Host blood meal identity modifies vector gene expression and competency
Handling Editor: Camille Bonneaud
Abstract
A vector's susceptibility and ability to transmit a pathogen—termed vector competency—determines disease outcomes, yet the ecological factors influencing tick vector competency remain largely unknown. Ixodes pacificus, the tick vector of Borrelia burgdorferi (Bb) in the western U.S., feeds on rodents, birds, and lizards. Rodents and birds are reservoirs for Bb and infect juvenile ticks, while lizards are refractory to Bb and cannot infect feeding ticks. Additionally, the lizard bloodmeal contains borreliacidal properties, clearing previously infected feeding ticks of their Bb infection. Despite I. pacificus feeding on a range of hosts, it is undetermined how the host identity of the larval bloodmeal affects future nymphal vector competency. We experimentally evaluate the influence of larval host bloodmeal on Bb acquisition by nymphal I. pacificus. Larval I. pacificus were fed on either lizards or mice and after molting, nymphs were fed on Bb-infected mice. We found that lizard-fed larvae were significantly more likely to become infected with Bb during their next bloodmeal than mouse-fed larvae. We also conducted the first RNA-seq analysis on whole-bodied I. pacificus and found significant upregulation of tick antioxidants and antimicrobial peptides in the lizard-fed group. Our results indicate that the lizard bloodmeal significantly alters vector competency and gene regulation in ticks, highlighting the importance of host bloodmeal identity in vector-borne disease transmission and upends prior notions about the role of lizards in Lyme disease community ecology.
1 INTRODUCTION
Vector competency — the ability of a vector to successfully acquire and transmit a pathogen — and the factors that modulate it are increasingly the focus of efforts to control the emergence and spread of vector-borne zoonotic diseases (Beard et al., 2002; Beerntsen et al., 2000; de la Fuente et al., 2017; Iturbe-Ormaetxe et al., 2011; Pais et al., 2008). Manipulation of vector competency has been discussed as a disease prevention strategy in mosquitoes, teste flies, and triatome bugs (Beard et al., 2002; Iturbe-Ormaetxe et al., 2011; Kean et al., 2015; Pais et al., 2008; Weiss & Aksoy, 2011). In these vectors, rearing of naturally resistant populations, modifications of vector endosymbionts, and gene editing have been studied and implemented as applications of biological control to alter vector competency and reduce disease transmission (Beard et al., 2002; Iturbe-Ormaetxe et al., 2011; Kean et al., 2015; Pais et al., 2008). While strides have been made in understanding and manipulating vector competency in many systems, these studies highlight the complexity of vector-pathogen interactions and suggest that a more mechanistic understanding of disease transmission holds promise for disease control. For tick-borne pathogen systems in particular, the plasticity of vector competency and responsiveness to environmental or biological inputs remains poorly understood.
Tick-borne diseases constitute 40% of the emerging vector-borne diseases worldwide (Jones et al., 2008; Schwartz et al., 2017; Swei et al., 2020) and are sensitive to changing abiotic and biotic interactions driven by land use change and increased globalization (van Baalen & Sabelis, 1995; de la Fuente et al., 2017; Keesing et al., 2010; Swei et al., 2020). In the northern hemisphere, Lyme disease is the most common vector-borne disease, causing an estimated 300,000 cases annually in the USA (Kilpatrick & Randolph, 2012; Mysterud et al., 2017; Swei et al., 2020). It is caused by the bacterial agent Borrelia burgdorferi (Bb) and vectored by Ixodes spp. ticks, whose life history involves blood-feeding on a wide range of hosts during each of their three life stages (larvae, nymph and adult) (LoGiudice et al., 2003). Tick blood-feeding induces physiological changes and challenges, including cuticular reconstruction (Gulia-Nuss et al., 2016; Perner et al., 2016) and antimicrobial activity (Smith et al., 2016). In addition, the identity of the blood meal host has important consequences for pathogen acquisition, tick survivorship, and microbiome composition (Landesman et al., 2019; Muturi et al., 2018; Sonenshine et al., 2005; Swei & Kwan, 2017).
Mounting evidence indicates that microbiome composition impacts vector competency through induced immunological responses, morphological changes, or direct competition between microbial components of the tick microbiome (Dong et al., 2009; Narasimhan et al., 2014, 2017; Ross et al., 2018; Wang et al., 2011). However, the precise relationship between microbiome composition and tick competency for Bb is not well understood (Eisen, 2020; Kwan et al., 2017). Bb acquisition by ticks is a complex process that requires the pathogen to evade numerous tick immune pathways and antimicrobial peptides (de la Fuente et al., 2017; Gulia-Nuss et al., 2016; Hajdušek et al., 2013; Hayes et al., 2020; Liu et al., 2012; Shaw et al., 2017; Smith et al., 2016) followed by successful colonization of the midgut (Pal et al., 2000; Zung et al., 1989). There is evidence that these interactions may be influenced by biotic interactions such as host bloodmeal identity or microbiome interactions (Landesman et al., 2019; Narasimhan et al., 2017; Swei & Kwan, 2017). Manipulation of the microbiome in laboratory-reared Ixodes scapularis found that lower microbiome diversity reduced Bb colonization through induced changes in tick midgut morphology (Narasimhan et al., 2014). In Ixodes pacificus, greater microbiome diversity was associated with Bb colonization in one study but not another (Kwan et al., 2017; Swei & Kwan, 2017). The life history of I. pacificus, the Lyme disease vector in the western U.S., provides a unique opportunity for natural microbiome manipulation. Juvenile I. pacificus feed predominantly on the western fence lizard, Sceloporus occidentalis, a Bb-refractory host that cannot infect ticks with Bb, but will also parasitize reservoir competent hosts, typically rodents such as Peromyscus spp. mice, western grey squirrels (Sciurus griseus), and dusky-footed woodrats (Neotoma fuscipes) (Eisen et al., 2003; Lane et al., 2005, 2009). In addition to being Bb-refractory, the blood of a lizard host contains complement proteins with borreliacidal properties that kill the Bb spirochetes in infected feeding ticks (Kuo et al., 2000). The broad variation in reservoir competency among bloodmeal hosts also leads to stark differences in tick microbiome composition (Swei & Kwan, 2017). Lizard-feeding results in a significant reduction in I. pacificus microbiome diversity relative to rodent-feeding (Swei & Kwan, 2017).
