Biogeographic inferences across spatial and evolutionary scales
Abstract
The field of biogeography unites landscape genetics and phylogeography under a common conceptual framework. Landscape genetics traditionally focuses on recent-time, population-based, spatial genetics processes at small geographical scales, while phylogeography typically investigates deep past, lineage- and species-based processes at large geographical scales. Here, we evaluate the link between landscape genetics and phylogeographical methods using the western fence lizard (Sceloporus occidentalis) as a model species. First, we conducted replicated landscape genetics studies across several geographical scales to investigate how population genetics inferences change depending on the spatial extent of the study area. Then, we carried out a phylogeographical study of population structure at two evolutionary scales informed by inferences derived from landscape genetics results to identify concordance and conflict between these sets of methods. We found significant concordance in landscape genetics processes at all but the largest geographical scale. Phylogeographical results indicate major clades are restricted to distinct river drainages or distinct hydrological regions. At a more recent timescale, we find minor clades are restricted to single river canyons in the majority of cases, while the remainder of river canyons include samples from at most two clades. Overall, the broad-scale pattern implicating stream and river valleys as key features linking populations in the landscape genetics results, and high degree of clade specificity within major topographic subdivisions in the phylogeographical results, is consistent. As landscape genetics and phylogeography share many of the same objectives, synthesizing theory, models and methods between these fields will help bring about a better understanding of ecological and evolutionary processes structuring genetic variation across space and time.
1 INTRODUCTION
Landscape genetic theory predicts that the degree of genetic differentiation experienced by populations distributed across a landscape is due to habitat, climate and topographic factors that influence dispersal and migration across these features (Balkenhol et al., 2015; Manel et al., 2003; Manel & Holderegger, 2013; Storfer et al., 2010). Landscape genetics tracks recent-time and fine-scale population structure and is useful for evaluating contemporary barriers to genetic connectivity (Landguth et al., 2010; Wang, 2010). Phylogenetic methods, on the other hand, are useful for describing deep-time evolutionary processes on the order of millions of years (Leaché & Oaks, 2017). It is possible that genetic differentiation and connectivity may be influenced by different factors at these broad scales, since the landscape elements influencing dispersal and migration may differ from those structuring population genetic structure across multiple generations. Importantly, it remains unclear how concordant landscape genetics and phylogeography may be within this broader context of biogeographical inference (Figure 1; Rissler, 2016).
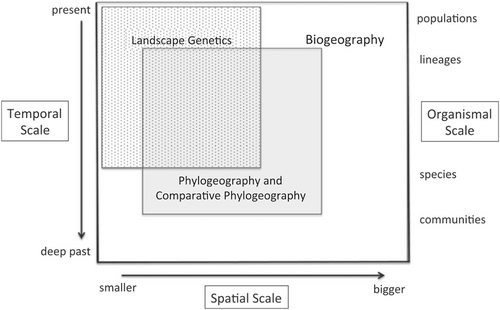
If landscapes exert a functionally similar effect on genetic variation across spatial and evolutionary scales, then we should see similarities in the areas of the landscape that promote and inhibit gene flow across landscape genetics and phylogeographical contexts. This may be expected for generalist or precocial species that disperse, migrate or otherwise interact with their physical environment in a consistent way across life-history stages and through evolutionary time. On the other hand, it may be that the landscape elements limiting gene flow at large spatial scales are distinct from those that limit gene flow at small spatial scales—as in the case of amphibians with distinct larval and adult life stages (Angelone et al., 2011; Trumbo et al., 2013; Wang et al., 2009). This could also be expected if the landscape features influencing dispersal processes (Wang et al., 2009) are distinct from those affecting range expansions and colonization of new environments, such as interglacial range expansions and contractions (Bouzid et al., 2022). Ultimately, developing a deeper understanding of the landscape elements that hinder or promote gene flow at various scales, and relating these patterns to what we understand about species ecology and physiology, will help inform a general understanding of the effect of landscape structure on genetic differentiation across space and time.
This study focuses on the western fence lizard (Sceloporus occidentalis), a generalist lizard widely distributed across Western North America that inhabits a diverse range of environments from sea level to elevations of ~3300 m (Baird & Girard, 1852; Leibold et al., 2022; Stebbins, 2003). This lizard species is an attractive model for this work because as ectotherms they are intimately dependent on several characteristics of their environment, including maximum and minimum temperatures, precipitation levels, and variation in seasonality—all of which we consider here. The Sierra Nevada is an excellent place to examine the role of spatial scale on patterns of genetic variation in a common environment. For example, similar plant communities can be found along the extent of the range (Schoenherr, 1992). The Sierra Nevada range also exhibits many similarities in terms of topographic variability and complexity along its extent. While the geological history of the Sierra Nevada is complex, the entirety of the Sierra Nevada mountain range is generally believed to have been formed by common geological processes (Hill, 2006; McPhee, 1993), and bears many similarities in the structure of river canyons that span predominantly east–west across the latitudinal extent of the range. Climate patterns are also largely similar across the range, characterized by cold, wet winters and warm, dry summers that often give way to afternoon thunderstorms in mid- to late summer (Schoenherr, 1992). These similarities make it possible to examine the effect of spatial scale without confounding effects of disparate environments.
Our first objective is to explicitly vary the spatial scale (i.e., “extent” of sampling area from 1225 to 6400 km2) to test the effect of different scales of analysis on landscape genetic inferences. This will inform if isolating mechanisms are similar across both large and small landscapes (i.e., are species interacting differently with their landscape at different spatial scales?). If the same barriers and facilitators to gene flow are identified at each scale, then this would suggest gene flow at large spatial scales is similar to that at small scales, and the scale of analysis does not appreciably change inferences in landscape genetics.
Our second objective is to compare landscape genetics results informed by population genetic data with phylogeographical results informed by phylogenetic data (i.e., biogeography; Rissler, 2016). As river valleys have been identified as important gene flow corridors for S. occidentalis around (Bouzid et al., 2022) and within (Wishingrad & Thomson, 2021) the Sierra Nevada range, we test the hypothesis that river canyons are major features structuring phylogenetic diversity as well. We evaluate this link by examining how diverse clades are in terms of their membership from different river canyons (i.e., canyons-per-clade) and how diverse canyons are in terms of their membership from distinct evolutionary groups (i.e., clades-per-canyon). We conduct this analysis at two timescales: with all major clades and major rivers in the Sierra Nevada, which are estimated to have diverged beginning ~700,000 years ago; and at a more recent scale, considering only minor clades that are estimated to have diverged within the last 100,000 years (Bouzid et al., 2022) in river valleys within the Sierra Nevada.
