassica rapa | Bra032894 | Dormancy-associated protein 1 | Lee et al. (2016) | |
| OSR1 | Arabidopsis | AT2G41230 | Organ growth and overall organ size | Feng et al., (2011) | |
| ARGOS | Arabidopsis | AT3G59900 | Response to auxin | Hu et al. (2003) | |
| Cytokinin (CK) | AHP5 | Arabidopsis | AT1G03430 | Signal transduction | Hutchison et al. (2006) |
| AHP4 | Arabidopsis | AT3G16360 | Signal transduction | Hutchison et al. (2006) | |
| AHP1 | Arabidopsis | AT3G21510 | Signal transduction | Hutchison et al. (2006) | |
| AHP2 | Arabidopsis | AT3G29350 | Signal transduction | Hutchison et al. (2006) | |
| AHP3 | Arabidopsis | AT5G39340 | Signal transduction | Hutchison et al. (2006) | |
| CKX3 | Arabidopsis | AT5G56970 | Cytokinin catabolic process | Bartrina et al., 2011 | |
| CKX5 | Arabidopsis | AT1G75450 | Cytokinin catabolic process | Bartrina et al., 2011 | |
| Brassinosteroid (BR) | BRI1 | Arabidopsis | AT4G39400 | Brassinosteroid receptor | Noguchi et al. (1999) |
| DWF4 | Arabidopsis | AT3G50660 | Brassinosteroid biosynthesis | Si et al. (1998) | |
| SMT2 | Arabidopsis | AT1G20330 | Brassinosteroid biosynthesis | Hwang et al. (2007) | |
| CYP72C1 | Arabidopsis | AT1G17060 | Brassinosteroid metabolic process | Takahashi et al. (2005) | |
| BjHMGS1 | Brassica juncea | AT4G11820 | Sterols biosynthetic process | Liao et al. (2014) | |
| Abscisic acid (ABA) | CSP2 | Arabidopsis | AT4G38680 | Regulation of cellular respiration | Nakaminami et al. (2006), Sasaki et al. (2013) |
| GRDP1 | Arabidopsis | AT2G22660 | Response to osmotic stress | Rodríguez-Hernández et al. (2017) | |
| SAT32 | Arabidopsis | AT1G27760 | Salt and ABA-responsive | Park et al. (2009) | |
| Ethylene (ETH) | EER5 | Arabidopsis | AT2G19560 | Ethylene-signalling | www.arabidopsis.org |
| – | Arabidopsis | AT1G48420 | 1-aminocyclopropane-1-carboxylic acid oxidase | Walton et al. (2012) | |
| Phytohormone | CYP78A9 | Arabidopsis/ Rapeseed | AT3G61880 | Fruit and seed development | Ito and Meyerowitz (2000); Shi et al. (2019) |
| Transcription factors | BoMF2 | Brassica oleracea | Bol029968 | Transcriptional regulatory factor | Kang et al. (2014) |
| MSH1 | Arabidopsis | AT3G24320 | Mitochondrial genome maintenance | www.arabidopsis.org | |
| CSP4 | Arabidopsis | AT2G21060 | Regulation of transcription | Nakaminami et al. (2006); Yang and Karlson (2011) | |
| YABBY family | CRC | Arabidopsis | AT1G69180 | Regulation of transcription | Prunet et al. (2008) |
| Zinc finger family | SUP | Arabidopsis, cucumber | AT3G23130 | Transcription factor | Zhao et al. (2012) |
| DOF4.2 | Arabidopsis | AT4G21030 | Regulation of transcription | Zuo et al. (2013) | |
| DOF4.4 | Arabidopsis | AT4G21050 | Regulation of transcription | Zuo et al. (2013) | |
| NTT | Arabidopsis | AT3G57670 | Zinc finger transcription factor | Chung et al. (2013) | |
| Tri-helix family | ASIL1 | Arabidopsis | AT1G54060 | Regulation of transcription | Gao et al. (2016) |
| AP2-ERF family | SlERF36 | Tomato, Arabidopsis | AT1G50640 | Regulation of transcription | Upadhyay et al. (2014) |
| ANT | Arabidopsis | AT4G37750 | Control of cell proliferation | Mizukami and Fischer (2000) | |
| MADS-box family | GbAGL2 | Arabidopsis, cotton | AT5G15800 | Plant ovule development | Liu et al. (2015) |
| SHP1/2 | Arabidopsis | AT3G58780 | Regulation of growth | Liljegren et al. (2004); Pinyopich et al. (2003) | |
| FUL | Arabidopsis | AT5G60910 | Fruit development | Liljegren et al. (2000) | |
| STK | Arabidopsis | AT4G09960 | Ovule development | Zhang et al. (2016) | |
| Homeobox | RPL | Arabidopsis | AT5G02030 | Homeodomain transcription factor | Roeder et al. (2003) |
| WOX14 | Arabidopsis | AT1G20700 | Vasculature development | Deveaux et al. (2008) | |
| bHLH family | AMS | Arabidopsis | AT2G16910 | Regulation of transcription | Sorensen et al. (2003) |
| IND | Arabidopsis | AT4G00120 | Regulation of transcription | Liljegren et al. (2000) | |
| ALC | Arabidopsis | AT5G67110 | Fruit development anddehiscence | Liljegren et al. (2000) | |
| B3 family | REM22 | Arabidopsis | AT3G17010 | Transcriptional factor B3 family protein | www.arabidopsis.org |
| Elongation factor | TaTEF-7A | Wheat, Arabidopsis | CJ655632.1/AT5G46030 | Transcript elongation factor | Zheng et al. (2014) |
| SPT4-1 | Arabidopsis | AT5G08565 | Chromatin organization | Dürr et al., (2014) | |
| SPT4-2 | Arabidopsis | AT5G63670 | Chromatin organization | Dürr et al., (2014) | |
| MicroRNA | MaEF1A | Arabidopsis, Banana | AT1G18070 | Translation elongation | Liu et al. (2008a) |
| miR172 | Arabidopsis/Peanut | AT2G28056 | Gene silencing by miRNA | José Ripoll et al. (2015) | |
| miR397b | Arabidopsis/Peanut | AT4G13555 | Gene silencing by miRNA | Wang et al. (2014) | |
| Md-miRNA156 | Arabidopsis/Apple | AT5G55835 | Flower and fruit development | Sun et al. (2013) | |
| Ubiquitin-proteasome pathway | UBP15 | Arabidopsis/Rice | AT1G17110 | Ubiquitin-specific proteases | Liu et al. (2016) |
| UBP26 | Arabidopsis | AT3G49600 | Ubiquitin-specific proteases | Luo et al. (2008) | |
| SWA1 | Arabidopsis | AT2G47990 | Embryo sac development | Shi et al. (2005) | |
| RHF1A | Arabidopsis | AT4G14220 | Regulation of cell cycle | Liu et al. (2008b) | |
| RHF2A | Arabidopsis | AT5G22000 | Regulation of cell cycle | Liu et al. (2008b) | |
