Hydrogen peroxide production and myo-inositol metabolism as important traits for virulence of Mycoplasma hyopneumoniae
Summary
Mycoplasma hyopneumoniae is the causative agent of enzootic pneumonia. In our previous work, we reconstructed the metabolic models of this species along with two other mycoplasmas from the respiratory tract of swine: Mycoplasma hyorhinis, considered less pathogenic but which nonetheless causes disease and Mycoplasma flocculare, a commensal bacterium. We identified metabolic differences that partially explained their different levels of pathogenicity. One important trait was the production of hydrogen peroxide from the glycerol metabolism only in the pathogenic species. Another important feature was a pathway for the metabolism of myo-inositol in M. hyopneumoniae. Here, we tested these traits to understand their relation to the different levels of pathogenicity, comparing not only the species but also pathogenic and attenuated strains of M. hyopneumoniae. Regarding the myo-inositol metabolism, we show that only M. hyopneumoniae assimilated this carbohydrate and remained viable when myo-inositol was the primary energy source. Strikingly, only the two pathogenic strains of M. hyopneumoniae produced hydrogen peroxide in complex medium. We also show that this production was dependent on the presence of glycerol. Although further functional tests are needed, we present in this work two interesting metabolic traits of M. hyopneumoniae that might be directly related to its enhanced virulence.
Graphical Abstract
Virulence and pathogenicity of Mycoplasma hyopneumoniae, the causative agent of enzootic pneumonia, has never been fully understood. We present in this work two interesting metabolic traits of M. hyopneumoniae that might be directly related to its enhanced virulence, especially regarding its ability to produce hydrogen peroxide.
Introduction
The notion that the lungs are sterile is frequently stated in textbooks; however, no modern studies have provided evidence for the absence of microorganisms in this environment (Dickson and Huffnagle, 2015). Several bacteria colonize the respiratory tract of swine. Mycoplasma hyopneumoniae, Mycoplasma flocculare and Mycoplasma hyorhinis are some of the most important species identified so far (Mare and Switzer, 1965; Meyling and Friis, 1972; Rose et al., 1979; Siqueira et al., 2017).
M. hyopneumoniae is widespread in pig populations and is the causative agent of enzootic pneumonia (Maes et al., 1996); M. hyorhinis, although not as pathogenic as M. hyopneumoniae, has already been found as the sole causative agent of pneumonia, polyserositis and arthritis in pigs (Kobisch and Friis, 1996; Davenport et al., 1970; Thacker et al., 2012; Whittlestone, 2012). M. flocculare, on the other hand, has high prevalence in swine herds worldwide, but up to now, is still considered a commensal bacterium (Kobisch and Friis, 1996).
Because of the genomic resemblance of these three Mycoplasma species (Stemke et al., 1992; Siqueira et al., 2013), it remains unclear why M. hyopneumoniae can become highly virulent if compared with the other two. It is also essential to understand that the simple presence or absence of each species is not in itself a determinant factor in the development of enzootic pneumonia: most piglets are thought to be vertically infected with M. hyopneumoniae at birth (Maes et al., 1996; Fano et al., 2006; Sibila et al., 2009) and many can become carriers of the pathogen throughout their entire life without developing acute pneumonia. Moreover, M. hyopneumoniae also persists longer in the respiratory tract, either in healthy animals or even after successful treatment of the disease (Thacker et al., 2001; Ruiz et al., 2002; Fano et al., 2005; Overesch and Kuhnert, 2017).
To make it even more complex, different strains of each species bear different levels (or even lack) of pathogenicity. For instance, M. hyopneumoniae has six sequenced strains, two of which are known to be attenuated by culture passages (Zielinski and Ross, 1990; Liu et al., 2013). These strains cannot cause the clinical symptoms of pneumonia in vivo and up to now it is not clear why.
In contrast to other pathogenic bacteria, and as revealed by the analysis of the sequenced genomes from several mycoplasmas (Himmelreich et al., 1996; Chambaud et al., 2001; Minion et al., 2004; Vasconcelos et al., 2005 Siqueira et al., 2013), pathogenic Mycoplasma species seem to lack typical primary virulence factors such as toxins, invasins and cytolysins (Pilo et al., 2005; Maes et al., 2017). For this reason, classical concepts of virulence genes are usually problematic and a broader concept for virulence is used for these species. In this way, a virulence gene in mycoplasmas is described as any nonessential gene for in vitro conventional growth, which is essential for the optimal survival (colonization, persistence or pathology) inside the host (Browning et al., 2014).
There have been many different types of virulence factors described so far in several Mycoplasma species, most of them related to adhesion (Razin and Jacobs, 1992), invasion (Sibylle et al., 2015), cytotoxicity (Vilei and Frey, 2001; Hames et al., 2009), host-evasion (Simmons and Dybvig, 2007) and host-immunomodulation (Katz et al., 1983; Waites and Talkington, 2004). As for M. hyopneumoniae and M. hyorhinis, adhesion factors such as antigen surface proteins and the ability of these organisms to produce a capsular polysaccharide have already been described in the literature (Tajima and Yagihashi, 1982; Citti et al., 1997; Djordjevic et al., 2004; Seymour et al., 2012; Whittlestone, 2012).
However, while the diseases caused by these swine mycoplasmas have been extensively studied, only recently their metabolism has been explored from a mathematical and computational point of view by our group (Ferrarini et al., 2016). We are well aware that metabolism does not fully explain the pathologies caused by either of them. However, adhesion proteins, classically related to virulence in mycoplasmas cannot be associated with the different levels of pathogenicity between M. hyopneumoniae and M. flocculare. Both species harbor similar sets of adhesion proteins (Siqueira et al., 2014) and have been shown to adhere to cilia in a similar way (Young et al., 2000). Thus, it remains unclear what prevents M. flocculare to cause disease in this context.