If a majority of canyons include lizards from a single clade, and if a majority of clades include lizards from a single canyon, then we would consider this evidence of high phylogeographical structure based on river canyons. However, if canyons tend to include lizards from several clades, and clades include lizards from several canyons, then this would be evidence of low phylogeographical structure based on river canyons and limited concordance in how dominant features of the landscape structure genetic variation between population genetic and phylogenetic scales.
2 MATERIALS AND METHODS
2.1 Lizard collection
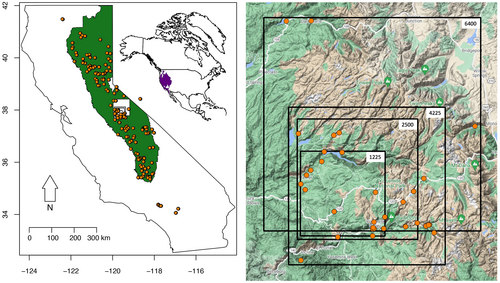
We captured lizards by hand or lizard lasso and recorded Global Positioning System (GPS) coordinates with a Garmin eTrex 10 GPS at the site of capture. We sampled lizards from June to August in 2016 and 2018 in California, USA, over a latitudinal range of >900 km and elevation range of 100–2800 m. For approximately half of the individuals collected, we followed a standard euthanasia protocol based on Conroy et al. (2009), with modifications specific for Sceloporus, and prepared specimens as vouchers, preserving liver samples in 95% ethanol. All voucher specimens and genetic samples are deposited at the Natural History Museum of Los Angeles County, section of Herpetology (see Supporting Information for sample details including voucher numbers). For the other half of the lizards collected, we removed <5 mm of the distal portion of the tail and preserved the tissue in 95% ethanol. We also obtained tissue samples from museum specimens at the Museum of Vertebrate Zoology at University of California, Berkeley, the California Academy of Sciences, the Yale Peabody Museum of Natural History, and from ongoing collaborations. Altogether we obtained a sample size of 220 lizards, retaining 49 individuals from the Yosemite region for the landscape genetics analysis, and a sample of 201 individuals with highest sequencing coverage over the greater sampling area for the phylogenetic portion of the study (Figure 2).
2.2 ddRAD library preparation and genomic sequencing
We followed Peterson et al.'s (2012) method for genomic library preparation, with some modifications. For each individual, we extracted high-molecular-weight genomic DNA using a standard phenol–chloroform extraction protocol (Tsai et al., 2019). We measured DNA concentrations using a Qubit fluorometer, and for each sample we digested 0.5 μg of DNA for 3 h with restriction enzymes SbfI and NIaIII. We then purified these fragments with Agencourt AMPure beads before ligation of barcoded Illumina adaptors onto the fragment ends. All barcodes differed by at least 2 bp to reduce duplexing error rates. We then pooled equimolar amounts of each sample before conducting size selection using a Pippin Prep to select fragments between 400 and 550 bp in length. We used proofreading Taq and Illumina's index primers for final library amplification for 8–10 cycles to reduce PCR (polymerase chain reaction) bias. We quantified the final library concentration using a Qubit fluorometer at high sensitivity. Samples were packed on dry ice and sent to the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley for sequencing. Quantitative PCR was used to determine the concentration of adapter-associated fragments, and a BioAnalyzer run confirmed fragment sizes as a quality control measure prior to sequencing. Final libraries were sequenced (100- or 150-bp, single-end runs) on Illumina HiSeq 4000 and NovaSeq SP lanes in a total of four sequencing lanes.
2.3 Bioinformatics protocols for SNP data
We inspected raw Illumina reads for sample quality using fastqc (Andrews, 2010). We used stacks version 1.48 (Catchen et al., 2013) for initial sequence data processing such as demultiplexing samples, steps to rescue barcodes with at most one mismatch, clean data and remove any read with an uncalled base, and truncate all reads to 95 bases (due to variable-length sequences from using both 100- and 150-bp sequencing platforms). Prior to analysis, we concatenated sequencing data for the same individual sequenced on different lanes. Then, we used a reference-based approach in the ipyrad version 0.9.42 (Eaton & Overcast, 2020) program to conduct individual-based analyses. Compared to de novo assembly approaches, reference-based approaches have much lower error rates, higher accuracy and less bias (Rochette & Catchen, 2017). We used the annotated S. occidentalis genome published by Harris et al. (2015) as the reference genome for the analysis.
2.4 ipyrad analysis protocol
We followed the analysis recommendations and default settings specified by the program authors with some modifications specific to our datasets. We filtered and edited demultiplexed reads by removing reads with five or more low-quality base calls (Q < 20), and trimmed bases from the 3′ end of reads if their quality scores fell below 20, which is 99% probability of a correct base call. Reads were then mapped to the reference genome using bwa (version 0.7.17-r1188) and clusters were aligned using muscle (version 3.8.31), requiring a minimum depth of 6, which is the minimum depth at which a heterozygous base call can be distinguished from sequencing error. We jointly estimated heterozygosity and error rate by specifying a maximum of two alleles per site in each consensus sequence and removed alignments with a high proportion (5%) of heterozygous base calls, as poor alignments tend to have an excess of heterozygous sites. To remove poor alignments in repetitive regions in the final data set, we allowed for a maximum of 20% single nucleotide polymorphisms (SNPs) per locus, as well as removed alignments with more than eight indels per locus. We set the maximum proportion of shared polymorphic sites in a locus to 0.5, as shared heterozygous sites across samples probably result from clustering of paralogues with fixed, rather than heterozygous, sites, and excluded any samples with fewer than 100,000 reads. We retained all loci shared by 75% of samples in the data set for analysis, as this provided a good balance between missingness and number of recovered SNPs in the analysis. Altogether for the landscape genetics analysis we retained a sample of 49 individuals with 11,215 loci in the assembly and 25.9% missing sites in the final SNP matrix. We output an SNP-based formatted file of one randomly selected variable site per locus to calculate individual-based pairwise genetic distances calculated as a proportion of shared alleles for individuals collected over the extent of our sampling range.
For the phylogenetic data set, we followed the same protocol as above for a sample of 200 S. occidentalis individuals that broadly encompass the Sierra Nevada range and one S. graciosus as an outgroup. We removed invariable sites from the alignment and retained all positions with data for at least 95% of the individuals (n = 191) in the data set. We obtained a data set for the phylogenetic analysis with 27,872 variable sites in the final sequence matrix with 17.1% missing sites.