| MMS21 | Arabidopsis | AT3G15150 | Regulation of meristem development | Liu et al. (2014a) | |
| UBC22 | Arabidopsis | AT5G05080 | Protein polyubiquitination | Wang et al. (2016) | |
| DA1 | Arabidopsis | AT1G19270 | Ubiquitin receptor | Li et al. (2018) | |
| SAP | Arabidopsis | AT5G35770 | E3 ubiquitin ligase complex | Wang et al. (2017c) | |
| G-protein signalling | XLG1 | Arabidopsis | AT2G23460 | G-protein γ-subunit | Wang et al. (2017b) |
| AGB1 | Arabidopsis | AT4G34460 | G-protein β-subunit | Lease et al. (2001) | |
| Arabinogalactan protein | AGP19 | Arabidopsis | AT1G68725 | Arabinogalactan protein | Yang et al. (2007) |
| AGP6 | Arabidopsis | AT5G14380 | Arabinogalactan glycoproteins | Levitin et al. (2008) | |
| AGP11 | Arabidopsis | AT3G01700 | Arabinogalactan glycoproteins | Levitin et al. (2008) | |
| HPGT1 | Arabidopsis | AT5G53340 | Hydroxyproline O-galactosyltransferase | Ogawa-Ohnishi and Matsubayashi (2015) | |
| HPGT2 | Arabidopsis | AT4G32120 | Hydroxyproline O-galactosyltransferase | Ogawa-Ohnishi and Matsubayashi (2015) | |
| HPGT3 | Arabidopsis | AT2G25300 | Hydroxyproline O-galactosyltransferase | Ogawa-Ohnishi and Matsubayashi (2015) | |
| FLA3 | Arabidopsis | AT2G24450 | Anchored component of membrane | Li et al. (2015) | |
| FLA4 | Arabidopsis | AT3G46550 | Mucilage biosynthetic process | Shi et al. (2019) | |
| RNA-binding protein | LSM1A | Arabidopsis | AT1G19120 | RNA metabolic process | Perea-Resa et al. (2012) |
| LSM8 | Arabidopsis | AT1G65700 | RNA metabolic process | Perea-Resa et al. (2012) | |
| LSM1B | Arabidopsis | AT3G14080 | RNA metabolic process | Perea-Resa et al. (2012) | |
| Receptor kinase signalling | SNF4 | Arabidopsis | AT1G09020 | Carbohydrate metabolic process | www.arabidopsis.org |
| ER | Arabidopsis | AT2G26330 | Regulation of cell adhesion | Zanten et al. (2010) | |
| RPK2 | Arabidopsis | AT3G02130 | Meristem maintenance | Mizuno et al. (2007) | |
| BAM3 | Arabidopsis | AT4G20270 | Regulation of meristem growth | DeYoung et al. (2006) | |
| CLV1 | Rapeseed/Arabidopsis | AT1G75820 | Shoot and floral meristem size | Xiao et al. (2018) | |
| CLV3 | Rapeseed/Arabidopsis | AT2G27250 | Shoot apical meristem size | Yang et al. (2018) | |
| Other proteins | HSP70 | Arabidopsis | AT3G12580 | Protein folding | Leng et al. (2016) |
| CINV1 | Arabidopsis | AT1G35580 | Sucrose catabolic process | Qi et al. (2007) | |
| CcCCOAOMT1 | Jute, Arabidopsis | AT4G34050 | Lignin biosynthetic process | Zhang et al. (2014) | |
| GhWBC1 | Cotton | AY255521.1 | ATP-binding cassette transporter | Zhu et al. (2003) | |
| LNG1 | Arabidopsis | AT5G15580 | Unidimensional cell growth | Lee et al. (2013) | |
| LNG2 | Arabidopsis | AT3G02170 | Unidimensional cell growth | Lee et al. (2013) | |
| AXY3/XYL1 | Arabidopsis | AT1G68560 | Glycoside hydrolase family 3 | Günl and Pauly (2011) | |
| CALS7 | Arabidopsis | AT1G06490 | Callose synthase 7 | Xie et al. (2011) | |
| BGAL10 | Arabidopsis | AT5G63810 | Glycoside hydrolase family 35 | Sampedro et al. (2012) | |
| FATA2 | Arabidopsis | AT4G13050 | Fatty acid biosynthetic process | Wang et al. (2016) | |
| HEMN1 | Arabidopsis | AT5G63290 | Oxidation–reduction process | Pratibha et al. (2017) | |
| GGT1 | Arabidopsis | AT4G39640 | Glutathione catabolic process | Giaretta et al. (2017) | |
| GGT2 | Arabidopsis | AT4G39650 | Glutathione transmembrane transport | Giaretta et al. (2017) | |
| BcRISP1 | Cabbage, Arabidopsis | AT5G13440 | Oxidation–reduction process | Liu et al. (2014a) | |
| BnaC9.SMG7b | Rapeseed | BnaC09g38310D | Meiotic cell cycle | Li et al. (2008) | |
| LPAT2 | Arabidopsis | AT3G57650 | CDP-diacylglycerol biosynthetic process | Kim et al. (2005) |
Auxins (AUX)
The action of auxins is demonstrated to be regulated in three layers: synthesis, transport and perception/signal transduction. In recent years, several genes involved in these processes have been shown to affect fruit growth and development.
The YUCCA flavin monooxygenase is a key enzyme in the simple two-step pathway that converts tryptophan to IAA (Zhao, 2012), the most abundant endogenous auxin in plants. Overexpression of a soya bean YUCCA gene, GmYUCCA5, in Arabidopsis resulted in higher plants with long and narrow leaves as well as few and short siliques (Wang et al., 2017b).
Auxin is perceived by the receptor of F-box family proteins (such as TIR1 and AFB), which recruit AUX/IAA proteins to the SCFTIR1/AFB complex for subsequent ubiquitination and proteasome-mediated degradation, leading to de-repression of ARFs that activate auxin-induced gene expression (Kong et al., 2016). The Arabidopsis PROTEASOME REGULATOR1 (PTRE1) is a homologue of the mammalian proteasome inhibitor 31 (PI31), which is involved in auxin-mediated AUX/IAA degradation by repressing 26S proteasome activity. The loss-of-function ptre1 mutant exhibited auxin-insensitive phenotypes: growth inhibition, including dwarf plants, small leaves and short siliques as well as arrested embryogenesis (Yang et al., 2011). The Arabidopsis MOB1A is involved in the auxin-activated signalling pathway and auxin-controlled cell division, which is uniformly expressed in embryos and suspensor cells during embryogenesis. The loss-of-function mob1a mutant displayed defects in organogenesis and growth, including reduced ovule number, shorter siliques and roots as well as smaller flowers (Cui et al., 2016).