Given recent findings that lizard-feeding significantly reduces microbiome diversity (Swei & Kwan, 2017) and experimental evidence of tick microbiome diversity affecting Bb colonization success (Narasimhan et al., 2014), we sought to determine the direct effect of blood meal identity on I. pacificus vector competency in a tick pathogen acquisition experiment. We fed larval ticks on either lizards or mice then subsequently fed those ticks on Bb-infected mice and found that the ticks with previous lizard bloodmeals were significantly more susceptible to Bb infection. We then investigated mechanisms by which host blood meal may alter vector competency by conducting the first RNA-seq analysis on whole-bodied I. pacificus nymphs and comparing gene expression profiles for I. pacificus ticks following mouse or lizard larval blood meals. We found significant differences in tick vector competency based on larval host blood meal identity and detect multiple immune and metabolic factors that may alter I. pacificus vector competency for the Lyme disease pathogen.
2 MATERIALS AND METHODS
2.1 Ixodes pacificus collection
Fed I. pacificus larvae were collected from either western fence lizards (S. occidentalis) or deer mice (Peromyscus maniculatus). As lizards have naturally high larval burdens of I. pacificus (mean = 25; Swei et al., 2012), we collected ticks from S. occidentalis by capturing and holding lizards in drop off cages suspended over water for 3–4 days in a temporary field laboratory to collect replete ticks. All S. occidentalis were collected at China Camp State Park in Marin, CA. We then transferred all collected, replete larvae to the laboratory facilities at San Francisco State University. Natural burdens of I. pacificus larvae on Peromyscus spp. are very low (Swei et al., 2012). Because of low natural tick burdens, we experimentally attached up to 200 larval I. pacificus (BEI Resources) to P. maniculatus (Peromyscus Genetic Stock Center) in the laboratory. Unfed colony ticks were received two weeks prior to their larval feeding and stored at standard conditions of 23°C and 90% relative humidity. Colonies of I. pacificus larvae from BEI resources were originally collected in the San Francisco Bay Area in 2000 (Troughton & Levin, 2007). We collected and stored all replete ticks from lizard and mouse drop-off procedures under standard rearing conditions until they molted eight weeks later. All larval tick collection and feedings occurred at San Francisco State University across three separate experimental trials. Molted nymphs from trial one were sent to Indiana University for their nymphal feeding, molted nymphs from trials two and three were maintained at San Francisco State University for their nymphal feedings. All fed larval ticks from both mouse and lizard bloodmeals were uninfected with Bb prior to their bloodmeal as nymphs. Laboratory ticks and P. maniculatis mice were pathogen free, while the borreliacidal property in the lizard blood guaranteed that feeding larvae would remain uninfected after completion of their bloodmeal.
2.2 Experiments involving animals
All experiments involving S. occidentalis, P. maniculatus, and C3H/HeJ mice at Indiana and San Francisco State University were preapproved by Institutional Animal Care and Use Committee (IACUC) under the protocol number AU19-01 and researchers were properly trained by the university veterinarian.
2.3 Host inoculation and tick Bb acquisition experiments
Across the three experimental trials, Bb cultures were grown until they reached a concentration of 105 spirochetes/ml for trial one, 107 spirochetes/ml for trial two and 106 spirochetes/ml for trial three (Polovinchik, 2002). Culture concentrations were determined via hemocytometer. Mice were inoculated intradermally with 100 μl of Bb culture, 1,000 total spirochetes for trial one, 100,000 spirochetes for trial two, and 10,000 total spirochetes for trial three. Eight weeks post inoculation, successful Bb acquisition in the mice was determined via nested PCR of ear tissue targeting the 5S-23S rRNA spacer region (Lane et al., 2004).
Seventeen, three, and two individual Bb inoculated C3H/HeJ mice were used to feed nymphs in the first, second, and third experimental trials, respectively (Jackson Laboratory, Bar Harbor, Maine). The number of C3H/HeJ mice used in each experimental trial was determined and approved by Indiana and San Francisco State University IACUC officers and attending veterinarians. Nymphs from both lizard and mouse larval blood meals were then placed on Bb-infected C3H/HeJ mice for nymphal feeding. A total of 36 lizard-fed and 46 mouse-fed I. pacificus nymphs were collected across the three experimental trials (Figure S1). All Bb inoculation of C3H/HeJ mice and tick feeding experimental protocols were consistent across all three trials. The number of fed nymphs collected from individual C3H/HeJ mice along with the identity of their larval blood can be found in Table S1.
In preparation for our downstream transcriptomic analyses additional nymphal tick feedings were conducted on uninfected C3H/HeJ in trial three to act as our uninfected control. Three uninfected C3H/HeJ mice were used to feed 12 mouse-fed and 12 lizard-fed larvae. Additionally, a subset of 12 mouse-fed and 12 lizard-fed molted nymphs from trial three were sacrificed prior to the nymphal feedings to examine the direct effect of the lizard or mouse larval bloodmeal on I. pacificus gene expression.