2.5 Vegetation, climate and environmental data layers
We obtained land cover and vegetation data from the GAP/LANDFIRE National Terrestrial Ecosystems data sets (usgs.gov). These data sets consist of detailed vegetation and land cover data for 584 unique classes at several levels of classification and a 30-m-resolution scale. For the purposes of our analyses, we used the “Class”-level classification, which partitions each unique class into 11 groups representing major land cover and vegetation categories (e.g., “Forest and Woodland”, “Shrub” and “Herb Vegetation”, etc.; Figure S1a–d). We obtained broad-scale climate data from the PRISM climate group (prism.oregonstate.edu). These data included: mean precipitation, mean temperature, and average monthly minimum and maximum temperatures over the most recent three full decades spanning the period 1981–2010, and are represented at 800-m resolution. We also obtained bioclimatic data from the WorldClim database (worldclim.org) for variables hypothesized to be potentially important in limiting distribution and migration in reptiles. These data are represented at 30-s resolution (0.93 × 0.93 = 0.86 km2 at the equator) and include several aspects of temperature variation: maximum and minimum temperature during the warmest and coldest quarters (3-month period) and warmest and coldest months, and two aspects of temperature variation, including temperature seasonality (standard deviation of temperature × 100) and temperature annual range (maximum temperature of warmest month – minimum temperature of coldest month). We included several aspects of precipitation variation in the environment, including maximum and minimum precipitation during the wettest and driest quarters (3-month period) and wettest and driest months, as well as precipitation seasonality (coefficient of variation in precipitation).
We obtained elevation data captured by the Advanced Land Observing Satellite (ALOS) DAICHI-2 (eorc.jaxa.jp/ALOS/en/aw3d30/), which is the most precise global-scale elevation data we could obtain, at a resolution of 30 m. Using the original 30-m elevation data, we generated slope and ruggedness spatial layers in qgis version 2.18 (qgis.org). Slope measures the angle of inclination of the terrain between adjacent cells, while ruggedness quantifies terrain heterogeneity as the change in elevation within a 3 × 3 cell grid (as described in Riley et al., 1999). Altogether we retained 19 layers for the analyses: one vegetation layer, four PRISM climate layers, 11 WorldClim bioclimatic layers and three topographic layers.
2.6 Spatial data processing
We generated spatial layers representing the axes of greatest variation in the environment from all layers comprising continuous data, which included all temperature, precipitation and topographic data. Specifically, we used r (version 3.6.1) and the package rstoolbox (version 0.2.6) to perform a principal components analysis (PCA) on stacks of raster layers. These principal component spatial layers represent different aspects of the environment in each spatial area (Figures S2–S4). Because the relationships between variables differed by degree in each of the study areas, we did not use a common set of PC axes to compare between regions, as a PC axis on one area may describe an environmental gradient that does not exist in another area. For each area we retained the first three principal components—which was sufficient to describe >90% of the variation in the environment in each area—along with vegetation layers in each landscape genetics analysis. The vegetation category layer was converted to integer values 1–11, with each integer representing a distinct vegetation category. In all cases, layers were converted using nearest neighbour interpolation to a common resolution of 0.0025, which is equivalent to a cell size of ~1 km2.
2.7 Description of study sites
We focused our landscape genetics studies in Yosemite National Park (YNP) (37.865, −119.538; Figure 2a). The location of each study area was positioned so as to maximize the number of samples within each extent, and each spatial scale evaluated exceeds dispersal distances for S. occidentalis (Massot et al., 2003). These landscape sizes were chosen arbitrarily such that the number of individuals within each landscape satisfied the requirements needed to accurately model landscape effects on dispersal and genetic connectivity using resistancega (Winiarski et al., 2020). We therefore carried out studies at four spatial scales: 35 × 35 km (1225 km2) with 26 individual locations; 50 × 50 km (2500 km2) with 35 individual locations; 65 × 65 km (4225 km2) with 46 individual locations; and 80 × 80 km (6400 km2) with 45 individual locations (Figure 2b), which was the largest scale we could evaluate given computational limitations. The analyses are focused around the centre of YNP, which includes Yosemite Valley and the Grand Canyon of the Tuolumne River; and at the largest extent includes Bridgeport in the northeast, Mt Lyell in the southeast, Yosemite West in the southwest, and the towns of Pinecrest and Dardenelle in the northwest. The landscape is dominated by forest and woodland habitat, including Jeffery pine (Pinus jeffreyi) and ponderosa pine (P. ponderosa) woodlands and mixed conifer woodlands; scrub, grassland and barren areas that include sagebrush (Artemesia spp.) shrublands, dwarf shrublands and sparsely vegetated tundra areas; and desert and semidesert areas to a lesser degree. Developed and human-use areas are found substantially in Yosemite Valley and near roads. Glacier-fed lakes and streams and granite outcrops are found throughout the landscape (Figure S1a–d).
2.8 Landscape genetics analysis
We used resistancega (Peterman, 2018; Peterman et al., 2014) for our landscape genetics analyses. resistancega uses a genetic algorithm to simultaneously estimate resistance to dispersal and genetic connectivity from continuous and categorical surfaces based on pairwise genetic data and effective geographical distances. The method is free from subjective a priori assumptions of the expected resistance values, and fully explores parameter space. This allows for bias-free estimations of gene flow and migration rates through different areas in the landscape. Here, we largely used default values in the genetic algorithm optimization settings, but selected maximum categorical and continuous values of 500, while evaluating all possible transformations for continuous surface values. We set the maximum number of layer combinations equal to 5, to evaluated models that include all combinations of layers together. To estimate variance around the parameter estimates and model fit, we implemented a resampling procedure that repeated the analysis for 1000 iterations of pseudoreplicated data sets comprising 75% of the samples in each analysis. We calculated pairwise cost–distance matrices using the commuteDistance function as implemented in the r package gdistance (Etten, 2017) because it is functionally equivalent to resistance distance while being more computationally efficient (McRae et al., 2008; Peterman, 2018). We evaluated the results using Akiake's corrected information criterion (AICc) where all models of delta AICc ≤2 of the top model were considered to have substantial support. We used circuitscape (version 4; Hall et al., 2021) to visualize optimized gene flow across the landscape, with sampling sites as the focal nodes and the output raster as the resistance surface. We compared similarities in these optimized gene flow surfaces with Pearson's correlations for all overlapping regions at each spatial scale.