The Arabidopsis AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE (ARGOS) is a positive regulator of lateral organ size, which is highly induced by auxin and can transduce signals downstream of AXR1 to regulate cell proliferation through ANT (Hu et al., 2003). Transgenic plants expressing sense or antisense cDNA of ARGOS display multiple phenotypic changes, including flowering time, plant height, leaf and flower size, as well as silique length and seed number per silique, which is resulted from the changes in cell number in these organs. The Arabidopsis ORGAN SIZE RELATED 1 (OSR1) is an endoplasmic reticulum-localized hormone-responsive gene, which acts redundantly with ARGOS and ARL to regulate organ growth. The overexpression of OSR1 in Arabidopsis delayed flowering time and increased the final size of many organs, including longer roots, larger leaves and flowers as well as longer siliques, which was resulted from increased cell number and size (Feng et al., 2011).
Recently, a major QTL for both silique length and seed weight has been cloned on the A9 chromosome of Brassica napus, which is resulted from a 55-amino acid deletion in an orthologue of the Arabidopsis auxin response factor 18 (designated as BnaA9.ARF18). The BnaA9.ARF18 acts as a negative regulator of fruit and seed size by restricting cell elongation in the silique wall through its inhibitory activity on downstream auxin-responsive genes (Liu et al., 2015). Overexpression of two auxin-repressed protein genes BrARP1 and BrDRM1 in Brassica rapa reduced vegetative and reproductive growth, including the number and length of siliques, possibly from inhibition of either cell elongation or expansion (Lee et al., 2013).
Gibberellins (GA)
Despite the identification of more than 130 types of gibberellin, only GA1, GA3, GA4 and GA7 are bioactive (Daviere and Achard, 2013). In recent years, several genes in the gibberellin metabolism and signalling pathway have been shown to play an essential role in regulating fruit growth/size in Arabidopsis.
GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox) are responsible for the last steps of the gibberellin biosynthetic and catabolic pathway, which catalyse consecutive reactions that convert GA intermediates to bioactive forms. The Arabidopsis contains four GA3ox genes (GA3ox1–GA3ox4): at the silique development stage, GA3ox1 is expressed mainly in the replum, funiculus and receptacle of silique, whilst the other three are expressed only in developing seeds (Hu et al., 2008). The mutants of GA3ox1 and its combination with GA3ox2–GA3ox4 showed GA-deficient phenotypes, including semi-dwarfism, smaller rosettes, shorter siliques, reduced male fertility and fewer seeds per silique. The Arabidopsis contains five GA20ox genes (GA20ox1–GA20ox5), knockout of GA20ox1–GA20ox3 also showed GA-deficient phenotypes, including delayed flowering, dwarfism, reduced male fertility, shorter siliques and few seeds per silique (Plackett et al., 2012; Rieu et al., 2008).
Gibberellin bind to its receptor, GIBBERELLIN-INSENSITIVE DWARF1 (GID1), which in turn interacts with the DELLA proteins. The Arabidopsis has three homologues of gibberellin receptors (GID1A, GID1B and GID1C) that are expressed in most tissues throughout the development but varied in expression level. A reverse genetic study showed that the combinatorial mutants of GID1A, GID1B and GID1C showed GA-insensitive phenotypes, including semi-dwarfism, short roots and siliques, reduced male fertility and fewer seeds per silique in Arabidopsis (Griffiths et al., 2006; Livne and Weiss, 2014). Arabidopsis also has five DELLA genes (GAI, RGA, RGL1—RGL3), and the knockout of five DELLA proteins resulted in reduced fertility, seed number per silique, and silique length (Fuentes et al., 2012). Further studies are required to identify the effectors or downstream components of gibberellin in fruit size control.
Cytokinins (CTK)
The cytokinin signal transduction pathway includes a His-Asp phospho-relay that is similar to bacterial two-component signalling systems, in which cytokinin binds to the CHASE domain of HKs (histidine kinases) receptors, and then AHPs (authentic histidine phosphotransferases) act as intermediates to transfer a phosphate from HKs to the downstream response regulators (Kieber and Schaller, 2018). The Arabidopsis has five AHP genes (AHP1–AHP5) that are redundant, positive regulators of cytokinin signalling, which can affect multiple developmental processes. The T-DNA insertion ahp quintuple mutant is less sensitive to cytokinin and has various abnormalities in growth and development, including reduced fertility and seed set, enlarged seeds, as well as shortened siliques and roots (Hutchison et al., 2006). The degradation of cytokinin is catalysed by cytokinin oxidase/dehydrogenase (CKXs), which is coded by seven homologous genes (CKX1–CKX7) in Arabidopsis. Of these, the ckx3 and ckx5 mutants had more and larger flowers, more and longer siliques, more ovules per ovary, leading to higher seed yield (Bartrina et al., 2011).
Abscisic acid (ABA)
Abscisic acid is well known to regulate seed germination, root and shoot development and abiotic stress responses (Humplik et al., 2017; Lin and Tan, 2011), whereas its role on fruit development is unclear. The Arabidopsis GRDP1 encodes the glycine-rich domain protein involved in the abscisic acid-activated signalling pathway. The Arabidopsis grdp1 mutant lines exhibited many developmental defects, including shortened siliques and aborted ovules as well as reduced seed number and weight (Rodríguez-Hernández et al., 2017). The Arabidopsis CSP2 encodes a glycine-rich protein that responds to cold stress through the ABA pathway, which is highly expressed in shoot apical meristems and siliques (Nakaminami et al., 2009). The overexpression of CSP2 in Arabidopsis resulted in later flowering, shorter siliques, and fewer seeds per silique (Sasaki et al., 2013). The Arabidopsis Salt-tolerance 32 (SAT32) encodes a protein similar to the human interferon-related development regulator, which is involved in salt resistance through the ABA signalling pathway. The T-DNA knockout mutant of SAT32 in Arabidopsis showed slightly longer roots but shorter siliques and fewer seeds per silique (Park et al., 2009).
Ethylene (ETH)
1-aminocyclopropane-1-carboxylic acid (ACC) is the direct precursor of ethylene biosynthesis, whilst ACC-deaminase can decrease the level of ACC. The ACC-deaminase transgenic canola line had reduced levels of ethylene in the siliques and seeds, as well as smaller siliques and seeds, and fewer seeds per silique (Walton et al., 2012). Interestingly, the contents of endogenous GA1, GA4 and IAA also declined in the siliques and seeds of transgenic lines, suggesting that ethylene can interact with other phytohormones to regulate fruit and seed development (Walton et al., 2012). The Arabidopsis ENHANCED ETHYLENE RESPONSE 5 (EER5) encodes a PAM domain protein involved in ethylene-activated signalling pathway (www.arabidopsis.org). The eer5 mutant showed hypersensitivity to ethylene with various developmental defects, such as shorter siliques, curly leaves, shorter primary roots and less lateral roots (Christians et al., 2008).