2.4 Nucleic acid isolation and pathogen testing
After completing their bloodmeal, nymphs were placed under standard rearing conditions for 24 h, flash frozen, and stored at –80°C until nucleic acid extraction. Prior to extraction, ticks were thoroughly surface sterilized with successive 1 ml washes of 3% hydrogen peroxide, 70% ethanol, and deionized H2O to remove surface contaminants. The tick was then lysed and homogenized using the Qiagen TissueLyser II (Qiagen). Tick samples were extracted simultaneously for total DNA and RNA using the Qiagen AllPrep DNA/RNA Micro Kit (Qiagen). DNA and RNA concentrations were measured using a Quibit fluorometer (ThermoFisher Scientific) in preparation for pathogen testing and library preparation. RNA content and quality were evaluated using a Bioanalyser (Agilent). DNA from engorged Bb-fed nymphs was tested for Bb infection in triplicate by qPCR using the protocol described in Barbour et al. (2009). Bb CA4 cultures were utilized to create a dilution series of Bb DNA standards for our qPCR testing ranging from 1–106 spirochetes per reaction. Ticks were determined negative if the calculated Ct score was greater than 40, which estimated a spirochete count of less than one per individual.
2.5 Statistical analyses
To determine if larval bloodmeal host was a predictor of nymphal pathogen acquisition, we used a generalized mixed-effect model (GLMM) with a binomial error distribution. We used larval host bloodmeal (lizard or mouse feeding) as a fixed effect with Bb infections status of the engorged nymphs as the response variable. With multiple nymphal ticks feeding on the same Bb-infected mouse, we included mouse ID (Table S1) as a random effect to account for pseudoreplication and any correlation between infection status and the individual nymphal host in our analyses. Trial number was nested with mouse ID as an additional random effect to account for host inoculum load variation among the experimental trials. The random effects were nested because trial and mouse ID were not independent covariates. Analyses were performed using the glmm package (v.1.4.2) in R (Knudson et al., 2021). All data and code for GLMM analyses are available at: https://github.com/kcring/Ring_et_al_2022_GLM
2.6 Tick transcriptome experimental design and analysis
Unfed, uninfected-fed, and Bb-fed nymphs with either mouse or lizard larvae bloodmeals from trial three of the host-feeding experiments were divided into six experimental groups and used in our transcriptomic analyses (Figure 1). Each experimental group will hereinafter be described with the following abbreviations: unfed nymphs are referred to as “UF” and fed nymphs are referred to by whether they were fed on a Bb-negative “-Bb,” or Bb-positive “+Bb” C3H/HeJ mouse. Additionally, the larval bloodmeal (lizard or mouse) in each experimental group is indicated in the subscript following the abbreviation. Groups one and two, which represent our unfed nymphs, “UFlizard” and “UFmouse” (Figure 1), were set aside to examine the effect of the lizard or mouse larval bloodmeal on I. pacificus gene expression. The remaining four groups represent the engorged nymphal ticks. Uninfected control groups three and four, “-Bblizard” and “-Bbmouse” were nymphs that fed on uninfected C3H/HeJ mice during their nymphal bloodmeal (Figure 1). Groups five and six, “+Bblizard” and “+Bbmouse” were nymphs that fed on Bb infected C3H/HeJ (Figure 1).
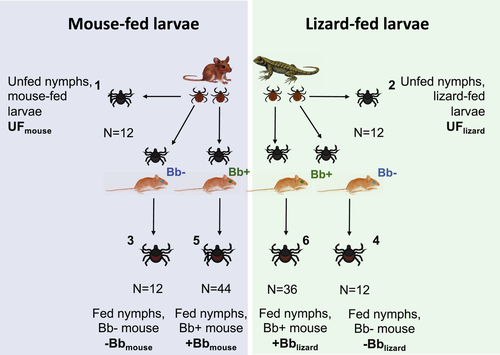
All engorged nymphs used in group five, +Bblizard, were tested and confirmed infected with Bb prior to library preparation. Conversely, engorged nymphs in group six, +Bbmouse, were tested and confirmed to be uninfected with Bb prior to library preparation. We used engorged nymphs with different infection statuses in groups 5 and 6 to examine gene expression differences associated with Bb acquisition success after different larval host bloodmeals.
We prepared three replicates from each of the six experimental tick feeding conditions (Figure 1) for a total of 18 libraries. We pooled three individual ticks for each experimental replicate. RNA-seq libraries were prepared from total RNA extracted from ticks followed by rRNA depletion using depletion of abundant sequences by hybridization (DASH) (Gu et al., 2016). The NEBNext Ultra II directional RNA library prep kit for Illumina (New England Biolabs, E7760S) was used to make RNA-seq libraries following the standard manual protocol. After constructing RNA-seq libraries from total RNA, reads containing rRNA sequences from Ixodes spp., Bb, and mouse were depleted using DASH, which targets Cas9 to abundant sequences in RNA-seq libraries. We utilized previously designed guide RNAs against mice and Ixodes spp. rRNAs (Dynerman et al., 2020), and we designed additional complementary guides to improve rRNA depletion of tick and Bb sequences using RNA-seq libraries made from total RNA from an I. scapularis nymph and Bb B-31 culture (Table S2).
2.7 rRNA depletion with DASH
Guide RNAs were designed using DASHit (http://dashit.czbiohub.org/), and prepared from DNA oligos as in Gu et al. (2016) following the protocol for in vitro transcription for dgRNA Version 2 (dx.doi.org/10.17504/protocols.io.3bpgimn). Both tracrRNA (5-AAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATGCTGTCCTATAGTGAGTCGTATTA) and pooled crRNA DNA oligo templates (Table S2) were annealed to equimolar amounts of T7 primer (5’-TAATACGACTCACTATAG) by heating to 95ºC for 2 min and slowly cooling to room temperature. Annealed templates were used in 1 ml in vitro transcription reactions with: 120 μl 10x T7 buffer (400 mM Tris pH 7.9, 200 mM MgCl2, 50 mM DTT, 20 mM spermidine (Sigma 85558)), 100 μl of T7 enzyme (custom prepped enzyme gifted from E. Crawford, diluted 1:100 in T7 buffer, final concentration: 100 μg/ml), 300 μl NTPs (25 mM each, Thermo Fisher Scientific, R0481), 4 μg of annealed crRNA template or 8 μg of annealed tracrRNA template, and water to 1 ml. In vitro transcription was performed for 2 h at 37°C. Reactions were split in half and purified using Zymo RNA Clean & Concentrator-5 Kit (Zymo Research R1015). To form the dgRNA complex for DASH, crRNA and tracrRNAs were diluted to 80 μM, mixed in equimolar amounts and annealed by heating to 95ºC for 30 s and cooling slowly to room temperature.