2.9 Phylogenetic inference
We conducted a Bayesian phylogenetic analysis using a data set of 27,872 variable sites to estimate phylogeographical-scale patterns of genetic groups distributed across geographical space. We used a sample of 200 S. occidentalis individuals that broadly encompass the Sierra Nevada range, and a sagebrush lizard (S. graciosus) we collected near the centre of this sampling region as an outgroup (Figure 2). We used jmodeltest version 2 (Darriba et al., 2012) to evaluate phylogenetic model fit of those implemented in mrbayes version 3.2.5 (Huelsenbeck & Ronquist, 2001). We used default priors for the analysis and ran mrbayes with the coding = variable option with two runs and four chains in a Metropolis-coupled Markov chain Monte Carlo (MC3) for 10,000,000 generations, discarding the first 25% as burn-in. We inspected the MC3 for convergence using tracer version 1.7 (Rambaut et al., 2018) to ensure both runs converged to apparently similar stationary distributions, that mixing was adequate and that a sufficient number of effectively independent samples had been sampled for each parameter. We then visualized the resulting phylogenetic tree using figtree version 1.4.4 (Rambaut, 2009).
2.10 Quantifying clade–canyon membership
We describe clade geographical diversity on the basis of unique river canyons each clade spans, which we term canyons-per-clade. A low value indicates that each clade contains sampled lizards from a few canyons, implying a high level of specificity in the clade–canyon relationship, while a high value would indicate higher clade diversity in terms of membership from several distinct river canyons. We also describe canyon diversity on the basis of unique clades that inhabit them, which we term clades-per-canyon. A low value would indicate that each canyon includes samples from a single lineage, while high values would indicate higher canyon diversity in terms of phylogenetic lineage diversity.
We examined the extent to which the major clades identified in the phylogenetic tree are distributed among the major rivers of the Sierra Nevada region. We defined major rivers based on river canyon depth and prominence within the Sierra Nevada (Clark et al., 2005; Schoenherr, 1992; Stock et al., 2004). These included the Kern River Canyon, Kaweah River, Kings River, San Joaquin River, Merced River, Tuolumne River, Stanislaus River, Mokelumne River, American River and Feather River. We also include the Owens River Valley and the California Central Valley to the west of the Sierra Nevada (including samples from the east and north of the Sierra Nevada). Altogether, we examined how these five major clades (Figure 4) were distributed among these 12 major river canyons. The southern California Transverse Range (Santa Ana River and San Gabriel River) samples were excluded because their phylogenetic position is emblematic of vicariance, and do not implicate “isolation by river canyon,” which we are exploring here. We also examined the extent to which the minor clades are distributed among river canyons of the Sierra Nevada region. This set included all major river canyons from above, with the addition of smaller river canyons identified in the U.S. National Atlas Water Feature Lines (Supporting Data).
3 RESULTS
3.1 Landscape genetics
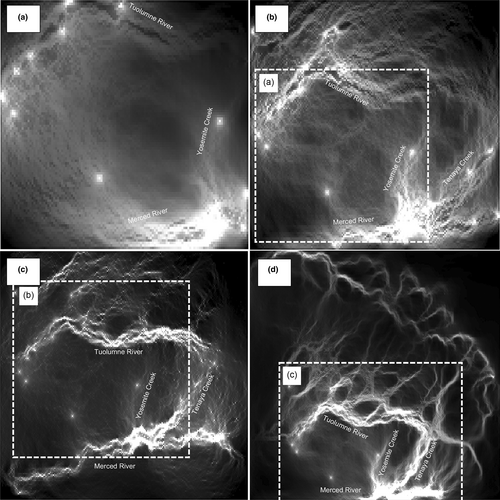
Landscape genetics analyses at several spatial scales in the Yosemite region were highly consistent in implicating canyon features as dispersal corridors for S. occidentalis (Figure 3; Figure S6). The Merced River canyon and the Tuolumne River canyon, in particular, were identified as areas of high gene flow. Furthermore, Yosemite River canyon and Tenaya Creek were identified as key features linking the Merced River and Tuolumne River canyons (Figure 3). Analyses at spatial scales of 1225, 2500 and 4225 km2 identified water bodies as barriers to gene flow from the climate and topographic PC axes (see Figure S1a–c for vegetation maps, including water bodies on the landscape) and Pearson's correlation values among the independent circuitscape layers were positive and significant (Table 1), while the analysis at a 6400-km2 spatial scale could not distinguish these low gene flow features such as water bodies from the high gene flow rates in adjacent canyons (Figure S1d; Figure 3d), and the circuitscape resistance layer at this spatial scale was poorly correlated with layers at the 1225-, 2500- and 4225-km2 scales (Table 1). Another difference is the 1225-km2 spatial scale identifies the western portion of the range in the vicinity of Bald Mountain as promoting a relatively higher gene flow, while the analyses at other spatial scales do not.
| 2500 | 4225 | 6400 | |
|---|---|---|---|
| 1225 | 0.91*** | 0.65** | −0.11 |
| 2500 | — | 0.59* | −0.07 |
| 4225 | — | — | 0.11 |
- Note: Correlations are statistically significant in all cases. ***Very strong association (>0.7); **strong association (>0.6); *moderate association (>0.5).
At the smallest spatial scale (1225 km2), PC components 1, 2 and 3 were substantially implicated in structuring genetic variation (Tables S1, S5, S9, S13 and S17; Figure S5a–c). Areas of high slope and temperature seasonality and high-elevation colder areas with more precipitation were identified as barriers to gene flow, while less rugged warmer low-elevation areas that exhibit less precipitation were facilitators of gene flow (Tables S13 and S17). Analyses at the 2500-km2 spatial scale shared many similarities with the 1225-km2 analyses, implicating PC components 1, 2 and 3 as well as distance in structuring genetic variation across this landscape (Tables S1, S6, S10, S14 and S17; Figure S5d–f). Rugged areas of high slope and high temperature seasonality, high-elevation cold areas, and areas with more dry-season precipitation inhibited gene flow. Low slope and ruggedness, low temperature seasonality and warmer low-elevation areas with less precipitation facilitated gene flow (Tables S14 and S17). Analyses at the 4225-km2 scale implicated PC component 2 in structuring genetic variation (Tables S3, S8, S11, S15 and S17; Figure S5g). Here, areas of high slope and ruggedness, high temperature seasonality and high precipitation inhibit gene flow (Tables S15 and S17). Finally, the largest spatial scale (6400 km2) likewise implicated PC component 2 with areas of high temperature seasonality and high precipitation inhibiting gene flow (Tables S4, S8, S12, S16 and S17; Figure S5h).