Brassinosteroids (BR)
Several genes involved in brassinosteroid synthesis and signal transduction affect fruit growth/size. The Arabidopsis HMGS/FKP1 encodes hydroxymethylglutaryl-CoA synthase, which is involved in the mevalonate pathway of sterols biosynthetic process (Ishiguro et al., 2010). The Arabidopsis HMGS/FKP1 is expressed strongly in floral buds, moderately in roots and weakly in leaves, and its T-DNA insertion mutant was male-sterile with short silique with few seeds, due to defect in pollen coat formation. Whereas, the overexpression of Brassica juncea BjHMGS1 in tobacco increased both vegetative growth (such as root, stem and leaf) and seed yield (pod size and seed number per pod), which was caused by the higher sterols content through regulating the expression of isoprenoid biosynthesis genes (Liao et al., 2014). The Arabidopsis HMG1 encodes a 3-hydroxy-3-methylglutaryl coenzyme A reductase, another enzyme of the mevalonate pathway involved in sterols biosynthetic process. The T-DNA insertion hmg1 mutant showed dwarfism, early senescence and male sterility with short siliques and few seeds, which was caused by suppression in cell elongation due to reduced sterol level (Suzuki et al., 2004). The Arabidopsis SMT2 encodes a sterol-C24-methyltransferase involved in sterol biosynthesis, and its T-DNA knockout mutant displayed reduced fertility, few seeds and shorter siliques (Hwang et al., 2007). The Arabidopsis DWF6/DET2 is similar to mammalian steroid-5-alpha-reductase that is involved in the brassinolide biosynthetic pathway, and its mutant also showed dwarfism, reduced male fertility, short siliques (Fujioka and Yokota, 2003). The Arabidopsis DWF3/CPD and DWF4 encode cytochrome P450 monooxygenase CYP90A1 and CYP90B1, respectively, which are the rate-limiting enzymes in the brassinosteroid biosynthetic pathway. Overexpression of Populus euphratica DWF4 or CPD in Arabidopsis increased plant height and silique length but decreased silique number (Si et al., 2016).
The Arabidopsis BRI1/BIN1/DWF2 encodes a plasma membrane-localized leucine-rich repeat receptor kinase, which is involved in brassinosteroid signal transduction. Its mutant showed multiple defects in growth and development, including dwarfism, reduced male fertility, few seeds and short siliques (Clouse et al., 1996). The Arabidopsis SHK1 encodes cytochrome P450 monooxygenase CYP72C1 similar to BAS1/CYP734A1 that regulates BR inactivation. The shk1-D mutant showed dwarfism, short siliques and smaller seeds along the longitudinal axis, which is caused by reduced cell elongation (Takahashi et al., 2005).
Recently, a major QTL for silique length and seed weight has been cloned on the A9 chromosome of Brassica napus, which is resulted from a CACTA-like transposable element inserted in the upstream region of an orthologue (designated as BnaA9.CYP78A9) of Arabidopsis CYP78A9 that acts as an enhancer to increase its expression (Shi et al., 2019). In fact, CYP78A9 has been long known to play an important role in reproductive development (Sotelo-Silveira et al., 2013), as its overexpression caused large flowers, siliques, and seeds but reduced fertility and seed number per silique in Arabidopsis (Ito and Meyerowitz, 2000). Further studies should be conducted to make clear the reactions catalysed by CYP78A9 so as to uncover its relationship with the known hormone pathways.
Transcription factors
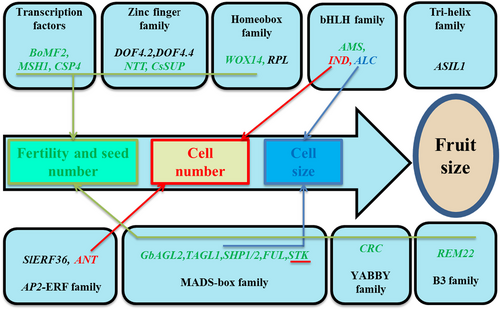
Transcription factors (TFs) are usually classified into different families based on their DNA-binding domains, which play a key role in plant development by temporally and spatially regulating the transcription of the corresponding target genes (Jin et al., 2017). A previous expression profiling study showed that most of the transcript factors were involved in the development of siliques in Arabidopsis (De Folter et al., 2004); their roles in regulating fruit size are summarized below (Figure 3; Table 1).
The Brassica oleracea BoMF2 encodes a nuclear-localized AT-hook DNA-binding protein homologous to Arabidopsis AHL16 (Kang et al., 2014), which is required for tapetum proliferation during anther development. The overexpression of BoMF2 led to reduced pollen viability, shorter siliques and fewer seeds per silique (Kang et al., 2014). The Arabidopsis MSH1 is a plant organelle DNA-binding and thylakoid protein that can influence genome stability and growth pattern (Xu et al., 2011). Inhibition of this gene by RNA interference causes multiple defects in the growth and development of several species, including slower growth, dwarfism, shorter siliques and reduced male fertility in Arabidopsis (Xu et al., 2012). The Arabidopsis cold shock domain proteins (CSPs) are highly conservative DNA-binding transcript factors, which are involved in the transition to flowering and silique development (Nakaminami et al., 2009). Of which, the overexpression of CSP4 in Arabidopsis reduces fruit length and induces embryo lethality (Yang and Karlson, 2011).
B3 family
The Arabidopsis Reproductive Meristem (REM) genes encode B3 family transcription factors (Swaminathan et al., 2008), which are preferentially expressed in flower and ovule/seed development (Mantegazza et al., 2014). Of these, the rem22 mutant exhibited reduced fertility and slow growth, including dwarf plants and short siliques (https://www.arabidopsis.org/).
Basic helix-loop-helix family
The ABORTED MICROSPORES (AMS) gene belongs to the MYC subfamily of basic helix-loop-helix (bHLH) superfamily, which is essential for microspore development. The ams mutant produced by T-DNA insertion showed a sporophytic recessive male-sterile phenotype as well as undeveloped silique (Sorensen et al., 2003). Also belonging to the bHLH superfamily, both IND and ALC genes are well known to involve in the regulation of fruit development and dehiscence (Ballester and Ferrándiz, 2017). Mutations in both IND and ALC can partly restore fruit elongation in flu mutant (Liljegren et al., 2004).
AP2-ERF family
The APETALA2-ETHYLENE RESPONSE FACTOR (AP2-ERF) is the major family of transcription factors with 140–280 members in several plants (Nakano et al., 2006). The Arabidopsis AINTEGUMENTA (ANT) is a member of the AP2 subfamily, which is well known to control the size of organs (including root, leaf, flower, fruit and seed) by regulating cell proliferation in plants (Mizukami and Fischer, 2000). SlERF36 is an EAR motif-containing ERF gene from tomato, and its overexpression reduced vegetative growth, including the size of rosettes, flowers and siliques (Upadhyay et al., 2014).
Homeobox family
The Arabidopsis WOX14 belongs to the WUSCHEL-related homeobox (WOX) subfamily, and its knockout mutant plants are partially male-sterile, with aborted and shorter siliques (Deveaux et al., 2008). The Arabidopsis REPLUMLESS (RPL) gene (also encodes a homeodomain protein) is required for the replum development, and its loss-of-function mutant showed shorter siliques (Roeder et al., 2003).
