Distinct routes to parallel adaptation in a mountain plant
Abstract
It is a common observation that distinct lineages adapt to similar environments through similar “parallel” changes in phenotype. Less clear is whether parallel phenotypic change evolves through genetic variation in the same or distinct genes. This question of redundancy—the number of genetic answers to the same evolutionary problem—is an important yet poorly understood feature of adaptive traits, and is a major hurdle in our transition from understanding evolutionary responses to predicting them. In this issue of Molecular Ecology, Szukala et al. (2023) examine the redundancy of local adaptation in montane and alpine ecotypes that has occurred in the flowering plant Heliosperma pusillum. By comparing gene expression in each of four montane or alpine-adapted H. pusillum population pairs, the authors observe distinct genetic responses underlying each case of parallel local adaptation.
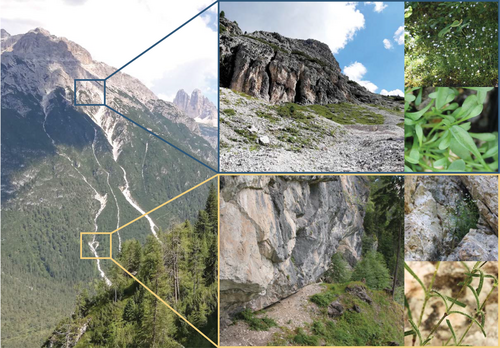
The prevailing model explaining the relationship between genetic variation and phenotypic variation (Fisher, 1919) is that phenotypes are determined by a large number of genetic variants with relatively small effects. This type of genetic architecture implies a high level of redundancy, with myriad combinations of small-effect alleles capable of producing the same phenotypic results. Numerous studies, however, have demonstrated diverse taxa adapting to similar environmental changes through mutation in the same genes and even codons for corresponding amino acids within these genes. Classic examples include pigmentation in mammals (Manceau et al., 2010) and pesticide resistance in insects (Ffrench-Constant, 2013). Such findings indicate low levels of redundancy in these particular traits, where evolution is observed to repeatedly follow the same path. However, conclusive counterexamples of traits with high redundancy have proven far more elusive, as they require a high genomic resolution and careful experimental design. Szukala et al.'s H. pusillum populations provide an intriguing system for the detection of such trait architectures. In four pairs of populations across Europe's southeastern Alps, locally adapted ecotypes of H. pusillum grow in either montane or alpine environmental conditions, which differ in altitude, temperature, humidity and light availability, and where H. pusillum exhibits morphological differences including presence or absence of glandular trichomes (Figure 1).
To explore this question of parallel ecotype divergence, Szukala et al. (2023) assembled a reference genome for H. pusillum and, after growing plants in a common garden, measured gene expression in representatives of each ecotype in each population pair through transcriptome sequencing. The sequencing provided insight into both genetic variation and gene expression differences between populations. Genetic relatedness analyses and modelling of the populations' demographic histories (as well as previous work by the group; Trucchi et al., 2017) supported the independence of each population pair, and therefore the hypothesis of parallel ecotype divergence central to the study's experimental design. Consistent with local adaptation in ecotypes, different genes were expressed by plants descended from alpine and montane populations. Perhaps surprisingly, however, the same genes rarely varied between different alpine–montane pairs. In fact, <2% of the gene expression differences between individuals could be explained by montane or alpine ecotype class. In contrast to the overall low parallelism in differentially expressed genes across the four population pairs, statistically significant signatures of parallel gene expression are observed in some pairwise comparisons of population pairs. This demonstrates how less extensive experimental designs may lead to an underestimation of redundancy in genetic responses.
However, while the genes differentially expressed in ecotype divergence were largely distinct between population pairs, the biological functions of the differentially expressed genes were convergent between pairs (Figure 2). These convergent functions included genes relevant to environmental differences such as temperature, light and water availability, as well as the glandular trichomes which are present in montane but not alpine ecotypes. Follow-up work by the group (Szukala et al., 2022) has demonstrated that derived montane ecotypes also exhibit increased plasticity in gene expression relative to ancestral alpine ecotypes.

Szukala et al. (2023) supplemented their gene expression analyses with scans of genetic signatures of local adaptation in two population pairs. They found no enrichment for selective signatures in differentially expressed genes, and even lower levels of parallelism at the genetic level than at the gene expression level between these populations, both results consistent with nonparallel, polygenic ecotype divergence in H. pusillum.
Despite strong theoretical evidence for polygenicity and hence high redundancy in the genetic architecture of adaptive traits, empirical studies regularly describe cases of monogenic, parallel adaptation. This is in part due to historical limitations in power to identify polygenic signals of adaptation with genome scans (Rockman, 2012); nevertheless, the relative contribution of high- and low-redundancy architectures to adaptive evolution remains unresolved. Szukala et al.'s (2023) interrogation of parallel ecotype divergence in H. pusillum joins a small but growing number of studies describing multiple distinct evolutionary pathways leading to similar adaptive traits.
ACKNOWLEDGEMENT
Open access publishing facilitated by Monash University, as part of the Wiley - Monash University agreement via the Council of Australian University Librarians.