3.2 Phylogeographical structure
The best fitting model was the general-time-reversible model of DNA substitution (GTR) with gamma-distributed rate variation among sites. The MC3 appeared to reach stationarity by visual inspection and convergence diagnostics. Average standard deviation of split frequencies (ASDF) reached 0.162, demonstrating that tree topologies are somewhat consistent across chains. While this is slightly higher than the typical ASDF target threshold of 0.1, ASDF is expected to be elevated in an intraspecific phylogenetic tree such as this where within-clade splits are more ambiguous because they are driven by a population genetic process. Mean effective sample size (ESS) for all parameters was >600, except the tree length parameter (TL) which was 124.50 and the alpha parameter which was 205.57. The potential scale reduction factor (PSRF; i.e., the Gelman and Rubin statistic or ) ranged from 0.999 to 1.070, indicating the continuous parameters from multiple chains have reached the same distribution. We rooted the tree using sagebrush lizard (S. graciosus) as an outgroup, and sorted the nodes in increasing order which roughly reflected latitudinal genetic structure (Figure S8).
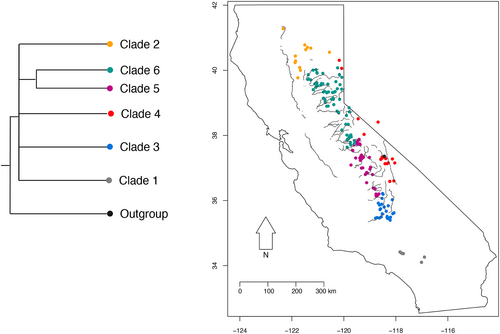
We identified major clades by grouping individuals into the six primary monophyletic groups in the phylogeny (Figure 4; Figure S8). We then identified minor clades by grouping individuals into groups based on high clade-credibility values, yielding 16 minor clades (Figure 5). We also identified a paraphyletic group (Group 6.4), which only includes samples from the American river (Figure 5; Figure S8). To maintain conservative estimates of clade–canyon specificity, we omit this paraphyletic group in our calculations of clade–canyon membership, as well as six samples that were not included in these clades (Figure S8). Therefore, our phylogeographical analysis and clade–canyon membership calculations include only samples that form distinct monophyletic clades at each of the scales of interest. Overall, we seek to investigate fine-scale phylogeographical patterns, with a specific focus on the degree to which river canyon topography is associated with phylogeographical structure within the Sierra Nevada range. We focus exclusively on how monophyletic groups are distributed among river drainages since these can be considered independent evolutionary lineages in a way that paraphyletic groups cannot.
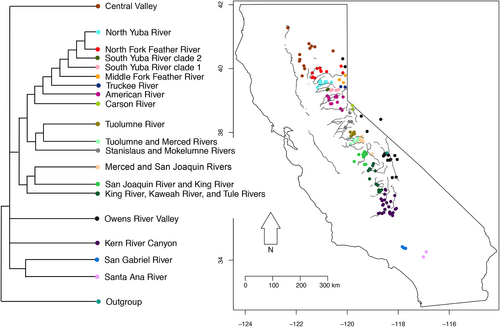
3.3 Phylogenetic membership among major clades and major river canyons
Across the five major clades, we find that Clade 2: California Central Valley, Clade 3: Kern River Canyon, and Clade 4: Owens River Valley each contain samples from a single canyon. These are the three largest river canyons and valleys across our sampling range. Clade 5 spans four major river canyons: Kaweah River, Kings River, Merced River and San Joaquin River. Clade 6 spans six major river canyons: the American River, Feather River, Merced River, Mokelumne River, Stanislaus River and Tuolumne River (Figure 4; Table 2; Figure S8). Overall, we find some degree of specificity with three of the clades each containing samples from single areas and drainages, while two of the clades span several drainages, albeit smaller than the California Central Valley, Owens River Valley and Kern River Canyons. Within each major river canyon, samples tend to be from the same clade—with the exception of Merced River samples, which are grouped in Clades 5 and 6. When examining the number of canyons-per-clade, 3/5 of these major clades (60%) include samples from a single river valley, while the remaining 2/5 (40%) from the interior Sierra Nevada include samples from more than three canyons. When examining the number of clades-per-canyon, 11/12 major river canyons include samples from a single clade (92%), with the exception of Merced River, which includes samples from Clades 5 and 6 (Figure 4; Table 3; Figure S8).
| Clade | Spanning canyons | No. of canyons |
|---|---|---|
| Clade 2 | California Central Valley | 1 |
| Clade 3 | Kern River Canyon | 1 |
| Clade 4 | Owens River Valley | 1 |
| Clade 5 | Kaweah River, Kings River, Merced River, San Joaquin River | 4 |
| Clade 6 | American River, Feather River, Merced River, Mokelumne River, Stanislaus River, Tuolumne River | 6 |
- Note: We also include the Owens River Valley and the California Central Valley to the west of the Sierra Nevada (including samples from the east and north of the Sierra Nevada).
| Canyon | Spanning clades | No. of clades |
|---|---|---|
| American River | Clade 6 | 1 |
| California Central Valley | Clade 2 | 1 |
| Feather River | Clade 6 | 1 |
| Kaweah River | Clade 5 | 1 |
| Kern River Canyon | Clade 3 | 1 |
| Kings River | Clade 5 | 1 |
| Merced River | Clade 5, Clade 6 | 2 |
| Mokelumne River | Clade 6 | 1 |
| Owens River Valley | Clade 4 | 1 |
| San Joaquin River | Clade 5 | 1 |
| Stanislaus River | Clade 6 | 1 |
| Tuolumne River | Clade 6 | 1 |
3.4 Phylogenetic membership among minor clades and river canyons
Here we evaluate the relationship between the minor clades and the minor river canyons within the Sierra Nevada region to assess the degree to which river canyons and phylogenetic clades are congruent. We excluded the paraphyletic group from the American River and six additional individuals (3% of samples) that do not group into clades. Overall, in terms of canyons-per-clade, we find 8/13 (62%) clades include lizards sampled from a single river canyon, 3/13 (23%) of clades include lizards sampled from two river canyons, and 2/13 (15%) clades include lizards sampled from three or more river canyons (Figure 5; Table 4; Figure S8). When examining the number of clades-per-canyon, we find 8/14 (57%) river canyons include samples from a single clade, while the remaining 6/14 (43%) river canyons include samples from two clades, and no river canyons included samples from three or more clades (Figure 5; Table 5; Figure S8). Inclusion of the American River paraphyletic group does not change the results in any meaningful way (e.g., a 2% increase in the clades-per-canyon statistic if included).