MADS-box family
Several members of the MADS-box family are involved in the development and dehiscence of fruits in Arabidopsis. SHATTERPROOF1 (SHP1) and SHP2 are two closely related and functionally redundant genes, which control the differentiation of dehiscence zone and promote the lignification of adjacent cells (Liljegren et al., 2000), by up-regulating IND and ALC (Ballester and Ferrándiz, 2017). As a negative regulator of SHP genes, the Arabidopsis FRUITFULL (FUL) is required for valve differentiation and expansion after fertilization, and its loss-of-function mutation results in fruit that fail to elongate (Ferrandiz et al., 2000; Zhang et al., 2016). The Gossypium barbadense AGL2 was an AGAMOUS (AG)-like gene, which was highly expressed in reproductive tissues (including ovules and carpels) but lowly in vegetative tissues. The overexpression of GbAGL2 in Arabidopsis resulted in longer siliques with more seeds per silique (Liu et al., 2009; Zhang et al., 2016). SEEDSTICK (STK) encodes a MADS-box transcription factor that is expressed in the carpels and ovules. STK is required for funiculus development by regulating cell expansion and division, and its loss-of-function mutant showed shorter siliques with rounder and smaller seeds (Pinyopich et al., 2003).
Tri-helix family
The Arabidopsis ASIL1 gene is a member of the tri-helix DNA-binding protein family, which is localized in the nucleus and belongs to the subfamily of 6b-interacting protein 1-like 1. ASIL1 is involved in the repression of early and seed mature genes via competitive binding to the GT-box-like element (Gao et al., 2009). The asil1 mutant in Arabidopsis had shorter siliques, smaller seeds and reduced seed weights per plant compared with the wild type.
YABBY family
The Arabidopsis CRABS CLAW (CRC), a member of the YABBY gene family (Bowman, 2000), is mainly expressed in nectary and carpel (Bowman and Smyth, 1999; Siegfried et al., 1999). In the loss-of-function mutants of CRC, carpels are smaller and unfused at their top (the so-called crab’s claw phenotype); siliques are also shorter (Alvarez and Smyth, 1999; Prunet et al., 2008).
Zinc fingers family
The SUPERMAN (SUP) gene encodes a C2H2-type zinc finger protein, which has a conserved function for the stamen and fruit development in plants. The Arabidopsis sup mutant showed low fertility, few seeds per silique and short silique (Zhao et al., 2014). Plant-specific DOF-type (DNA-binding with one finger) transcription factors control various biological processes, of which the Arabidopsis dof4.2 mutant showed increased silique length and seed yield (Zou et al., 2013). The Arabidopsis NO TRANSMITTING TRACT (NTT) gene encodes a C2H2/C2HC zinc finger transcription factor that is specifically expressed in the transmitting tract and responsible for replum development (Chung et al., 2013). NTT loss function in Arabidopsis leads to reduced male fertility, seed set and silique length (Marsch-Martínez et al., 2014).
Elongation factors
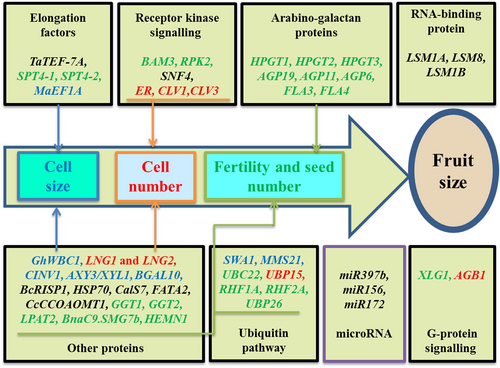
Eukaryotic elongation factors can be divided into transcript and translation elongation factors according to their respective roles in biological processes. Recently, several genes belonging to the elongation factors have been shown to play a role in fruit size regulation (Figure 4; Table 1).
The transcript elongation factors (TEFs) can facilitate efficient mRNA synthesis and perform diverse functions during transcription (including the modification of histone and RNA polymerase II activity), which can regulate growth and development by participating in various processes (Van Lijsebettens and Grasser, 2014). Overexpression of the wheat TaTEF-7A gene in Arabidopsis has multiple effects on asexual and reproductive traits, including increased silique number and length as well as grain length (Zheng et al., 2014). The heterodimeric SPT4/SPT5 complex is a TEF that interacts with RNA polymerase II to regulate mRNA synthesis in the chromatin context. Each subunit is encoded by two genes in Arabidipsis, and the RNAi-mediated down-regulation of SPT4-1/2 showed reduced cell elongation and vegetative and reproductive defects, including short roots and stems, small leaves, flowers and siliques with fewer seeds (Dürr et al., 2014).
Plant translation elongation factor 1 alpha (EF1A) is not only involved in protein synthesis but also a core part of plant protein trafficking, signal transduction, immune responses and apoptosis. Overexpression of banana MaEF1A in Arabidopsis greatly increased plant height, root length, as well as rhachis and silique length by promoting cell expansion and elongation (Liu et al., 2016).
MicroRNA
The microRNAs (miRNAs), which are about 21 nucleotides (nt) in length, are key components within the gene regulatory networks of eukaryotes (Bao et al., 2014). In recent years, several miRNAs have been shown to involve in fruit size regulation across several plant species (Figure 4; Table 1). The overexpression of orange Pt-miR156a in Arabidopsis leaded to late flowering, short siliques and small leaves (Wang et al., 2017a). Similarly, the overexpression of apple Md-miRNA156 in Arabidopsis resulted in late flowering, dwarfism, more leaves, short siliques with partially aborted seeds, by down-regulating its target SPL genes (Sun et al., 2013). As the downstream of FUL and ARF6/8, the miR172 plays a key role in regulating fruit development, as it is required for valve growth by restricting AP2 and TOE3 activity (José Ripoll et al., 2015). The reduced lignin deposition and increased silique number/length and seed size/yield was observed in transgenic Arabidopsis overexpressing miR397b, which regulated a laccase gene LAC4 that can polymerize monolignols into lignin (Wang et al., 2014). In peanut, a small RNA profiling and degradome analysis revealed several active modules during early pod development, including AP2 (miR172) and GRF (miR396), NAC (miR164), PPRP (miR167/miR1088), SPL (miR156/157), respectively (Gao et al., 2017).
Ubiquitin-proteasome pathway
Ubiquitin-specific protease (UBP) is a highly conserved protein family in eukaryotes, which plays an important role in protein de-ubiquitination (Liu et al., 2008b). Of these, the Arabidopsis ubp15 mutants have defect in cell proliferation and display late flowering and short root, stem, leaf, flower and fruit, as well as reduced fertility, whilst its overexpression shows opposite phenotypes (Liu et al., 2008b). The Arabidopsis UBP26 is essential for heterochromatin silencing by catalysing the de-ubiquitination of histone H2B, and its T-DNA insertion mutant showed short siliques, reduced fertility and few seeds (Luo et al., 2008).
The Arabidopsis ubiquitin-conjugating enzyme 22 (UBC22) is the only member of the Arabidopsis E2 subfamily, which can catalyse ubiquitin dimer formation in vitro in a Lys11-dependent manner. The knockout mutants of UBC22 in Arabidopsis had reduced silique length and seed number per silique, which was caused by ovule sterility due to severe defects in embryo sac that often contained no gamete nuclei (Wang et al., 2016).