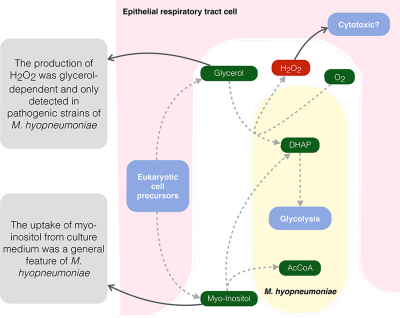
In our previous work (Ferrarini et al., 2016), we compared the reconstructed metabolic models of these three Mycoplasma species, and pointed out important metabolic differences that could partly explain the different levels of pathogenicity between the three species. The most important trait was related to the glycerol metabolism, more specifically the turnover of glycerol-3-phosphate into dihydroxyacetone-phosphate (DHAP) by the action of glycerol-3-phosphate oxidase (GlpO, EC 1.1.3.21), which was only present in the genomes of M. hyorhinis and M. hyopneumoniae. This would allow the usage of glycerol as a primary energy source, with the production of highly toxic hydrogen peroxide in the presence of molecular oxygen. The metabolism of glycerol and the subsequent production of hydrogen peroxide by the action of GlpO are essential for the cytotoxicity of lung pathogens Mycoplasma pneumoniae (Hames et al., 2009) and Mycoplasma mycoides subsp. mycoides (Vilei and Frey, 2001). Moreover, the Mycoplasma hominis group is not the only one where hydrogen peroxide production via glpO has been reported. In some Spiroplasma species (specifically Spiroplasma taiwanense) and within the pneumoniae group (for instance in Mycoplasma penetrans), the presence of this enzyme was also associated with virulence (Kannan and Baseman, 2000; Lo et al., 2013).
Another major difference between our previous models was related to the presence of a complete transcriptional unit (TU) encoding proteins for the uptake and metabolism of myo-inositol in M. hyopneumoniae (with the exception of one enzyme). This could be another important trait for the enhanced virulence of this species if compared with the other two. Here, we studied this pathway in more detail to try to find this missing enzyme and the possible reasons as to why natural selection kept these genes only in this Mycoplasma species.
In a recent review, Maes et al. (2017) emphasize the need for the further investigation of the role of glycerol and myo-inositol metabolism and their contribution to virulence in M. hyopneumoniae. Here, we experimentally tested these two traits to show how they might be related to the different levels of pathogenicity, by comparing not only the species themselves but different strains of M. hyopneumoniae. Contrary to what we anticipated, only the two pathogenic strains of M. hyopneumoniae were able to produce hydrogen peroxide in complex medium, and we confirmed that this production was dependent on the presence of glycerol. The myo-inositol metabolism, in turn, was tested with the aid of deuterated myo-inositol in Friis medium. We were able to detect by mass spectrometry (MS) a slight decrease in the marked myo-inositol concentration throughout time, indicating the ability of M. hyopneumoniae to uptake such carbohydrate. We also show here that only the M. hyopneumoniae strains remained viable when myo-inositol was the primary energy source.
We present here two metabolic traits specific to M. hyopneumoniae that might be directly related to its enhanced virulence, especially in its ability to successfully overgrow the other two Mycoplasma species in the respiratory tract of swine, persist longer in this environment and possibly cause disease.
Results
Comparative genomics of glpO from glycerol metabolism
Highly conserved homolog genes to glpO from M. mycoides subsp. mycoides (EC 1.1.3.21) were found only in the genomes of M. hyopneumoniae and M. hyorhinis. Despite the annotation as a dehydrogenase in both M. hyopneumoniae and M. hyorhinis, we propose this enzyme to act as glycerol-3-phosphate oxidase (GlpO), using molecular oxygen as the final electron acceptor and producing DHAP and hydrogen peroxide. We therefore refer to the encoded protein in M. hyopneumoniae and M. hyorhinis as GlpO, rather than GlpD. The high similarity between these predicted proteins (Supporting Information Fig. S1A) may be an indication that this trait might be essential for the pathogenicity of these Mycoplasma species.
Particularly, the cytotoxicity of M. mycoides subsp. mycoides is considered to be related to the translocation of the hydrogen peroxide into the host cells (Bischof et al., 2008). This is presumably possible because of the close proximity to the host cells along with the integral membrane GlpO (Pilo et al., 2005, 2007). Different transmembrane prediction softwares (Hofmann, 1993; Combet et al., 2000; Krogh et al., 2001; Kahsay et al., 2005) identified putative transmembrane portions in the GlpO proteins from M. hyopneumoniae and M. hyorhinis (Supporting Information Fig. S1B). Similar results were reported for the homolog enzyme in M. mycoides subsp. mycoides (Pilo et al., 2005), and a recent proteomic study has detected GlpO from M. hyopneumoniae in surface-enriched extracts through LC-MS/MS (Personal communication from H. B. Ferreira, (Machado et al., 2018).
Pathogenic M. hyopneumoniae strains produce hydrogen peroxide from glycerol
Contrary to what we had anticipated, we were only able to detect the production of hydrogen peroxide from the two pathogenic strains of M. hyopneumoniae (7448 and 7422) in Friis medium, as can be seen in Fig. 1A. The attenuated strain from the same species (M. hyopneumoniae strain J), along with M. hyorhinis and M. flocculare did not produce detectable quantities of this toxic product. In order to verify if the amount of hydrogen peroxide produced was comparable between strains, we also counted the number of cells for each replicate. In this way, the two pathogenic strains produced approximately the same amount of hydrogen peroxide and had cell counts of the same order of magnitude (available in Supporting Information Table S1).
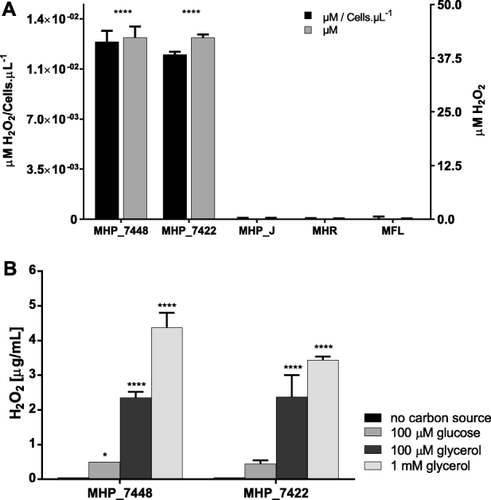
Hydrogen peroxide production by swine mycoplasmas.
A. In Friis medium after bacterial growth: Hydrogen peroxide was only detected in growth media from pathogenic strains (field isolates) of M. hyopneumoniae 7448 (MHP_7448) and 7422 (MHP_7422). Neither the attenuated strain J (MHP_J) nor the other species M. hyorhinis (MHR) and M. flocculare (MFL) produced detectable amounts of this toxic product. The concentration was also standardized based on the average number of cells from each culture. Data are presented as mean and standard deviation of three independent samples and statistical analysis was performed considering M. flocculare as a control strain (since it lacks the glpO gene).