| Clade | Spanning canyons | No. of canyons |
|---|---|---|
| Clade 3.1 | Kern River Canyon | 1 |
| Clade 5.1 | Kings River, Tule River, Kaweah River | 3 |
| Clade 5.2 | San Joaquin River, Kings River | 2 |
| Clade 5.3 | Merced River, San Joaquin River | 2 |
| Clade 6.1 | Sanislaus River, Mokelumne River | 2 |
| Clade 6.2 | Tuolumne River, Merced River, Stanislaus River | 3 |
| Clade 6.3 | Carson River | 1 |
| Clade 6.5 | Truckee River | 1 |
| Clade 6.6 | Feather River (Middle Fork) | 1 |
| Clade 6.7 | South Yuba River (clade 1) | 1 |
| Clade 6.8 | South Yuba River (clade 2) | 1 |
| Clade 6.9 | Feather River (North Fork and Middle Fork) | 1 |
| Clade 6.10 | North Yuba River | 1 |
| Canyon | Spanning clades | No. of clades |
|---|---|---|
| Carson River | Clade 6.3 | 1 |
| Feather River | Clade 6.6, Clade 6.9 | 2 |
| Kaweah River | Clade 5.1 | 1 |
| Kern River | Clade 3.1 | 1 |
| Kings River | Clade 5.1, Clade 5.2 | 2 |
| Merced River | Clade 5,3, Clade 6.2 | 2 |
| Mokelumne River | Clade 6.1 | 1 |
| North Yuba River | Clade 6.10 | 1 |
| San Joaquin River | Clade 5.2, Clade 5.3 | 2 |
| South Yuba River | Clade 6.7, Clade 6.8 | 2 |
| Stanislaus River | Clade 6.1, Clade 6.2 | 2 |
| Truckee River | Clade 6.5 | 1 |
| Tule River | Clade 5.1 | 1 |
| Tuolumne River | Clade 6.2 | 1 |
4 DISCUSSION
4.1 Landscape genetics at multiple spatial scales
We find a great deal of concordance among landscape genetics inferences at the 1225-, 2500- and 4225-km2 spatial scales in both our visual assessment of the circuitscape surfaces and based on correlation tests, where river canyons appear to be important features maintaining genetic connectivity. In this case, it appears that the abiotic environment within river canyons—higher temperatures, less seasonality in temperature and precipitation, less rugged areas—are the landscape features that promote connectivity, rather than the rivers within the canyons themselves, as is presumably the case for amphibians such as the foothill yellow-legged frog (Rana boylii; Lind et al., 2011) and the aquatic paradoxical frog (Pseudis tocantins; Fonseca et al., 2021). The larger spatial scale circuitscape layer at 6400 km2 also largely identified river canyons as facilitators to gene flow, but demonstrated a low correlation with the other spatial layers (Table 1). Here, although the general landscape features that strongly determine genetic similarity (e.g., river canyons) are similar at different scales including at the 6400-km scale, there is a notable lack of resolution around features known to inhibit gene flow across the landscape (such as water bodies—including lakes and rivers; Figures S1d, S3d and S6d). We believe this result exemplifies a tradeoff in landscape genetics analyses at different spatial scales. At smaller spatial scales, the model correctly identifies canyons as facilitators to gene flow and water bodies as barriers to gene flow. However, at the largest spatial scale, while river canyons are found to facilitate gene flow, there is a loss of resolution in the fine details of the landscape features that promote and inhibit gene flow. This may be partially attributed to the large flat area in the vicinity of Bridgeport in the northeastern corner of the largest 6400-km2 scale, which should not restrict movement for S. occidentalis in the manner in which water bodies (which are also flat low-ruggedness regions of space) are known to (Figure 2; Figures S1–S4d). As this region should not restrict connectivity, and even may promote connectivity, it may be that the large-scale analyses are confounded by seemingly identical environments that exert dissimilar effects on population connectivity. This issue may be exacerbated at larger spatial scales where such environments are increasingly common. This result highlights the importance of conducting landscape genetics studies at a relevant scale (Anderson et al., 2010; Jackson & Fahrig, 2014). In sum, although we found a great deal of concordance between results at several smaller spatial scales spanning 1225–4225 km2, the larger spatial scale (i.e., 6400 km2) failed to capture some of the nuances of landscape effects on genetic connectivity that are apparent at smaller scales.
4.2 Phylogeography and river canyons
Perhaps the most striking pattern from the phylogeographical data is the major clade structure along the largest river valleys in the vicinity of the Sierra Nevada range. Most notably, Clade 4 extends across the broad Owens River Valley ~500 km along the eastern side of the Sierra Nevada, adjacent to several other major clades with individuals sampled from adjacent but distinct river canyons. Similarly, Clade 2, present in the California Central Valley, covers an area of ~20,000 km2 and includes samples from the Central Valley, north Sierra Nevada, and a sample from the eastern Sierra Nevada adjacent to the Owens River Valley Clade 4. This may exemplify the pattern of secondary contact at the northern extent of the Sierra Nevada range as the species moved north following Quaternary glacial cycles, as illustrated by Bouzid et al. (2022). Our phylogenetic tree is broadly consistent with recent range-wide studies on the phylogenetic structure of S. occidentalis, where Southern California and West Sierra Nevada populations form distinct clades (Bouzid et al., 2022; Wishingrad & Thomson, 2023). However, because our focus was the Sierra Nevada range, our sampling did not extend to the central and southern California Central Valley, which we expect would group with Clade 2. More generally, broad comparative phylogeographical studies have identified the California Central Valley as a common geographical break between lineages spanning east to west, and less so north to south, suggesting that large river drainages such as these are common conduits maintaining broad-scale distributions of lineages (Rissler et al., 2006).