The Arabidopsis SWA1 encodes a transducin family nucleolar protein that is involved in the E3 ubiquitin ligase complex, which is required for the normal progression of mitotic division cycles by regulating cell metabolism. The SWA1 mutation causes ovule abortion, and short silique in Arabidopsis (Shi et al., 2005). The RING-H2 group F1a (RHF1a) and RHF2a encode two RING-finger E3 ubiquitin ligases, of which RHF1a can directly interact with a cyclin-dependent kinase inhibitor ICK4/KRP6, leading to proteasome-mediated degradation. The rhf1a rhf2a double mutant showed reduced fertility and short siliques, which was caused by the defect in the gametophytes formation due to arrested mitotic cell cycle (Liu et al., 2008a). The Arabidopsis MMS21 encodes a SUMO E3 ligase, which is highly conserved in eukaryotes and essential for DNA repair and chromosome stability (Liu et al., 2014a). The mutation of MMS21 in Arabidopsis caused dwarfism and semi-sterility with short siliques and few seeds per silique, due to defect in gametogenesis (Liu et al., 2014a). The Arabidopsis STERILE APETALA (SAP) encodes an F-box protein that is involved in SCF (Skp1/Cullin/F-box) E3 ubiquitin ligase complex, which can promote meristemoid cells proliferation to control organ size by interacting with and targeting PPD proteins for degradation (Wang et al., 2016). The sap mutant displayed small leaves, flowers and siliques, due to decreased cell number in these organs.
The Arabidopsis DA1 encodes an ubiquitin-activated peptidase that acts as a putative ubiquitin receptor and functions cooperatively with the E3 ubiquitin ligases DA2 and EOD1/BB to negatively regulate organ size by restricting cell proliferation. The mutation of DA1 in Arabidopsis showed thicker stems, larger leaves and flowers, wider siliques and higher seed weight and yield due to increased cell numbers in these organs (Li et al., 2008).
G-protein signalling
The heterotrimeric GTP-binding proteins (G proteins) are highly conservative signalling components in eukaryotes, which consist of three subunits—Gα, Gβ and Gγ. In Arabidopsis thaliana, there is one canonical Gα (GPA1), one Gβ (AGB1) and three Gγ genes (AGGs). In addition to GPA1, there are three Gα-like genes (named XLGs) in Arabidopsis, which can interact with E3 ligases PUB4 and PUB2 and act in cytokinin signalling (Wang et al., 2017c). The xlg1/2/3 triple knockout mutant showed reduced male fertility and short siliques in Arabidopsis (https://www.arabidopsis.org/). The Arabidopsis AGB1 gene is involved in the regulation of organ shape, and its mutation increased the width but decreased the length of leaf, flower and silique, by promoting cell proliferation (Lease et al., 2001).
Receptor kinase signalling
The fruit size is largely determined by the classical CLAVATA–WUSCHEL (CLV-WUS) pathway, firstly identified in Arabidopsis and appeared to be conserved in other higher plants (Somssich et al., 2016), which can control locule number by regulating the size of shoot apical meristem (SAM). This pathway included three CLAVATA genes in Arabidopsis, of which CLV1 encodes a leucine-rich repeat receptor-like kinase (LRR-RLK), CLV2 encodes an LRR receptor-like protein (RLP) that lacks a kinase domain, and CLV3 encodes a stem cell-specific protein that can be further processed into a 12–amino acid peptide ligand for the CLV1 receptor (Kitagawa and Jackson, 2019). The loss-of-function clv mutants in Arabidopsis showed similar phenotypic changes, including larger inflorescence meristem, leading to fasciated stems and multilocular siliques. The BAM1 (derived from barely any meristem 1), BAM2, and BAM3 encode CLV1-related receptor-like kinases, which are required for ovule specification and male gametophyte development (DeYoung et al., 2006). The double and triple mutants of BAM1/2/3 genes displayed many defects in organ development, including reduced fertility and smaller leaf, flower and silique (DeYoung et al., 2006). Recently, the BjLn1 and BjMc1 gene (responsible for multilocular siliques) were successfully cloned, which was caused by the natural mutations in the CLV1 orthologues, respectively, on the A and B genome of Brassica juncea (Xiao et al., 2018; Xu et al., 2017). In recent years, the CLV-WUS pathway has become an attractive target of genome editing for crop improvement (Rodriguez-Leal et al., 2017), which has produced a series of exciting achievements. The genome editing of CLV3 orthologues can produce multilocular siliques in several dry fruit crops, including Brassica rapa and B. napus (Fan et al., 2014; Yang et al., 2018). In addition, the genome editing of CLV1/2/3 also produced multiple locular fruits with increased size in fleshy fruit crops, such as tomato and groundcherry (Lemmon et al., 2018; Li et al., 2018; Rodrı´guez-Leal et al., 2017; Xu et al., 2015; Zsögön et al., 2018).
In addition to the known CLV-WUS pathway, several genes encoding receptor kinase are also shown to involve in the fruit size regulation. As a plant-specific subunit of the SNF1-related protein kinase 1 complex, the Arabidopsis KINβγ is required for the pollen germination on the stigma surface (Gao et al., 2016), and its mutant had short stature and silique (https://www.arabidopsis.org/). ERECTA (ER) encodes receptor protein kinases containing a cytoplasmic protein kinase catalytic domain, an extracellular leucine-rich repeat, and a transmembrane region, which is required for the specification of organs originating from shoot apical meristem. The er mutant showed reduced plant height, increased inflorescence thickness and shorter but wider silique (Torii et al., 1996). The RECEPTOR-LIKE PROTEIN KINASE2 (RPK2) encodes a leucine-rich repeat receptor-like kinase, which functions as a regulator of meristem maintenance. The rpk2 mutants caused male sterility with more and short siliques failed to produce seeds due to defects in anther dehiscence and pollen maturation (Mizuno et al., 2007).
Arabinogalactan proteins
Arabinogalactan proteins (AGPs) are plant-specific extracellular glycoproteins, which are involved in various processes in growth and development, including cell division, expansion and differentiation (Pereira et al., 2015). Arabinogalactan proteins are classified into several types: classical AGPs, lysine-rich AGPs, AGP peptides, fasciclin-like AGPs and chimeric AGPs.
The Arabidopsis AGP6 and AGP11 encode classical AGPs, both of which are specifically expressed in stamens, pollen grains and tubes and required for male reproductive function. Loss of function in AGP6 and AGP11 caused reduced pollen tube growth, leading to lower male fertility, fewer seeds per silique and shorter silique (Levitin et al., 2008).The Arabidopsis AGP19 encodes a lysine-rich AGP, which was highly expressed elongating siliques and floral buds, moderately expressed in young seedlings and lowly expressed in roots and leaves (Yang et al., 2007). The T-DNA knockout mutant of AGP19 displayed multiple defects, including slow growth and reduced fertility, fewer and shorter siliques and fewer seeds per silique. The Arabidopsis FLA3 encodes a fasciclin-like AGP, which is involved in embryogenesis and microspore development. Both the Arabidopsis FLA3 RNAi and overexpression transgenic plants showed reduced fertility, fewer seeds and shorter siliques (Li et al., 2010). Similarly, the Arabidopsis FLA4 encodes a fasciclin-like AGP required for normal cell expansion, and its mutant showed short silique with fewer seeds per silique (Shi et al., 2003). The Arabidopsis HPGT1, HPGT2 and HPGT3 encode hydroxyproline O-galactosyltransferases that are required for AGPs biosynthesis (Ogawa-Ohnishi and Matsubayashi, 2015). The loss-of-function hpgt1, hpgt2, hpgt3 mutant exhibited reduced growth in vegetative organs, including small leaves, short stems and siliques.