After transcription of dgRNAs, we performed DASH Protocol Version 4 (dx.doi.org/10.17504/protocols.io.6rjhd4n). All our RNA-seq libraries were pooled in equimolar amounts to a final concentration of 2.8 nM. Cas9 and transcribed dgRNAs were prepped by mixing: 2.5 μl 10x Cas9 buffer (500 mM Tris pH 8, 1 M NaCl, 100 mM MgCl2, 10 mM TCEP), 2.5 μl 40 μM Cas9 (gift from E. Crawford, prepared as in Gu et al., 2016), and 5 μl of 40 μM transcribed dgRNAs. The mix was incubated at 37°C for 5 min before 10 μl of pooled libraries were added. The mixture was incubated at 37°C for 1 h and then purified with Zymo DNA Clean & Concentrator-5 (Zymo Research D4014) following the PCR product protocol and eluting DNA into 10.5 μl of water. During cleanup, Cas9 was again mixed with buffer and dgRNAs as above and incubated at 37°C for 5 min. Following cleanup, eluted DNA was added to the second Cas9-dgRNA mix, and mixture was incubated at 37°C for 1 h for a second time. Then, 1 μL of proteinase K (New England Biolabs, P8107S) was added, and the mixture was incubated at 50°C for 15 min. The libraries were then purified with 0.9x volume of sparQ PureMag Beads (QuantaBio 95196) following the standard protocol, eluting in 23 μl of water. Libraries were amplified in a BioRad CFX96 using the Kapa HiFi Real-Time Amplification Kit in a 50 μl reaction with 25 μl master mix, 21 μl of the DASHed library pool, and 2 μl of 25 μM mix of Illumina P5 (5’-AATGATACGGCGACCACCGAGATCT) and P7 (5’- CAAGCAGAAGACGGCATACGAGAT) primers. The qPCR program for amplification was as follows: 98°C for 45 s (1 cycle), (98°C for 15 s, 63°C for 30 s, 72°C for 45 s, plate read, 72°C for 20 s) for 13 cycles. The libraries were removed from cycling conditions before leaving the exponential phase of amplification then purified with 0.9X volume of sparQ PureMag Beads according to the standard protocol.
Following DASH, RNA-seq libraries were sequenced on an Illumina NextSeq with paired-end 75 base pair reads. Fastq files and raw read counts have been deposited in the gene expression omnibus (GEO) under accession number GSE173109.
2.8 RNA-seq sequence processing and analysis
RNA-seq reads were trimmed of adapters and bases with quality score lower than 20 using Cutadapt (Martin, 2011) via Trim Galore! v0.6.5 and then mapped to the I. scapularis ISE6 genome (assembly GCF_002892825.2_ISE6_asm2.2_deduplicated) accessed from NCBI RefSeq (Miller et al., 2018), using STAR (v2.7.3a; Dobin et al., 2013). The I. scapularis transcriptome was used as a reference because genomic references for I. pacificus were not readily available at the time of this study. Reads mapping to predicted genes (gtf-version 2.2, genome build: ISE6_asm2.2_deduplicated, NCBI genome build accession: GCF_002892825.2, annotation source: NCBI I. scapularis Annotation Release 100) were tabulated using Subread FeatureCounts (v2.0.0; Liao et al., 2013), counting primary hits only.
DESeq2 v1.26.0 (Love et al., 2014), was used to determine differential expression between groups. DeSeq2 is a program in R that organizes sequence reads into a count matrix with one row for each gene and one column for each sample, a GLM is then fit for each gene determining whether the “reference” and “comparison” model coefficients are significantly different than zero. A Wald's test is utilized in the DeSeq2 GLM to test if a gene is significantly differentially expressed between experimental groups (Love et al., 2014). Additional information about the pairwise comparisons used in our DeSeq2 analyses are described in Table S3. Volcano plots from the EnhancedVolcano package in R, were used to visualize significant differential gene expression between experimental groups (Blighe et al., 2019).