The Kern River Canyon (Clade 3) is the largest river canyon of the Sierra Nevada and lies at the southern end of the mountain range. All lizards sampled from this river canyon clustered in a single clade extending due north along the canyon, including samples that lie further north than Clade 5 samples from the Tule River in a nearby but distinct river canyon. Interestingly, the northern Kern River Canyon also coincides with a phylogeographical break in several other reptiles and amphibians, including the California newt (Taricha torosa) and the Sierra newt (T. sierrae) (Kuchta, 2007; Kuchta & Tan, 2006), the mountain yellow-legged frog (Rana muscosa) and the Sierra Nevada frog (R. sierrae) (Vredenburg et al., 2007), and northeastern and coastal subclades of the California mountain kingsnake (Rodríguez-Robles et al., 1999; but see Myers et al., 2013). While this area seems to be potentially important in structuring phylogeographical lineages for some species, a broader study of 22 reptile and amphibian species in California representing 75 phylogeographical lineages did not identify the northern Kern River Canyon as a prominent region generating this pattern (Rissler et al., 2006). It may be that this area represents a comparatively less effective phylogeographical break than the California Central Valley, Tehachapi Mountains and San Francisco Bay, or is idiosyncratic with respect to the species that encounter this region as a barrier to gene flow and genetic connectivity.
The Kern River Canyon clade, along with the sister Clades 5 and 6 which span several more, though comparatively small river canyons, constitute the remainder of the Sierra Nevada range. Interestingly, these three clades are roughly situated within distinct hydrological regions that account for significant levels of genetic structure in Rana boylii (Lind et al., 2011). Clades 5 and 6 have an abrupt break in the YNP region (Figure 4). This finding agrees with an early mitochondrial DNA study of S. occidentalis phylogeography in the Yosemite region, in which a deep phylogenetic break separates the Tuolumne River and east Merced River samples (Leaché et al., 2010). At a finer phylogeographical scale, we also find a striking association between phylogenetic structure and geography in terms of clustering by river canyons (Figure 5). The majority of river canyons surveyed comprised samples from a single clade, indicating that distinct river canyons tend to contain evolutionarily distinct groups.
4.3 Inferences in landscape genetics and phylogeography
One of our primary objectives was to evaluate the degree of concordance between landscape genetics and phylogeographical inferences. Landscape genetics relies distinctly on population genomic data—in this case, allelic similarity between individuals distributed across a landscape—and represents recent timescales (Wang, 2010). Phylogeographical inference, on the other hand, relies on phylogenetic sequence data and groups individuals by evolutionary history, therefore representing relatively more ancient timescales (Wang, 2010). This distinction is reasonably expected to lead to different inferences, since the ecological processes structuring recent differentiation may differ from those structuring long-term evolutionary differentiation (Angelone et al., 2011; Jackson & Fahrig, 2014; Trumbo et al., 2013). Nevertheless, the broad-scale pattern of river canyons as key features linking populations in the landscape genetics results, and isolation by river canyon in the phylogeographical results, is consistent across the spatial and evolutionary timescales examined here.
Landscape genetics approaches are useful to estimate the cost of gene flow through various dimensions of environmental space along with the contribution of vegetation types on dispersal and genetic connectivity (Balkenhol et al., 2015; Manel & Holderegger, 2013; Peterman, 2018; Peterman et al., 2019; Storfer et al., 2007). These methods summarize which aspects of the landscape inhibit or promote gene flow, estimate the degree to which a particular feature affects gene flow relative to others, and highlight specific areas of a species range that are important for maintaining genetic connectivity across space (Balkenhol et al., 2009; Storfer et al., 2010). A drawback, however, is that very large extents (such as the entire Sierra Nevada range) are not computationally tractable, and current implementations of landscape genetics methods do not jointly evaluate population structure (Peterman, 2018; Richardson et al., 2016). In the Yosemite region, for example, landscape genetics methods identify water bodies as barriers to gene flow and river canyons as facilitators to gene flow. However, the method does not give any indication that the east Merced River is a distinct lineage from the Tuolumne River with an area of overlap between distinct clades along the western portion of the Merced River and following north along Yosemite River canyon and Tenaya Creek. Phylogeographical approaches, on the other hand, can be used to summarize patterns of evolutionary history and similarity over deep time across a large range (such as the entirety of the Sierra Nevada range), as we have done here (Avise, 2000; Rissler, 2016). These phylogeographical methods can also help elucidate genetic structure of a species over a large range, with details about areas of the landscape where lineages are evolutionarily unique. A limitation, however, is that phylogeographical methods do not inherently link isolation by landscape features to evolutionary dissimilarity (Rissler, 2016). While we have focused here on the union of landscape genetics and phylogeography, other spatial population genetics models such as Estimated Effective Migration Surfaces (FEEMS; Marcus et al., 2021), which visualizes areas of the landscape relative to isolation by distance (IBD), and Migration and Population Site Surfaces (MAPS; Al-Asadi et al., 2019) methods, which uses identity-by-descent tracks from genomic data to model migration rates and population sizes, may both further contribute to biogeographical inferences in uniquely advantageous ways.
This study illustrates the power of landscape genetics methods when used to generate a hypothesis about the effect of prominent landscape features on gene flow and genetic connectivity, and phylogeographical methods to test a directed hypothesis about the effect of a landscape feature on evolutionary history and genetic structure. This approach using complementary methods represents a powerful way to link both small- and large-scale evolutionary and spatial processes. In the case of S. occidentalis, river canyons appear to be highly relevant in structuring populations at both small and large geographical scales as well as ancient and recent evolutionary timescales. As predicted for polygynous species, S. occidentalis dispersers are usually males that may attempt to seek out other suitable habitats (Massot et al., 2003). Our data suggest encountering unfavourable environmental conditions experienced at higher elevations leads to dispersal along valleys, which tend to be warmer, less rugged topographically, and experience less seasonality in temperature and precipitation. River canyon valleys have remained important gene flow corridors today, and tend to be more genetically homogeneous within, rather than between, river valleys.
While our current study provides insights about a terrestrial lizard, we believe a much stronger understanding of how the environment structures genetic variation across space and time relies on data from more species. Indeed, even closely related species can exhibit different patterns of population structure depending on the spatial scale (Dudaniec et al., 2016; Mcgreevy et al., 2021). The degree of specialization, environmental tolerances and other traits have been shown to play a role in spatial genetics processes (Moritz et al., 2012; Papadopoulou & Knowles, 2016; Paz et al., 2015). We believe examining these processes across an increasing number of species will be critical to understanding the general rules governing spatial patterns of genetic variation.