RNA-binding proteins
RNA-binding proteins can bind to RNA molecules and play an important role in the post-transcriptional regulation of RNAs (Hentze et al., 2018), which are known to involve in plant growth, development and stress response (Lee and Kang, 2016). The SM-like proteins (LSMs) are a large family of RNA-binding proteins that involve in multiple aspects of RNA metabolism. The Arabidopsis contains 11 genes that encode the eight highly conserved SM-like proteins in yeast and animals (Perea-Resa et al., 2012). Of which, the LSM1A, LSM1B and LSM8 mutant plants showed severe development defects, including smaller leaves and shorter siliques with fewer seeds, which was caused by altering development-related gene expression through the regulation of mRNA splicing and decay (Perea-Resa et al., 2012).
Other proteins
The heat shock proteins (HSPs) functions as molecular chaperones to maintain cellular homeostasis and facilitate plants to adapt to environmental stimuli. Of the Arabidopsis HSP70 family, the knockout/down mutants of hsp70-1/4 and hsp70-2/4/5 displayed multiple phenotypic changes, including accelerated development, smaller leaves, thinner stems and shorter siliques (Leng et al., 2016). The Arabidopsis CINV1 encodes alkaline/neutral invertase that breaks sucrose down into fructose and glucose, which is highly expressed in leaf vasculature, shoot stipules, root tip and vascular cylinder, playing multiple roles in plant development. The EMS mutant of CINV1 in Arabidopsis showed earlier floral transition, smaller rosette leaves and siliques (Qi et al., 2007). The Caffeoyl-CoA 3-O-methyltransferase is a key enzyme in the lignin biosynthetic pathway, which is encoded by CCoAOMT genes found in many plant species. The overexpression of jute CCoAOMT1 gene in Arabidopsis led to taller plants and longer siliques, as well as higher lignin content (Zhang et al., 2014). The WBC1 encodes an ATP-binding cassette transporter of the white/brown complex subfamily. Overexpression of the cotton GhWBC1 gene in Arabidopsis led to 13% transformants producing short siliques with shrivelled embryos and few seeds (Zhu et al., 2003). The Arabidopsis LONGIFOLIA1 (LNG1) and LNG2 encode novel proteins that can regulate longitudinal cell elongation, which is expressed in various tissues (Lee et al., 2006). The overexpression and loss-of-function of LNG1/LNG2 in Arabidopsis could, respectively, increase and decrease the length of many organs, including leaves, flowers, siliques and seeds. The Arabidopsis AXY3/XYL1 encodes a bifunctional alpha-l-arabinofuranosidase/beta-d-xylosidase that belongs to glycoside hydrolases family 3, which can affect the structure and accessibility of the hemicellulose xyloglucan in cell walls. The mutations in axy3 led to reduced silique length and seed number per silique, likely due to the altered xyloglucan metabolism and cell wall structure (Günl and Pauly, 2011). The Arabidopsis BGAL10 encodes a member of glycoside hydrolase family 35, whose expression pattern and functions are similar to XYL1. The bgal10 mutant displayed unusual xyloglucan subunits and growth defects, especially shorter sepals and siliques (Sampedro et al., 2012). The Arabidopsis CALS7 encodes a phloem-specific callose synthase 7 required for callose deposition, which is specifically expressed in the phloem of vascular tissues (Xie et al., 2011). The T-DNA insertion cals7 mutants showed growth and reproduction defects, including short root, stem and silique as well as reduced male fertility and seed number per silique (Xie et al., 2011) The Arabidopsis FATA2 gene encodes Acyl-ACP thioesterase involved in fatty acid biosynthetic process, which was expressed d in roots, seedlings, leaves, stems, flowers, with especially high abundance in siliques. The fata2 T-DNA insertion mutants produced longer siliques with more but smaller seeds as well as increased oil content (Wang et al., 2013). Lysophosphatidyl acyltransferase (LPAT) is a key enzyme for adjusting the metabolic conversion of lysophosphatidic acid into exclusive phosphatidic acids in numerous tissues (Kim et al., 2005). The Arabidopsis LPAT2 is ubiquitously expressed in diverse tissues and is required for female but not male gametophyte development. The homozygous lpat2 mutant was lethal, and its heterozygous mutant showed shorter siliques with aborted ovules (Kim et al., 2005). The Arabidopsis HEMN1 encodes coproporphyrinogen III oxidase involved in the tetrapyrrole biosynthesis pathway, which is mainly expressed in anthers, ovules and endosperm of developing seeds. The T-DNA insertion hemen1 mutant showed defects in gametophyte development, leading to reduced fertility, seed number per silique and silique length (Pratibha et al., 2017). The Arabidopsis GGT1 and GGT2 encode apoplastic gamma-glutamyl transferases responsible for glutathione degradation, of which GGT1 is expressed in the vascular system and inside leaves, whilst GGT2 is expressed in trichomes, seeds, and roots. The knockdown of GGT1/GGT2 by RNAi showed reduced vegetative growth rate (such as smaller leaves and roots) and lower seed yield due to fewer siliques with lower length (Giaretta et al., 2017). The Brassica cabbage RISP1 gene is highly homologous to the Arabidopsis AT5G13440, which encodes ubiquinol-cytochrome C reductase iron-sulphur subunit involved in mitochondrial electron transport. The overexpression of BcRISP1 in Arabidopsis showed reduced seed set and short siliques, which is caused by the reduced pollen formation and impaired pollen tube elongation due to the interruption of the mitochondrial electron transport chain by affecting the expression of mitochondrial breathing chain-related genes (Liu et al., 2014b). A major QTL qSS.C9 for seeds per silique has been cloned on the C9 chromosome of Brassica napus (Li et al., 2015), which encodes a predicted small protein with 119 amino acids homologous to the Arabidopsis SMG7 gene that is involved in meiotic cell cycle. The BnaC9.SMG7b was mainly expressed in the vascular tissue of various organs, including cotyledons, rosette leaves, roots, young pedicels and pistils, but not in stamens, petals, stems and mature siliques. Natural loss or artificial knockdown of BnaC9.SMG7b resulted in decreased seed number per silique, silique length and seed yield but increased seed weight, which was caused by the reduced ovule fertility due to the developmental defects in the formation of functional female gametophytes.