A heat map and principal coordinate analysis plot were generated using Deseq2 and were utilized for exploratory data analysis and to visualize the effect of the experimental conditions on overall gene expression in each group (Love et al., 2014). The heatmap — which shows an overview over similarities and dissimilarities between samples — was generated from sample-to-sample distances and is based on normalized read counts for all genes. The principal coordinate analysis was also based on a distance matrix generated from normalized read counts and shows the overall effect of experimental covariates by displaying all samples in a 2D plane spanning their first two principal components. All code in our Deseq2 analysis is available at: https://github.com/choulabucsf/Ipac_DE_Ring_et_al_2021
3 RESULTS
3.1 Host-to-tick Bb acquisition experiment
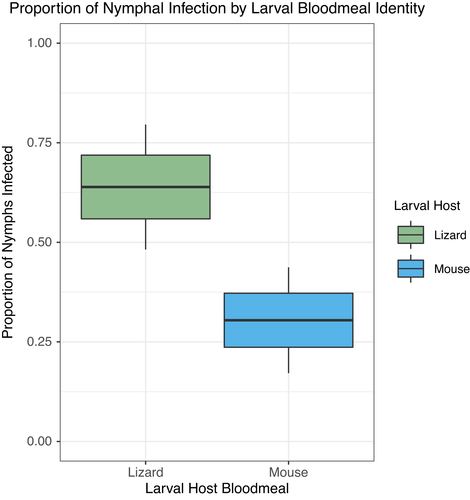
The effect of larval host bloodmeal on I. pacificus nymphal vector competency was examined in a host-to-tick pathogen acquisition experiment where replete larval I. pacificus were obtained from mice or lizards and then subsequently fed on Bb-infected C3H/HeJ mice (Figure 1). I. pacificus nymphs that fed on Bb-inoculated C3H/HeJ mice were significantly more likely to become infected if they previously fed on lizards as larvae than if they fed on mice (Table 1). During their nymphal bloodmeal, 64% of lizard-fed ticks (3.2 = 23/36) became infected with Bb compared to 30% of the mouse-fed ticks (N = 14/46; Figure 2). Even after accounting for C3H/HeJ mouse ID and trial as random effects, our GLMM analyses found that the lizard larval bloodmeal is a significant, positive predictor of Bb acquisition in I. pacificus (Table 1). These results support our hypothesis that the host bloodmeal source may shape intrinsic vector competency of ticks across at least one life stage transition.
| Response | Fixed effect | Estimate | Standard error | z value | p-value |
|---|---|---|---|---|---|
| Infection status | (Intercept) | −2.45 | 1.12 | −2.19 | <.05* |
| Infection status | Lizard larval bloodmeal | 4.67 | 1.51 | 3.09 | <.01** |
3.2 Experimental transcriptomic differences
To investigate potential explanations for the differences in vector competency between lizard-fed and mouse-fed ticks and I. pacificus gene expression patterns in general, we conducted an RNA-seq analysis to compare gene expression between ticks with different blood histories, engorgement status, and pathogen exposure. A total of 18 RNA-seq libraries were prepared from the six experimental groups, each represented by three replicates and resulting in over 370 million total reads (Figure 1). Mapping rates among replicates averaged at 45% with 15 million reads per library. Sequencing statistics for each replicate are presented in Table S4.
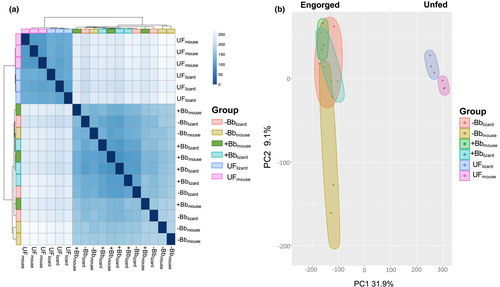
The heatmap showed that tick engorgement status (unfed vs. engorged) induced significant changes in I. pacificus gene expression (Figure 3a). Additionally, the PCA plot indicated significant distinction of overall gene expression between unfed ticks of either bloodmeal type, UFlizard and UFmouse (Figure 3b). The engorged experimental groups (groups three to six) had similar gene expression profiles and did not distinctly cluster together by experimental condition (Figure 3b).
3.3 Differential gene expression
To investigate the mechanism through which host blood alters tick vector competency, we took a global transcriptomic approach to identify key genes or pathways modulated by mouse or lizard hosts. Differential gene expression analyses focused on several pairwise comparisons to examine transcriptomic differences between (1) the lizard versus mouse bloodmeal in the unfed group (2) unfed versus fed ticks, and (3) bloodmeal identity distinctions between Bb exposed groups.
The comparison between our unfed nymphs (UFlizard vs. UFmouse), demonstrated that the lizard bloodmeal induced distinct transcriptomic changes in I. pacificus with 468 significantly differentially expressed genes (DEGs) (Table S5). While many of the DEGs remain undescribed, some of the highest upregulated genes induced by the lizard bloodmeal in the unfed group included antioxidants and antimicrobial peptides (Figure 4a). The antioxidant glutathione peroxidase was the most significant DEG and was upregulated 48.5-fold after the lizard bloodmeal compared to mouse bloodmeal. Other tick antioxidants that were upregulated after the lizard bloodmeal include peroxidase (upregulated 21-fold) and glutathione-S-transferase (upregulated four-fold; Figure 4a). We also found several DEGs that are related to the regulation of antimicrobial peptides but have never been described in I. pacificus ticks, such as acanthoscurrin-1, acanthoscurrin-2-like, micropulsin and micropulsin isoform, which were upregulated by 27.9, 104, 4, and 22.6-fold, respectively (Figure 4a; Table S5).
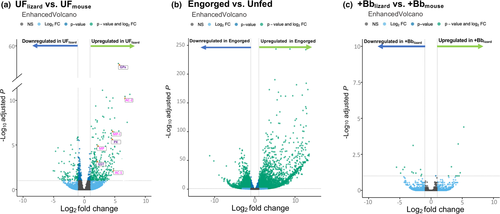
To analyse the DEGs between engorged and unengorged ticks, we combined the two unfed groups (UFlizard & UFmouse) as our reference ‘unfed’ group and compared this group to the engorged nymphs (i.e., -Bblizard, -Bbmouse, +Bblizard, & +Bbmouse). Engorgement-induced significant gene expression differences, producing 6730 significant DEGs with most of the difference in expression being upregulated genes in the engorged groups (Figure 4b). Of the top 100 most significant DEGs, 25% were related to cuticle formation (Table S6). Other notable genes that were differentially expressed include the antioxidant and detoxifying genes, glutathione peroxidase and sulphotransferase, which were both upregulated 4096-fold in the engorged group (Table S6). Over a hundred DEGs between unfed and engorged ticks remain uncharacterized.