B. In the presence of different carbon sources: Pathogenic M. hyopneumoniae strains were used to test hydrogen peroxide production in incubation buffer supplemented with either glycerol or glucose after 2 h of incubation. Both strains were able to produce significant amounts of the toxic product when glycerol was present. Data are represented as mean and standard deviation of four independent samples (*p < 0.05; **** p < 0.0001).
We also show (Fig. 1B) that the hydrogen peroxide produced by the M. hyopneumoniae strains 7448 and 7422 was dependent on the presence of glycerol in the incubation buffer.
Levels of glpO transcripts do not differ from pathogenic to attenuated strains of M. hyopneumoniae
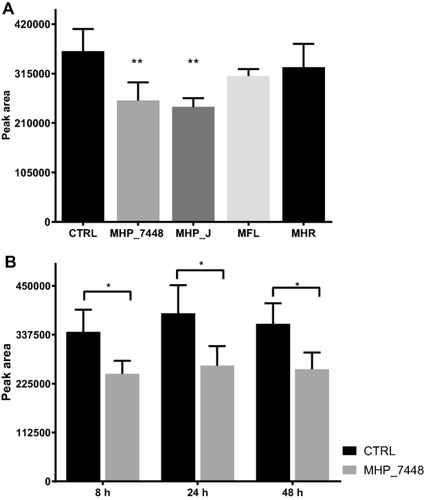
We tested the three M. hyopneumoniae strains (7448, 7422 and J) in order to compare the mRNA expression levels of glpO gene by RT-qPCR. Since the transcript levels of normalizer genes were not comparable between strains, we used relative quantification normalized against unit mass; in our case, the initial amount of RNA. We chose one of the replicates from strain 7448 as the calibrator, and we were able to show (Fig. 2 and Supporting Information Table S2) that there was no significant difference in the transcript levels of glpO in all tested strains from M. hyopneumoniae.
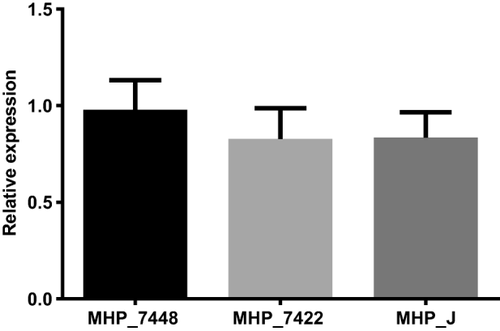
Expression levels of glpO gene in M. hyopneumoniae strains.
We did not find any significant difference on the transcript levels of glpO from all tested strains. Bars show the average relative quantification normalized against unit mass (500 ng of total RNA) and replicate 2 from strain 7448 was used as the calibrator. Average expression levels were calculated with independent biological triplicates (p < 0.05).
Enzymes for the uptake and catabolism of myo-inositol are specific to M. hyopneumoniae strains
M. hyopneumoniae is the only reported species among the Mollicutes that contains genes involved in the catabolism of myo-inositol. Since Mycoplasma species seem to maintain a minimum set of essential metabolic capabilities, we decided to further investigate this pathway and the influence of its presence on the metabolism and pathogenicity of M. hyopneumoniae. The degradation of inositol can feed glycolysis with DHAP and also produces an acetyl coenzyme-A (AcCoA) (Fig. 3). A TU for the myo-inositol catabolism is present in all M. hyopneumoniae strains, with the exception of the gene that codes for the enzyme 6-phospho-5-dehydro-2-deoxy-d-gluconate aldolase (IolJ, EC 4.1.2.29), responsible for the turnover of 6-phospho-5-dehydro-2-deoxy-d-gluconate (DKGP) into malonate semialdehyde (MSA).
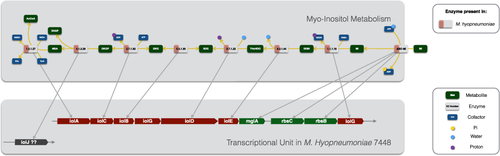
Myo-inositol catabolism pathway in all M. hyopneumoniae strains and its transcriptional unit in M. hyopneumoniae strain 7448.
Metabolites are depicted in dark green and enzymatic activities present in M. hyopneumoniae can be seen in pink. Metabolite abbreviations are as follows: MI (myo-inositol), 2KMI (2-keto-myo-inositol), THcHDO (3D-(3,5/4)-trihydroxycyclohexane-1,2-dione), 5DG (5-deoxy-d-glucuronate), DKG (2-deoxy-5-dehydro-d-gluconate), DKGP (6-phospho-5-dehydro-2-deoxy-d-gluconate), MSA (malonate semialdehyde), AcCoA (acetyl coenzyme-A), DHAP (dihydroxyacetone phosphate). EC 1.2.1.27: MSA dehydrogenase; EC 4.1.2.29: DKGP aldolase; EC 2.7.1.92: DKG kinase; EC 5.3.1.30: 5DG isomerase; EC: 3.7.1.22: THcHDO hydrolase; EC 4.2.1.44: Myo-inosose-2 dehydratase; EC 1.1.1.18: MI-2-dehydrogenase; ABC-MI: ABC transporter for myo-inositol.
The gene encoding IolJ in other organisms is similar to the one coding for enzyme fructose-bisphosphate aldolase (Fba) from glycolysis (EC 4.1.2.13). There are two annotated copies of the gene fba in M. hyopneumoniae (fba and fba-1, Supporting Information Table S3). We performed homology and gene context analyses (with the use of the MGcV software (Overmars et al., 2013)), 3D comparative modelling and protein–ligand interaction analysis to check if either of them would be a suitable candidate for this activity.
The gene context and protein sequence alignment for 15 selected Fba homologs in Mollicutes can be seen in Supporting Information Figs S2 and S3. Comparative models for both copies of Fba from M. hyopneumoniae and previously characterized IolJ and Fba from Bacillus subtilis (Yoshida et al., 2008) were constructed based on available structures of Fba in PDB (Berman et al., 2000) (Fig. 4 and Supporting Information Table S4). Fba structures from Escherichia coli and Giardia intestinalis were used to gather more information about substrate binding (Supporting Information Fig. S3). The alignment shows a highly conserved zinc binding site (residues marked as ‘*’), essential for substrate binding and catalysis. Positions ‘a’, ‘b’, ‘c’, ‘d’ and ‘e’ surround the substrate cavity. The structural analysis suggests that the interaction mode of DKGP (substrate of IolJ) with the zinc ion of the active site is similar to that observed for FBP (fructose-1,6-bisphosphate, substrate of Fba). Nevertheless the substrate specificity is strongly dependent on the residues that form the substrate cavity.