Conclusions and future prospects
In recent years, many genes that regulate fruit size have been identified, mostly from Arabidopsis and tomato (Table S1), the two model plants for fruit development studies. Although several reviews have discussed genetic and epigenetic regulation of fruit size in the fleshy fruit type tomato (Pesaresi et al., 2014; Seymour et al., 2013; Tanksley, 2004), current knowledge of fruit size control in the dry fruit type Arabidopsis and related crops is limited. Such information is essential if a comprehensive genome editing approach is to be applied. Some initial targets for fruit size have been targeted using editing approaches, with some success (Lemmon et al., 2018; Li et al., 2018; Rodríguez-Leal et al., 2017; Zsögön et al., 2018), suggesting that, with a more comprehensive understanding of the genetic and signalling pathways underlying this trait, major gains could be achieved. In addition to improving fruit size in elite germplasm, genome editing can be used for improvement of orphan crops (Lemmon et al., 2018) or de novo domestication of crop wild relatives (Zsögön et al., 2018), which often harbour agronomically valuable disease resistance traits (Dangl et al., 2013).
In this review, we summarized those genes that have been identified as regulating fruit size, with emphasis on their genetic and molecular mechanisms. In addition, we revealed the complex genetic regulatory networks for the first time (Figure 5), based on an examination of the current knowledge on fruit size control in Arabidopsis and other dry fruits. These include phytohormone (e.g. AUX, GA, CTK, ABA, ETH and BR), transcription factors (e.g. tri-helix, YABBY, AP2-ERF, MADS-box, bHLH, zinc finger, homeobox and B3), transcription/translation elongation factors, ubiquitin-proteasome and microRNA pathways, G-protein and receptor kinase signalling, AGPs, RNA-binding proteins (Figure S2) and indicate the complexity of fruit size regulation in plants. Interestingly, many of these genes have a conserved function in regulating fruit development in both dry and fleshy fruit types, suggesting broad applicability of common genome edits. For instance, CYP78A9 and CLV-WUS pathway genes can regulate both silique size in Arabidopsis/rapeseed (Fan et al., 2014; Shi et al., 2019; Xiao et al., 2018; Xu et al., 2017; Yang et al., 2018) and fruit size in cherry/tomato (Lemmon et al., 2018; Li et al., 2018; Qi et al., 2007; Rodriguez-Leal et al., 2017; Xu et al., 2015). Besides, many of these genes have similar functions in regulating fruit size in Arabidopsis and other crops. For example, ARF18, ARP1, BoMF2, DRM1, CYP78A9, CLV1, CLV3 and SMG7 can regulate silique length in Arabidopsis and rapeseed (Kang et al., 2014; Lee et al., 2013; Li et al., 2015; Liu et al., 2015; Shi et al., 2019; Xiao et al., 2018; Yang et al., 2018). More importantly, many of the fruit size regulatory genes have expression activity and additional effects on other organs, such as roots, stems, leaves, flowers and seeds, indicating a cooperative/synergistic regulation. These results strongly suggest the conserved function of these genes in regulating the size of different organs among plant species, and so can be targeted for molecular improvement of size for fruit and other plant organs through genome editing approaches such as CRISPR/Cas.
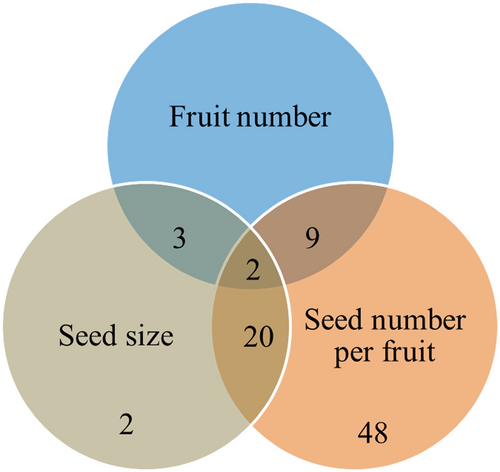
The most complicated issue in terms of regulatory networks is the relationship between fruit size and other seed yield components, including fruit number, seed number per fruit and seed size (Figure 6). Analyses of mutants with phenotypic changes in both fruit size and fruit number show an inverse relationship between the two characters/traits. This indicates a trade-off between them, which can be explained by the competition among sink organs and which is entirely consistent with many decades of agronomic research in crop plants. However, there are also some exceptions, such as miR397b, GGT1, GGT2 and TaTEF-7A can affect silique size and silique number per plant, in the same direction, which provides a unique opportunity to simultaneously improve both traits. Another important question involves the relationships between fruit size and seed number per fruit and seed size. Analyses of mutants with phenotypic changes in fruit size and seed number per fruit showed that changes in fertilization rate and seed number per fruit are usually associated with changes in fruit length, but the reverse is not true. For example, AHP1-5, BRI1, FATA2, GbAGL2, GhWBC1, GID1A, GRDP1, HEMN1, SWA1 and WOX14 are genes that control fruit size and seed number per fruit, always in the same direction. Therefore, the fertilization rate and seed number per fruit are likely upstream of fruit size. As expected, mutants with reduced fertility and seed number per fruit are usually accompanied by reductions in fruit growth/size, which indicates feedback or cross-talk between seed development and fruit growth. However, the relationship between fruit size and seed size is rather confused: in many cases (e.g. ANT, ARF18, ASIL1, CYP78A9, DA1, UBP15), they are regulated in the same direction; whereas in other cases (e.g. FATA2, AHP1-5, STURDY), they are affected in the opposite direction. Therefore, attention should be paid to the relationships between fruit size and fruit number and between seed number per fruit and seed size (i.e. the three components of seed yield) when selecting targets for editing.
Identification of these regulatory pathways is still in the early stages: the identified fruit size genes are limited, and only a little is known about the relationships between different genes within the same pathway and between pathways (such as phytohormone → signalling transduction/ubiquitin-proteasome degradation → transcription factors → response genes). The major goal for the future is to fully demonstrate the molecular mechanisms underlying fruit size regulation and construct its genetic networks. The application of modern biotechnologies, such as genome-wide association, genome editing, omics and bioinformatics, will accelerate the identification and confirmation of the fruit size regulators in plants and provide the basis for the accelerated improvement of these crops.
Acknowledgements
This research was supported by the National Key/Basic Research and Development Program (2016YFD0100305/2015CB150203), the Natural Science Foundation of Hubei Province (2018CFA075), Wuhan Youth Science and Technology Morning Project (2017050304010286), the Natural Science Foundation (31101181 and 31771840), the Rapeseed Industry Technology System (CARS-13), the Agricultural Science and Technology Innovation Project (CAAS-ASTIP-2013-OCRI), the Core Research Budget of the Non-profit Governmental Research Institution (1610172017001).
Conflict of interest
The authors declared that they have no conflict of interest.
Author Contributions
J.Q. Shi, H.Z. Wang, G.H. Liu and X.F. Wang jointly developed the conceptual structure of manuscript. Q. Hussain, J.Q. Shi and J.P Zhang collected the fruit size genes from the published literatures and analysed and integrated the relevant information. J.Q. Shi and Q. Hussain wrote the manuscript, including all the figures and tables. D. Edwards, G.J. King, A. Scheben and G.J. Yan provided a critical feedback and revised the manuscript.