Despite significant differences in pathogen acquisition success between the +Bblizard and +Bbmouse in pathogen transmission experiments (Table 1), only 25 genes were differentially expressed between the two groups (Figure 4c). The two most significantly DEGs included exonuclease V-like (upregulated 32-fold) and 4-coumarate–CoA ligase (downregulated 8-fold) in +Bblizard (Figure 4c). No genes that are known to be related to immune function were detected as differentially expressed in the +Bblizard vs. +Bbmouse comparison, with 7 of the 25 differentially regulated genes classified as uncharacterized (Table S7).
4 DISCUSSION
Vector competency is considered an intrinsic property of a vector that determines its ability to acquire, maintain, and transmit pathogens (Beerntsen et al., 2000), but the extent to which it is modulated by biotic or abiotic factors is poorly understood, especially in tick-borne pathogen systems. Here, we conducted a tick Bb acquisition experiment and transcriptome analysis on I. pacificus to determine if and by what potential mechanisms host bloodmeal history affects I. pacificus vector competency for the Lyme disease pathogen, Bb. Through Bb feeding experiments, we found that larval bloodmeal history significantly affects I. pacificus pathogen acquisition, a key component of vector competency. When ticks that fed on either lizards or mice as larvae fed on Bb-infected mice as nymphs, the previously lizard-fed ticks were twice as likely to acquire the pathogen. Further, significant transcriptomic signatures were detected between ticks with different bloodmeal histories. Gene expression analysis identified an upregulation of tick antioxidants and antimicrobial peptides in I. pacificus that fed on lizards, which may play a role in altering tick vector competency for Bb. Our results begin to uncover a mechanistic understanding of how host blood meal affects I. pacificus gene expression and the ecological factors that control I. pacificus susceptibility to Bb.
Recent studies suggest that tick microbiome composition can impact vector competency (Narasimhan et al., 2017) and that host bloodmeal source can shape microbiome community structure (Landesman et al., 2019; Swei & Kwan, 2017). These two recent findings motivated this study to test whether the lizard bloodmeal host that has been previously shown to reduce tick microbiome diversity (Swei & Kwan, 2017) can have subsequent effects on vector competency. Across three separate experimental trials, we found that a previous lizard bloodmeal significantly increased the acquisition of Bb in nymphal I. pacificus. These results were surprising, especially given that infected I. pacificus that feed on S. occidentalis are cleared of their infection (Kuo et al., 2000; Lane & Quistad, 1998). The Bb-refractory nature of S. occidentalis has long been held as evidence of the lizard's importance in maintaining lower prevalence of Lyme disease in the western U.S. and it probably contributes to lower disease risk relative to the northeastern U.S. However, whether this Bb-refractory property could be sustained transstadially in I. pacificus was unknown. Our results indicate that lizard-feeding does not preclude Bb infection in future life stages of I. pacificus, but rather enhances pathogen acquisition success relative to ticks with a prior mouse bloodmeal (Table 1). These results indicate that the acute and long-term consequences of a lizard bloodmeal on pathogen transmission are divergent.
The role of the microbiome in tick vector competency is unresolved (Kwan et al., 2017; Landesman et al., 2019; Narasimhan et al., 2014). In a prior study, lower microbiome diversity in I. scapularis was associated with lower Bb colonization success due to decreased expression of genes involved in gut epithelium renewal, which enhances Bb colonization (Narasimhan et al., 2014). Therefore, we predicted that lizard-fed ticks, previously shown to have significantly lower microbiome species diversity than mouse-fed ticks (Swei & Kwan, 2017), would similarly have lower Bb infection prevalence. However, our Bb acquisition experiment found that I. pacificus microbiome diversity, resulting from lizard-feeding (Swei & Kwan, 2017), and pathogen transmission success are negatively correlated. This finding may be due to species-specific differences between I. scapularis and I. pacificus or be driven using different experimental procedures used to manipulate the vector microbiome. Additionally, lizard feeding may affect tick vector competency through altering specific microbes rather than altering overall microbial diversity. Ultimately, the role of microbiome diversity and composition on pathogen acquisition success in Ixodes spp. remains uncertain and future studies are needed to disentangle the complicated interactions of these microbes.
Our RNA-seq analysis of I. pacificus with different bloodmeal histories revealed potential mechanisms that could be driving the differences observed in tick pathogen acquisition. Larval bloodmeal identity and engorgement have large impacts on I. pacificus gene expression (Figure 3). Unfed nymphs clustered significantly by larval bloodmeal type (lizard vs. mouse; Figure 3b), indicating that larval bloodmeal source induced distinct transcriptomic alterations in I. pacificus. We analysed gene expression profiles in unfed nymphs right after they molted from larvae to nymph. Our analysis suggests that the effect of the larval bloodmeal on I. pacificus gene expression is carried through the trans-stadial molt and is present prior to the initiation of the nymphal bloodmeal. Among the unfed ticks, bloodmeal history drove divergence of 468 significantly expressed genes between unengorged lizard and mouse fed ticks (Table S5). The most significant DEG between unfed ticks with different bloodmeal histories was glutathione peroxidase being upregulated in the lizard-fed group (Figure 5a). Glutathione peroxidase is an important antioxidative enzyme, that works by reducing H2O2 and detoxifying OH radicals and prevents oxidative stress and cell damage in the tick (Galay et al., 2017; Lubos et al., 2011). Two other known antioxidative enzymes, peroxidase and glutathione S-transferase were significantly upregulated after the lizard bloodmeal (Figure 5b,c). A nutritional dependence on blood has required ticks to evolve and produce antioxidants to digest an inherently toxic meal containing high levels of iron and pro-oxidant levels (Galay et al., 2017). Notably, glutathione peroxidase is homologous to SALP25d, a tick antioxidant produced in the salivary glands that protects Bb from harmful hydroxyl radicals in vitro (Narasimhan et al., 2007). A previous study silenced the SALP25d gene in ticks feeding on a Bb-infected mouse. They found that ticks lacking the SALP25d gene experienced a significant reduction in Bb colonization in their midgut compared to control ticks, indicating that SALP25d facilitates Bb colonization in the tick midgut (Narasimhan et al., 2007).