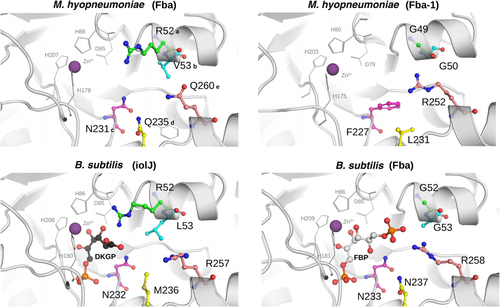
Substrate cavity prediction for Fba and Fba-1 from M. hyopneumoniae strain 7448.
Cavities from the comparative models of Fba and Fba-1 from M. hyopneumoniae in comparison to the models constructed for Fba and IolJ from B. subtilis. The specificity for DKPG in IolJ seems to be strongly associated to the presence of a conserved arginine in position ‘a’ (R52 in Fba-1 from M. hyopneumoniae). In contrast, Fbas generally bear glycines in this position (for complete explanation see Supporting Information Figs S3 and S4). While Fba-1 from M. hyopneumoniae resembles more the experimentally solved Fba enzymes from B. subtilis, E. coli and, G. intestinalis, the predicted structure of Fba from M. hyopneumoniae is more similar to the IolJ structure from B. subtilis.
While there seems to be several common features between Fba and IolJ (residues ‘c’, ‘d’, ‘e’ and ‘*’), residue ‘a’ appears to be essential for the substrate interaction with IolJ. This residue is generally occupied by an arginine (R52) in several putative IolJs from other organisms (Supporting Information Fig. S4), and absent in all predicted Fbas analysed in this study. From the predicted structures, the presence of this positively charged arginine in IolJ seems to disfavour the interaction with the phosphate group of FBP whilst it is complementary to the carboxyl group from DKGP.
In this way, the predicted structure of Fba-1 from M. hyopneumoniae resembles more the Fba structures from the experimentally solved Fbas in B. subtilis, E. coli and G. intestinalis. The annotated Fba from M. hyopneumoniae, on the other hand, seems to be more similar to the IolJ structure from B. subtilis. Although functional studies are needed to test this hypothesis, we propose that all enzymes needed for the myo-inositol catabolism are present in M. hyopneumoniae.
M. hyopneumoniae is able to uptake myo-inositol from the culture medium
In order to ascertain the ability of different bacteria to uptake myo-inositol, we used two different approaches. The first was the use of marked myo-inositol in complex medium and analysis by MS, and the second was to check the viability of cells (through ATP production) whenever myo-inositol was used as primary energy source.
When we tested if cells were able to uptake the marked myo-inositol, over the course of 48 h, we found no significant difference in M. flocculare and M. hyorhinis when compared to the control medium (CTRL), as observed in Fig. 5A. As expected, the concentrations of myo-inositol for both strains of M. hyopneumoniae after 48 h of growth were lower than the control medium. We also collected two extra time points for M. hyopneumoniae strain 7448 and CTRL: 8 h and 24 h of growth (Fig. 5B). In all time points, there is significant difference between the residual marked myo-inositol and the control medium, which implies that M. hyopneumoniae is able to uptake such carbohydrate from the medium. MS peak data is available in Supporting Information Table S5.
Deuterated myo-inositol-1,2,3,4,5,6-d6 uptake in complex medium.
A. Comparison after 48 h of growth of M. hyopneumoniae J ATCC 25934 (MHP_J) and field isolate 7448 (MHP_7448), M. flocculare ATCC 27716 (MFL) and M. hyorhinis ATCC 17981 (MHR). While there is no significant difference in the concentrations between MFL and MHR and the control medium (CTRL), both M. hyopneumoniae strains seem to be able to uptake myo-inositol.
B. We also collected two extra time points for MHP_7448 and CTRL: 8 h and 24 h of growth. In all time points there is significant difference between residual marked myo-inositol and the control medium. Data are presented as mean and standard deviation of four independent biological replicates. Asterisks indicate statistically significant differences in residual marked myo-inositol (*p < 0.05; **p < 0.01).
Since we had glucose and glycerol present in this complex medium analysed by MS, we also wanted to check if the viability of the different strains and species altered when myo-inositol was the primary energy source. For this, we incubated cells in myo-inositol defined medium (depleted with glucose and glycerol) for 8 hours and measured the amount of ATP these cells were able to produce. This in turn was directly related to the amount of viable cells after cultivation in the specific medium tested. Considering we do not know the energetic yield and efficiency of each strain and species, we could not directly compare the amount of ATP produced between different organisms. For this reason, growth in regular defined medium (with glucose) for each strain was used as a normalization control and the ratio of ATP production in both media was used to compare the viable cells between strains. Since there was no other energy source available in the medium and in accordance with our previous predictions and results, only M. hyopneumoniae cells remained viable (ranging from 75% to 280%) when compared to their control growth in the regular defined medium (Fig. 6 and Supporting Information Table S6). The viability of the other species in this medium was 11.5% for M. hyorhinis and 0.2% for M. flocculare. We also achieved similar results when comparing the growth in myo-inositol defined medium versus Friis medium (Supporting Information Fig. S5).
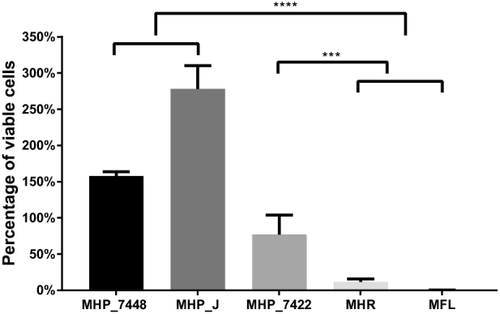
Viability of M. hyopneumoniae, M. hyorhinis and M. flocculare after 8 hours of incubation in myo-inositol defined medium.
The viability of cells in myo-inositol defined medium was measured by ATP production in comparison to inoculation in regular defined medium (glucose-containing medium). Data are represented as the ratio between ATP production in each media. There is a significant decrease of ATP production in M. hyorhinis and M. flocculare whereas at least 75% of the cells from M. hyopneumoniae remained viable after cultivation in the myo-inositol defined medium (***p < 0.001; ****p < 0.0001).