While the geographical scale of spatial genetic structure and different aspects of a species' life history are predicted to play a role in how genetic variation is distributed across space (Anderson et al., 2010), most landscape genetic studies have not carefully considered the effect of spatial scale on relationships between population genetic structure and landscape features (Balkenhol et al., 2015). However, some early studies have examined the question of scale to various degrees, with mixed results. An important early landscape genetics study on American black bears (Ursus americanus) identified different factors influencing gene flow across study areas of different sizes (Short Bull et al., 2011). Trumbo et al. (2013) found that stream dispersal was not an important landscape variable facilitating gene flow in Cope's giant salamander (Dicamptodon copei) in the Cascade region. This contrasts with the inferences drawn by Steele et al. (2009), who found evidence for stream-based gene flow in D. copei when sampling at a finer spatial scale. We hypothesize that these contrasting results may be attributed to an effect of study extent and that different landscape feature can be important barriers or corridors to gene flow at different scales (Trumbo et al., 2013). In another study, Angelone et al. (2011) explicitly considered spatial scale informed by movement frequency in the European tree frog (Hyla arborea) at three different spatial scales, corresponding to distances they regularly move (<2 km), less often move (2–4 km) and rarely move (4–8 km). Their results demonstrated scale-dependent landscape genetic effects, with different landscape elements hindering gene flow at different scales. Importantly, a simulation study showed that multiple generations of dispersal and gene flow linked local populations, suggesting that scale-dependent inferences should be examined at scales exceeding those over which animals regularly disperse (Jackson & Fahrig, 2014). Recent studies have been approaching this question more deliberately. For example, a study examining the effect of area size, or extent, on landscape genetics inferences in the dispersal-limited Mississippi slimy salamander (Plethodon mississippi) revealed that the effect of land-use class on gene flow was relatively consistent between two study areas of different sizes (Burgess & Garrick, 2021). Finally, another study on Texas bobcats (Lynx rufus) identified herbaceous rangeland as an important feature structuring genetic variation at small scales, whereas agriculture was more important at broader geographical scales, indicating scale-dependent effects (Cancellare et al., 2021). The present study adds to the growing body of evidence that scale-dependent effects are important to consider, and helps clarify how consistent this finding is across landscapes when also considering climate and topography, in species with different life-history strategies, and across landscapes that span even larger spatial scales informed by both landscape genetics and phylogeographical approaches.
5 CONCLUSIONS
New advances continue to reveal an increasing number of details about how evolutionary history is structured and influenced by the landscape. Conceptual frameworks that expand on early ideas in spatial population genetics, such as Wright's IBD (Sharbel et al., 2000; van Strien et al., 2015; Wright, 1943), are now commonplace. Isolation by environment (IBE), for example, describes the relationship between different environments and genetic variation (Cancellare et al., 2021; Kozakiewicz et al., 2020; Lee & Mitchell-Olds, 2011; Wang & Bradburd, 2014), while isolation by resistance (IBR) aims to quantify gene flow through distinct landscape features separating populations or individuals across space (Cushman et al., 2006; Goldberg & Waits, 2010; Kozakiewicz et al., 2020; McRae, 2006; Short Bull et al., 2011), both of which organize genetic structure across landscapes at different scales and different biological contexts (Wang & Bradburd, 2014). Phylogeographical methods have similarly contributed to our understanding of spatially explicit population structure (Zamudio et al., 2016). As these two frameworks share many of the same objectives, synthesizing theory, models and methods between these fields will help bring about a better understanding of ecological and evolutionary processes structuring genetic variation (Rissler, 2016). Here we evaluate the degree to which landscape genetics recapitulates phylogenetics, the spatial scales at which each is best applied, and the relative benefit of each method in the context of understanding spatial population structure. We look forward to continued progress in the field of spatially explicit population genetics, especially studies focused on integrating methods from across fields and across species, which hold promise for generating increasingly rich insights into the genetic structure of populations in the context of environmental variation and landscape composition across space and time.
AUTHOR CONTRIBUTIONS
V.W. was responsible for project conceptualization, field sampling, data collection, data analysis, data visualization, and writing the manuscript. V.W. and R.C.T. contributed to review and editing of the manuscript.
ACKNOWLEDGEMENTS
We are grateful to G. Pauly contributing supplies needed to preserve museum specimens, and to A. Leaché and S. Bouzid for contributing samples. We thank A.B. Musgrove for assistance with fieldwork, A.J. Barley for providing guidance with lab work, M. Hadfield and the Kewalo Marine Lab for providing access to Pippin Prep equipment for DNA size-selection, three anonymous reviewers for insightful comments that improved this study, and L. Risser for permission to reuse her figure. We are grateful to M. Kakimoto for her keen attention to detail as an editor and first reader. V.W. is deeply indebted to his PhD committee members: R. Toonen, F. Reed, A. Wright, and D. Rubinoff for their intellectual guidance and expert advice. The technical support and advanced computing resources from the University of Hawaiʻi Information Technology Services – Cyberinfrastructure are gratefully acknowledged. This study was funded by the Theodore Roosevelt Memorial Grant from the American Museum of Natural History, the RCUH Fellowship from the University of Hawaiʻi, the Watson T. Yoshimoto Fellowship from University of Hawaiʻi Ecology, Evolution and Conservation Biology program, the Systematic Research Fund from the Linnean Society of London and the Systematics Association, the Jones-Lovich Research Grant in Southwestern Herpetology from the Society for the Study of Amphibians and Reptiles, a Grant-in-Aid of Research from the Society of Integrative and Comparative Biology, a Grant-in-Aid or Research from Sigma Xi, a University of Hawaiʻi GSO grant, and an NSF grant DEB 1754350 awarded to R.C. Thomson. Euthanasia methods followed those in protocol 16-2384, approved by the UH Institutional Animal Care and Use Committee. All research was conducted under Scientific Collecting Permit SC-13472 issued to V. Wishingrad by the California Department of Fish and Wildlife. We acknowledge the Indigenous lands on which this study took place, including those of the Newe, Kawaiisu, Numu, Yokuts, Tübatulabal, Monache, Chukchansi, Miwok, Washoe, Nisenan, Kojomk'awi, Mechoopda, Maidu, Yana, Atsugewi, Achumawi, Winnimem Wintu, and Shasta.
6 CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Open Research
OPEN RESEARCH BADGES
This article earned an Open Data Badge for making publicly available the digitally-shareable data necessary to reproduce the reported results. For details see the Data Availability statement.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available at Dryad: https://doi.org/10.5061/dryad.2ngf1vhsw, and raw sequencing reads are archived at NCBI: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA914787.