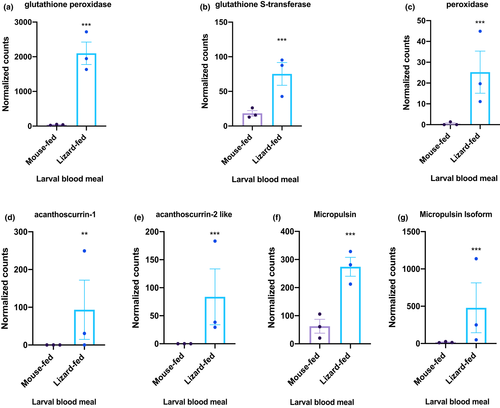
The exact mechanism by which the lizard blood initiates the production of antioxidants in feeding ticks is unknown, but unlike mammals, reptiles have nucleated red blood cells (Claver & Quaglia, 2009). Enucleation — the evolutionary loss of a nucleus in red blood cells — is unique to mammals, and is thought to have evolved to elevate haemoglobin levels to improve oxygen transport (Ahmed et al., 2020; Ji et al., 2011). Additionally, fence lizards are known to have significantly lower glutathione peroxidase activity within their tissues when compared mice (Tappel et al., 1982). We speculate that the upregulation of antioxidants in the lizard-fed ticks may be a result of oxidative stress in response to lower oxygen levels, lower glutathione peroxidase activity, and potentially higher reactive oxygen species in the nucleated lizard bloodmeal compared to the enucleated mammalian bloodmeal (Salin et al., 2015). The upregulation of glutathione peroxidase and other antioxidants in lizard-fed ticks has the potential to directly benefit Bb colonization from host to tick during the nymphal bloodmeal by increasing antioxidant concentration and protecting Bb from the harmful oxidative components of blood.
There was also a strong signal of microbial defense signals in unfed tick comparison. The antimicrobial peptides (AMPs) acanthoscurrin-1, acanthoscurrin-2, micropulsin, and a micropulsin isoform were all significantly upregulated in the unfed nymphs with prior lizard bloodmeals relative to prior mouse bloodmeals (UFlizard; Figure 5d–g). Acanthoscurrin is a glycine-rich cationic AMP, known to be expressed in the haemocytes of tarantula spiders, Acanthoscurria gomesiana, and has activity against the yeast, Candida albicans, and gram-negative bacteria (Lorenzini et al., 2003). Micropulsin is a cysteine-rich AMP with histidine-rich regions, found in the hemolymph of the cattle tick, Rhipicephalus microplus, with high activity against gram-positive bacteria and fungi (Silva et al., 2009). Neither of these AMPs have been detected in I. pacificus prior to this study, but these results indicate that they may play an important role in pathogen acquisition and warrant further study. While lizard blood feeding contributes to the expression of AMPs, it is unclear what upstream components initiate their production. The antimicrobial activity may be an outcome of an initiated humoral immune response or derived from host immune effector molecules, as demonstrated when I. scapularis feeds on a Bb-infected mouse (Smith et al., 2016). To understand how the expression of these AMPs occur, future gene expression studies should examine the tick immune response to the lizard larval bloodmeal at multiple time points to track the immune response at different stages of feeding. Interestingly, the upregulation of AMPs with broad activity against microbes coincides with a previously described study showing that a lizard bloodmeal significantly reduces I. pacificus microbiome diversity after feeding (Swei & Kwan, 2017). We are unsure if these two patterns are linked or if tick microbiome composition and richness have downstream alterations on vector competency. Future studies should examine (1) the activity of these AMPs on common microbes in I. pacificus, and (2) how microbiome composition facilitates or competes with Bb acquisition.
To further characterize the physiological changes that occur during I. pacificus host feeding, we analysed unfed versus fed ticks, 24 h after ticks completed their nymphal bloodmeal. Our study confirmed that engorgement induces a large number of transcriptional changes to the physical structure of the tick (Perner et al., 2016; Figure 3b). The greatest number of DEGs was between fed and unfed ticks (Figure 4). Genes related to cuticle formation, antioxidant production, and detoxification were all significantly upregulated in fed ticks (Table S6) and are consistent with structural reformation that occurs during the engorgement process when ticks must rapidly synthesize a new cuticle over the course of taking a large bloodmeal (Gulia-Nuss et al., 2016). Glutathione peroxidase and sulphotransferase were highly upregulated during engorgement and are critical for detoxifying the massive host bloodmeal and protect ticks from harmful oxidative stress inherent in blood feeding (Gulia-Nuss et al., 2016; Perner et al., 2016). These results, while unsurprising, indicate that transcriptomic changes during I. pacificus engorgement are like the physiological alterations found in I. scapularis and Ixodes ricinus (Gulia-Nuss et al., 2016; Perner et al., 2016).
Gene expression of I. pacificus is heavily shaped by engorgement status and bloodmeal history in unfed ticks but among the engorged nymphal ticks (groups 5 and 6; Figure 1), there was not a strong signal of bloodmeal history or infection status (Figure 3b). Despite the significant differences in pathogen acquisition between host bloodmeal experimental groups (Figure 2), only 25 genes with no known pathogen or immune function were differentially expressed between these groups. Comparing these results to our gene expression analysis from unfed nymphs, the strongest divergence in gene expression is present in the unfed ticks. This suggests that the physiological changes induced by the larval bloodmeal has lasting effects into the nymphal stage.