Discussion
In this study, we wanted to find possible differences between pathogenic and attenuated strains of M. hyopneumoniae and also compare them with M. hyorhinis and M. flocculare and assess possible links to the enhanced virulence of M. hyopneumoniae. While M. hyopneumoniae strains 7422 and 7448 are considered pathogenic, strain J became attenuated after serial passages of in vitro culture; M. hyorhinis strain ATCC 17981 was isolated from swine but, to our knowledge, its level of pathogenicity has not been tested in vivo; and even though M. flocculare is not considered pathogenic, strain ATCC 27399 was isolated from a case of swine pneumonia (strain ATCC 27716 is derived from this strain). In our previous study (Ferrarini et al., 2016), through mathematical modelling, we predicted two traits of M. hyopneumoniae in silico that could be associated with its enhanced virulence: the myo-inositol catabolism and the link between the glycerol and the glycolysis metabolism, with the production of highly toxic hydrogen peroxide (by the activity of the GlpO enzyme). In this work, we tested whether these species indeed differed from each other regarding their ability (i) to produce hydrogen peroxide in vitro and whether this was related to the availability of glycerol, (ii) to uptake myo-inositol and (iii) to remain viable in a defined medium with myo-inositol as the primary energy source. While the uptake of myo-inositol might be a general feature of M. hyopneumoniae, the production of hydrogen peroxide in complex medium seems to be specific to pathogenic strains of this species.
Glycerol metabolism and hydrogen peroxide production
Even though the GlpO enzyme was previously detected in proteomes from both pathogenic and attenuated strains of M. hyopneumoniae (232 and J) (Pinto et al., 2009; Pendarvis et al., 2014), only the pathogenic strains tested in our study (7448 and 7422) were able to produce detectable amounts of hydrogen peroxide in Friis medium (Fig. 1). To our knowledge, no other study up to now was able to show that M. hyopneumoniae strains were able to produce this toxic product in vitro (Maes et al., 2017). We also show here that the production of hydrogen peroxide in the pathogenic strains of M. hyopneumoniae is dependent on the presence of glycerol (Fig. 1B).
The metabolism of glycerol and the formation of hydrogen peroxide were described as essential for the cytotoxicity of lung pathogens M. mycoides subsp. mycoides (Vilei and Frey, 2001) and M. pneumoniae (Hames et al., 2009). Moreover, although both M. hyopneumoniae and M. flocculare can adhere to the cilia of tracheal epithelial cells in a similar way, only the adhesion of M. hyopneumoniae causes tissue damage (Young et al., 2000).
We showed that the difference in enzyme activity was not related to the expression levels of glpO gene from the strains tested (Fig. 2). We did not find any extreme differences in their aminoacid sequences either (Supporting Information Fig. S3). This could be an indication that either this enzyme undergoes post-translational modifications in order to be active and/or the availability of the substrate (glycerol) intracellularly might be a limiting step for its activity. Post-translational modifications have been extensively reported experimentally in several proteins of M. hyopneumoniae (Djordjevic et al., 2004; Burnett et al., 2006; Pinto et al., 2007; Seymour et al., 2010; Tacchi et al., 2016). From transcriptomic and proteomic literature data, we were not able to find any enlightening differences in this pathway between strains or species (Supporting Information Table S7).
As for the availability of intracellular glycerol, in our previous metabolic models, we predicted differences in the metabolism of glycerol among the three Mycoplasma species (Supporting Information Fig. S6). While M. hyopneumoniae has five different ways of uptaking glycerol (dehydrogenation of glyceraldehyde, ABC transport of glycerol and glycerol-phosphate, import of glycerophosphoglycerol and glycerophosphocholine), the other two species lack at least two reactions. This might also limit the rate of production of hydrogen peroxide in each species. In this way, the enhanced pathogenicity of M. hyopneumoniae over M. hyorhinis and M. flocculare may therefore also be due to hydrogen peroxide formation resulting from a higher uptake of glycerol as an energy source. Similarly, one reason that could partially explain why M. mycoides subsp. mycoides is highly pathogenic in comparison with the less pathogenic M. pneumoniae might be the greater intracellular availability of glycerol due to the presence of a specific and very efficient ABC transporter in M. mycoides subsp. mycoides (Hames et al., 2009).
Since the production of hydrogen peroxide was not reported as essential to the in vivo virulence of Mycoplasma gallisepticum (Szczepanek et al., 2014), more studies are needed to better understand the importance of this metabolism in M. hyopneumoniae. Moreover, future biochemical and functional studies are needed to prove that GlpO is indeed responsible for the activity proposed here and to check if the enzyme in attenuated strains/species is functional.
Myo-inositol uptake and catabolism
M. hyopneumoniae is the only Mycoplasma species with sequenced genome that has the genes for the catabolism of myo-inositol. Myo-inositol is an essential precursor for the production of inositol phosphates and inositol phospholipids in all eukaryotes (Gonzalez-Salgado et al., 2012). Myo-inositol is also widespread in the bloodstream of mammalians (Reynolds, 2009), which would make it a suitable energy source for bacteria in the extremely vascularized respiratory system. Previously, Mycoplasma iguanae was described to produce acid from inositol (Brown et al., 2006), but the methods used in that paper are not clear and there is no complete genome from this organism for us to draw any conclusions. Based on sequence homology, orthology, synteny and tridimensional analyses, we proposed a possible candidate for the missing enzyme IolJ in M. hyopneumoniae, namely a duplication of the fba gene from glycolysis. This functional divergence after duplication is particularly interesting in bacteria for which evolution was mostly driven by genome reduction. Another reported example of this event is the duplication of the trmFO gene in Mycoplasma capricolum and more recently in Mycoplasma bovis. The duplicated TrmFO in M. capricolum was reported to catalyze the methylation of 23S rRNA (Lartigue et al., 2014) while the duplicated copy in M. bovis has been described to act as a fibronectin-binding adhesin (Guo et al., 2017).
We showed here that M. hyopneumoniae was able to uptake marked myo-inositol from a complex culture medium (Fig. 5); in addition this was the only species that remained viable whenever myo-inositol was used as the primary energy source (Fig. 6). From our metabolic model predictions (Ferrarini et al., 2016), the use of myo-inositol would be much more costly than the uptake and metabolism of glucose, which corroborates the small uptake of myo-inositol in Friis medium (glucose-rich) (Fig. 5). This basal uptake of myo-inositol could also be an indication that this pathway is important not only for energetic yield. Supporting this idea, microarray studies on strain 232 showed that several genes (if not all) from the myo-inositol catabolism were differentially expressed during stress treatments: heat shock (downregulated) (Madsen et al., 2006), iron depletion (upregulated) (Madsen et al., 2006) and norepinephrine (downregulated) (Oneal et al., 2008). Moreover, a previous transcriptome profiling of M. hyopneumoniae (Siqueira et al., 2014) showed that all genes from the myo-inositol catabolism were transcribed under normal culture conditions. Furthermore, three genes from the pathway (iolB, iolC and iolA) belonged to the list of the 20 genes with the highest number of transcript reads. Besides the transcription of these genes, proteomic studies of M. hyopneumoniae strains 232 (Pendarvis et al., 2014), 7422, 7448 and J (Pinto et al., 2009; Reolon et al., 2014) (Supporting Information Table S7) showed that several enzymes from this pathway were present in normal culture conditions.
Indeed, myo-inositol has been extensively reported in several organisms as a signaling molecule (Downes and Macphee, 1990; Gillaspy, 2011). Moreover, the myo-inositol catabolism has been experimentally described as a key pathway for competitive host nodulation in the plant symbiont and nitrogen-fixing bacterium Sinorhizobium meliloti (Kohler et al., 2010). Host nodulation is a specific symbiotic event between a host plant and a bacterium. Kohler et al. (2010) showed that whenever inositol catabolism is disrupted (by single gene knockouts from the inositol operon), the mutants are outcompeted by the wild type for nodule occupancy. This means that genes for the catabolism of inositol are required for a successful competition in this particular symbiosis. Moreover, the authors were not able to find a suitable candidate for the IolJ activity. In our case, we proposed that the activity of the missing enzyme IolJ is taken over by a duplication of fba. We were able to find a similar duplication (also not inside the myo-inositol cluster) in the genome of S. meliloti 1021 (SM_b21192 and SM_b20199, both annotated as fructose-bisphosphate-aldolase, EC 4.1.2.13). This means that in at least one other symbiont that has the myo-inositol catabolism genes, there could exist a putative IolJ not close to the myo-inositol cluster, just as we proposed here.
Whether this entire pathway is functional in M. hyopneumoniae is yet to be tested and further experiments should take place to support this hypothesis. However, the ability of M. hyopneumoniae to persist longer in the swine lung if compared to the other two mycoplasmas might come from the fact that this species is able to uptake and process myo-inositol. Furthermore, the ability of M. hyopneumoniae to grow in diverse sites (Le Carrou et al., 2006) if compared to M. flocculare might also be due to this specific trait.
Concluding remarks
It is important to remember that even though M. hyopneumoniae is considered highly pathogenic, the three Mycoplasma species studied here are widespread in pig populations and can easily be found in healthy hosts (Fano et al., 2005; Pieters et al., 2010). However, the main question permeating this fact is: what causes the switch from a nonpathogenic Mycoplasma community to a pathogenic one? And what makes some strains pathogenic while others inflict no harm to the host cells?
Some strains of M. hyopneumoniae become less pathogenic in broth culture and, after serial passages, they lose their ability to produce gross pneumonia in pigs (Whittlestone, 2012). In a proteomic study comparing strains 232 and J, researchers have described that the attenuated strain J switches its focus to metabolism and therefore has developed better capabilities to profit from the rich culture medium while the ability to infect host cells becomes less important so that adhesion-related genes are downregulated (Li et al., 2009). This might be related to the fact that here we detected a higher production of ATP in this attenuated strain when compared to the pathogenic strains 7448 and 7422. Liu et al. (2013) have investigated genetic variations between M. hyopneumoniae strains 168 and attenuated 168-L and found out that almost all reported Mycoplasma adhesins were affected by mutations. Tajima and Yagihashi (1982) reported that capsular polysaccharides from M. hyopneumoniae play a key role in the interaction between pathogen and host. Indeed in several bacterial species it has been reported that the amount of capsular polysaccharide is a major factor in their virulence (Corbett and Roberts, 2009) and it decreases significantly with in vitro passages (Kasper et al., 1983). In this way, it is likely that the difference in pathogenicity between strains in M. hyopneumoniae does not solely depend on their metabolism, but also on their ability to adhere to the host.
A recent metagenomic analysis of community composition (Siqueira et al., 2017) has described that M. hyopneumoniae is by far the most prevalent species in both healthy and diseased hosts. The difficult isolation of Mycoplasma species from diseased lung extracts is due to the fact that, in culture, fast-growing bacteria will overcome the slow-growth of mycoplasmas (McKean et al., 1979). This means that, in vitro, the competition for an energy source between fast and slow-growing bacteria usually ends with an overpopulation of the fast growing ones. Given the fact that mycoplasmas survive for longer periods inside the host even in competition with other bacteria (Fano et al., 2005; Overesch and Kuhnert, 2017), we must assume that other factors exist and are usually not mimicked in cell culture.
While M. hyopneumoniae might cause no harm, depending mostly on the environment, the characteristics of the host and the composition of this dynamic lung microbiome, any unbalance in this system is probably capable of turning a nonpathogenic community into a pathogenic one. The final conclusion is that the disease is a multifactorial process depending on several elements that include intraspecies mechanisms, community composition, host susceptibility and environmental factors. One possibility is that the competition with fast-growing species could result in a lower carbohydrate concentration and that M. hyopneumoniae might have to overcome this environmental starvation with the uptake of glycerol or myo-inositol. Since the uptake of myo-inositol does not lead to the production of any toxic metabolite, it is more interesting for its persistence in the long run. Other bacteria will strongly compete for glucose and other related carbohydrates, while M. hyopneumoniae will have the entire supply of myo-inositol for itself. The uptake of glycerol as an energy source, on the other hand, will probably lead to the production of toxic hydrogen peroxide as reported in other Mycoplasma species. This toxic product combined with other toxins from the external bacteria in the system would most probably recruit immune system effectors. Since M. hyopneumoniae has efficient mechanisms of host evasion (Fano et al., 2005; Maes et al., 2017), the newly introduced and fast-growing bacteria might be eliminated faster and M. hyopneumoniae, in this way, would be able to persist longer than other species inside the host (as it is reported in vivo).