Our study documents a correlation between host blood meal and vector competency. We designed our experiments to mitigate limitations caused by logistical constraints in acquiring both mouse- and lizard-fed ticks. We were able to source laboratory colony reared ticks (BEI Resources, Manassas, VA) which originated in the same geographic region (San Francisco Bay Area, CA) as the field lizard-fed ticks, reducing the likelihood of them being genetically distinct populations (Troughton & Levin, 2007). This was necessary because field Peromyscus have naturally low tick burdens (Swei et al., 2012) and use of field-collected larvae is problematic because it is difficult to verify whether a tick had a previous, incomplete blood meal (Eisen, 2020). Importantly, laboratory reared ticks are routinely used in laboratory transmission studies and show no impairment to acquire pathogens (Hart et al., 2018; Koci et al., 2018; Yang et al., 2009). Laboratory reared ticks have been found to be less diverse than wild ticks (Kwan et al., 2017; Narasimhan et al., 2014). Although we did not quantify microbiome composition in this study, previous studies indicate that the effect of lizard feeding results in a larger decrease in I. pacificus microbiome diversity than laboratory rearing (Kwan et al., 2017; Swei & Kwan, 2017). These patterns suggest that the reduction in microbiome species richness that results from lizard-feeding should, in theory, reduce rather than enhance the observed differences in microbiome species richness and pathogen acquisition between the colony-reared mouse-fed and wild lizard-fed ticks. While we cannot fully rule out the possibility that tick source contributes to observed differences, the significance of our transmission data and the directionality of our observed findings suggests that there are other factors, such as blood meal source, that probably contribute to these results.
The genes described above were significantly differentially expressed based on host blood meal and represent biologically sensible mechanisms through which host blood meal may alter vector competence. However, we note that further investigation is needed to confirm the functional role of these genes. RNA interference experiments could be used to measure pathogen transmission rates after silencing specific genes highlighted here, which would clarify the mechanisms linking tick transcriptional changes to vector competence and aid in identifying candidate genes for disease control applications. Additionally, our study highlights the importance of a more complete annotation of the reference transcriptome for Ixodes spp. ticks. A large proportion of DEGs remain uncharacterized indicating additional investigation into tick molecular function and transcriptomics is needed. The lack of differentially expressed genes in our comparison of Bb-exposed nymphs may be due to gene expression associated with alterations in vector competency being rare and missed in sequencing compared to gene expression involved in blood digestion of the nymphal bloodmeal. Additionally, these results could be attributed to the timing of RNA sampling (24 h after completed bloodmeal). Deeper sequencing and examination of gene expression before, during, and immediately after feeding, would improve insight into the mechanism of pathogen colonization into I. pacificus.
Future experiments should focus on understanding the role of antioxidants and the AMPs identified in this study in modifying tick vector competency. We found a strong association between lizard bloodmeal history and antioxidant activity as well as AMP production. These responses coupled with naturally high natural tick burdens and preferential feeding on lizards may suggest an evolutionary benefit to feeding on the western fence lizard for I. pacificus. The broader ecological implications that lizard feeding has on tick vector competency and overall prevalence of Bb on a population level remains unclear. Because lizard feeding affects pathogen acquisition in the nymphal bloodmeal, this may affect infection prevalence in the adult tick stage or perhaps lead to changes in epigenetic regulation at the population level (De et al., 2021) but further studies are needed to explore this question.
The public health burden of Lyme disease is increasing, and diagnosis and treatment are expensive and imperfect. The complexity of tick-host-pathogen interactions involve many competing interactions, making intervention or prevention of disease transmission very difficult. A better understanding of the molecules, microbes, and antigens involved in vector competency presents a different approach to prevention (Bhowmick & Han, 2020; Galay et al., 2017; Iturbe-Ormaetxe et al., 2011; Pais et al., 2008). Identifying molecular and microbial drivers of tick survival and vector competency are attractive targets for novel control methods (de la Fuente et al., 2017). Using the perturbation of natural host bloodmeal, our study identified multiple molecular components that may be important in the successful acquisition of Bb in I. pacificus and identifies potential new targets for manipulating and preventing the transmission of tick-borne diseases.
ACKNOWLEDGEMENTS
We thank the Chan Zuckerberg Biohub for sequencing. KR and AS want to acknowledge funding support from the Pacific Southwest Regional Center of Excellence for Vector-Borne Diseases funded by the U.S. Centers for Disease Control and Prevention (Cooperative Agreement 1U01CK000516. This work was also funded by NSF (nos. 1427772, 1745411, 174037 to AS) and NIH (R01AI132851 to SC). Additional support for SC was provided by Chan Zuckerberg Biohub and the Pew Biomedical Scholars Program. FY is supported by the HHMI Gilliam Fellowship and ALS by the Life Sciences Research Foundation.
AUTHOR CONTRIBUTIONS
Kacie Ring, Lisa I. Couper, Seemay Chou, and Andrea Swei conceived the study. Kacie Ring and Lisa I. Couper conducted fieldwork. Kacie Ring, Lisa I. Couper, Keith Clay, and X. Frank Yang performed experimental tick feedings. Anne L. Sapiro and Kacie Ring executed library preparation, created the bioinformatic pipeline, implemented DASH, and analysed the RNA-seq data. Chase Mateusiak, Anne L. Sapiro, and Kacie Ring wrote and edited the bioinformatic code. Kacie Ring wrote the manuscript and Andrea Swei, Seemay Chou, Lisa I. Couper, Anne L. Sapiro, and Fauna Yarza edited and approved the final manuscript.
Open Research
OPEN RESEARCH BADGES
This article has earned an Open Data, for making publicly available the digitally-shareable data necessary to reproduce the reported results. The data is deposited in the Gene Expression Omnibus (GEO) under accession number GSE17 3109.
DATA AVAILABILITY STATEMENT
Fastq files and raw read counts have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE173109.