As mentioned before, virulence factors in Mycoplasma species cover a broader concept if compared to other species: they are genes not essential for in vitro conventional growth that are instead essential for optimal survival in vivo. From our M. hyopneumoniae metabolic models, neither the GlpO activity nor the uptake and metabolism of myo-inositol seem to be essential features for in vitro growth. However, we were able to show that they might be two metabolic traits important for the enhanced virulence of M. hyopneumoniae when compared to M. hyorhinis and M. flocculare and could be essential for its survival in vivo and directly affect its pathogenicity.
Experimental procedures
Mycoplasma cultivation
We used the following strains for experimental validation: M. hyopneumoniae strains 7448, 7422 (field isolates) and J (ATCC 25934), M. hyorhinis ATCC 17981 and M. flocculare ATCC 27716. Cells were cultivated in Friis medium (Friis, 1975) at 37°C for varying periods of time with gentle agitation in a roller drum.
Hydrogen peroxide detection
Hydrogen peroxide was detected in culture medium by the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit (Invitrogen Cat. No A22188), according to the manufacturer's manual. M. hyopneumoniae, M. hyorhinis and M. flocculare were cultivated for 48 h in modified Friis medium (with no Phenol Red) and thereafter centrifuged. The supernatant was used for the hydrogen peroxide readings compared to a standard curve (Supporting Information Fig. S7). The medium without bacterial growth was used as negative control. We used biological and technical triplicates to infer the average amount of hydrogen peroxide produced, and the concentration was standardized based on the average number of cells from each culture. Statistical analyses were performed using GraphPad Prism 6 software by one-way ANOVA followed by Dunnett's multiple comparison test considering M. flocculare as a control (p < 0.05).
In order to determine if the hydrogen peroxide production was dependent on the glycerol metabolism, we used the Merckoquant Peroxide Test (Merck Cat. No 110011) with detection range of 0.5 to 25 μg of peroxide per ml of solution (as described in Hames et al. (2009). Fifteen ml of M. hyopneumoniae 7448 and 7422 strains were grown for 48 h in Friis medium, harvested by centrifugation at 3360 g and washed twice in the incubation buffer (67.7 mM HEPES pH 7.3, 140 mM NaCl, 7 mM MgCl2). Cells were resuspended in 4 ml of incubation buffer and aliquots of 1 ml were incubated for 1 h at 37°C. To induce hydrogen peroxide production, either glycerol or glucose (final concentration 100 μM or 1 mM) was added to the cell suspension and samples were incubated at 37°C for additional 2 h. Hydrogen peroxide levels were measured using colorimetric strips according to the manufacturer's instructions. Aliquots without any added carbon source served as an incubation control. The statistical significance of the results was calculated using one-way ANOVA followed by Dunnett's multiple comparison test (p < 0.05). The results represent four biological replicates with at least two technical replicates each.
Mycoplasma cell count with flow cytometry
Mycoplasma cells cultivated for hydrogen peroxide detection were sedimented at 3360 g for 20 min at 4°C and washed three times with NaCl 0,9% (1x 3360 g for 20 min and 2x 3360 g for 4 min). Cells were resuspended in 1 ml of NaCl 0,9% and diluted 1:30 for flow cytometry readings in a Guava EasyCyte cytometer (Millipore, USA). Cells were characterized by side-angle scatter (SSC) and forward-angle scatter (FSC) in a four-decade logarithmic scale. Absolute cell counting was performed up to 5000 events and the samples were diluted until the cell concentration was below 500 cells/μl. The number of counts obtained was then converted to cells/ml.
Transcript levels of glpO with the use of real-time quantitative RT-PCR
Total RNA was isolated from 20 ml culture of M. hyopneumoniae strains 7448, 7422 and J grown at 37°C for 24 h. Cells were harvested by centrifugation at 3360 g for 15 min, resuspended in 1 ml of TRizol (Invitrogen, USA) and processed according to the manufacturer's instructions followed by DNA digestion with 50 U of DNaseI (Fermentas, USA). Absence of DNA in the RNA preparations was monitored by PCR assays. The extracted RNA was analysed by gel electrophoresis and quantified with the QubitTM system (Invitrogen, USA). A first-strand cDNA synthesis reaction was conducted by adding 500 ng of total RNA to 500 ng of pd(N)6 random hexamer (Promega, USA) and 10 mM deoxynucleotide triphosphates. The mixture was heated for 65°C for 5 min and then incubated on ice for 5 min. First-strand buffer (Invitrogen, USA), 0.1 M dithiothreitol and 200 U M-MLV RT (Moloney Murine Leukemia Virus Reverse Transcriptase – Invitrogen, USA) were then added to a total volume of 20 μl. The reaction was incubated at 25°C for 10 min and at 37°C for 50 min followed by 15 min at 70°C for enzyme inactivation. A negative control was prepared in parallel, differing only by the absence of the RT enzyme. Quantitative PCR (qPCR) assay was performed using 1:2.5 cDNA as template and Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen, USA) with specific primers for glpO (5′GGTCGGGAACCTGCTAAAGC3′ and 5′CCAGACGGAAACATCTTAGTTGG3′) on StepOne Real-Time PCR Systems (Applied Biosystems, USA). The qPCR reactions were carried out at 90°C for 2 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. A melting curve analysis was done to verify the specificity of the synthesized products and the absence of primer dimers. The amplification efficiency was calculated with the LinRegPCR software application (Ruijter et al., 2009). A relative quantification normalized against unit mass (500 ng of total RNA) was used to analyse the expression data with the equation:
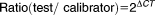 , where
, where
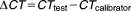 (Calviño et al., 2008) and MHP_7448 (Replicate 2) was chosen as calibrator. Statistical analyses were performed using GraphPad Prism 6 software by one-way ANOVA followed by Tukey's multiple comparison test (p < 0.05).
(Calviño et al., 2008) and MHP_7448 (Replicate 2) was chosen as calibrator. Statistical analyses were performed using GraphPad Prism 6 software by one-way ANOVA followed by Tukey's multiple comparison test (p < 0.05).
Comparative modelling and protein–ligand interaction analysis of Fba and IolJ
The SWISS-MODEL server (Schwede et al., 2003; Biasini et al., 2014) was used for template search and the comparative modelling for all Fba and IolJ proteins in this study. The best homology models were selected according to coverage, sequence identity, Global Model Quality Estimation (GMQE) and QMEAN statistical parameters (Benkert et al., 2009; Benkert et al., 2011). The Fba from M. hyopneumoniae along with IolJ and Fba from B. subtilis were modeled using the crystal structure of fructose 1,6-bisphosphate aldolase from Bacillus anthracis in complex with 1,3-dihydroxyacetonephosphate (PDB 3Q94) while Fba-1 from M. hyopneumoniae was modeled using the fructose-1,6-bisphosphate aldolase from Helicobacter pylori in complex with phosphoglycolohydroxamic acid (PDB 3C52). Both selected templates have the same resolution range (2.30 Å). Fba structures experimentally solved from E. coli (Hall et al., 1999) and G. intestinalis (Galkin et al., 2009) were used to include information about substrate binding in the active site. The DKGP and FBP ligands were drawn in the Avogadro version 1.1.1 (Hanwell et al., 2012) by editing the tagatose-1,6-biphosphate (TBP) molecule complexed with the Fba structure of G. intestinalis (PDB 3GAY). Each model was submitted to 500 steps of an energy minimization protocol using the universal force field (UFF). The DKGP and FBP molecules were inserted into the substrate binding sites of the acquisition models obtained by superposition of the models with the Fba structure of G. intestinalis.
Detection of marked myo-inositol through mass spectrometry
Solvents and reagents
Acetonitrile and formic acid (Optima LC/MS Grade) were purchased from Fisher Scientific (Loughborough, UK). MilliQ water was obtained from a Direct-Q 5UV system (Merck Millipore, Billerica, MA, USA). Deuterated myo-inositol-1,2,3,4,5,6-d6 was purchased from CIL (C/D/N Isotopes Inc. Cat No. D-3019, Canada).
Cultivation in the presence of marked myo-inositol
Cells were cultivated in Friis medium supplemented with 0.25 g/l of deuterated myo-inositol-1,2,3,4,5,6-d6 (C/D/N Isotopes Inc. Cat No. D-3019). Cultures were interrupted after 8 h, 24 h and 48 h of cultivation for mass spectrometry analysis.
Sample preparation
All samples were filtered and concentrated with the use of Amicon Ultra 3 kDa (Merck Millipore Cat. No. UFC200324). After this step, samples were dried in a miVac sample concentrator (Genevac, Ipswich, UK) for approximately 45 min at 50°C. All samples were ressuspended in ultrapure water to a final concentration of 10 g/l and were subsequently submitted to mass spectrometry.
Mass spectrometry
Aqueous extracts of Mycoplasma sp. and commercial deuterated myo-inositol-1,2,3,4,5,6-d6 were analysed using an Accurate-Mass Q-TOF LCMS 6530 with LC 1290 Infinity system and Poroshell 120 Hilic column (3 × 100 mm, 2.7 μm) (Agilent Technologies, Santa Clara, USA). The extracts were dissolved in water (10 g/l) and injection volume was 3 μl. A binary mobile phase system (A: 0.4% formic acid in milliQ-water and B: acetonitrile) was pumped at a flow rate of 0.9 ml/min at the following gradient: 0–3.5 min, 90% B; 3.5–7 min, 90% to 0% B; 7–9.5 min, 0% B; 9.5–10 min 0% to 90% B; 10–15 min, 90% B (total run: 15 min). MS and MS/MS spectra were obtained in negative mode, with the following conditions: nebulization gas (nitrogen) at 310°C, at a flow of 10 l/min and 40 psg pressure. The capillary tension was 3600 V and gave ionisation energy of 100 eV. In targeted MS/MS mode, collision energy was set at 18 eV. Acquisition range was m/z 50–500. MassHunter Qualitative Analysis Software (version B.07.00) was used for data analysis.
Data analysis
Deuterated myo-inositol-1,2,3,4,5,6-d6 was quantified in all aqueous extracts by HPLC-MS. For that, a calibration curve (based on peak area) of a commercial myo-inositol was performed from 0.001 g/l to 0.05 g/l in replicate (4 times during the batch analysis). Statistical analyses were performed using GraphPad Prism 6 software. One-way ANOVA followed by Dunnett's multiple comparison test was used to test for differences in residual marked myo-inositol in culture after bacterial growth of all tested strains for 48 h (p < 0.05). A two-tailed unpaired t-test was used to compare the residual marked myo-inositol between M. hyopneumoniae 7448 and the control medium with two extra timepoints: 8 h and 24 h (p < 0.05).
Determination of cell viability of M. hyopneumoniae in myo-inositol defined medium
All available strains were grown in Friis medium at 37°C for 48 h, sedimented by centrifugation at 3360 g for 20 min at 4°C, washed twice with ice cold PBS and inoculated in glucose regular defined medium (described in Ferrarini et al. (2016), supplemented with 5 g/l of succinate) or myo-inositol defined medium (regular defined medium depleted with glucose and glycerol and supplemented with 0.5 g/l of myo-inositol and 5 g/l of succinate). Viability of cells was measured by ATP production with live cells recovered after 8 h of growth in either media with a BacTiter-GloTM Microbial Cell Viability Assay Kit (Promega, USA) according to the manufacturer's manual. Luminescence was recorded in a SpectraMax MiniMax 300 Imaging Cytometer (Molecular Devices, USA) with an integration time of 0.5 s in an opaque-walled multiwell plate. Average ATP production was calculated with biological duplicates and technical triplicates. The ATP production of each strain was compared between regular defined medium and myo-inositol defined medium to determine the ratio of viable cells and to allow a comparison between strains. A 10-fold serial dilution of ATP was used as a standard curve (Supporting Information Fig. S8). Statistical analyses were performed using GraphPad Prism 6 software by one-way ANOVA followed by Tukey's multiple comparison test (p < 0.05).
Acknowledgements
This work was supported by grants from CAPES-COFECUB 782/13 and Inria. MGF was granted postdoctoral fellowship funded by the European Research Council under the European Community's Seventh Framework Programme (FP7/2007–2013)/ERC grant agreement no. [247073]10. SGM was the recipient of a CAPES doctoral fellowship. DP was granted postdoctoral fellowship funded by the European Union Framework Program 7, Project BacHbERRY number FP7–613793. JFRB is a recipient of a CAPES postdoctoral fellowship. The mass spectrometry analysis was carried out in the Centre d'Etude des Substances Naturelles at the University of Lyon.
Authors’ contributions
MGF, SGM, MFS and AZ conceived and designed the work. MGF and SGM performed most of the experimental work. DP, GM and GC collaborated in the mass spectrometry experiments and analysis. JFB performed tridimensional analysis of proteins. All authors collaborated in the analysis of all data. MGF and SGM wrote the manuscript with inputs from the other authors. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.