Effect of torpor on host transcriptomic responses to a fungal pathogen in hibernating bats
Abstract
Hibernation, the use of prolonged torpor to depress metabolism, is employed by mammals to conserve resources during extended periods of extreme temperatures and/or resource limitation. Mammalian hibernators arouse to euthermy periodically during torpor for reasons that are not well understood, and these arousals may facilitate immune processes. To determine whether arousals enable host responses to pathogens, we used dual RNA-Seq and a paired sampling approach to examine gene expression in a hibernating bat, the little brown myotis (Myotis lucifugus). During torpor, transcript levels differed in only a few genes between uninfected wing tissue and adjacent tissue infected with Pseudogymnoascus destructans, the fungal pathogen that causes white-nose syndrome. Within 70–80 min after emergence from torpor, large changes in gene expression were observed due to local infection, particularly in genes involved in pro-inflammatory host responses to fungal pathogens, but also in many genes involved in immune responses and metabolism. These results support the hypothesis that torpor is a period of relative immune dormancy and arousals allow for local immune responses in infected tissues during hibernation. Host–pathogen interactions were also found to regulate gene expression in the pathogen differently depending on the torpor state of the host. Hibernating species must balance the benefits of energy and water conservation achieved during torpor with the costs of decreased immune competence. Interbout arousals allow hibernators to optimize these, and other, trade-offs during prolonged hibernation by enabling host responses to pathogens within brief, periodic episodes of euthermy.
1 INTRODUCTION
Bouts of prolonged torpor enable hibernating mammals to conserve energy and/or water by suppressing metabolism, heart rate and body temperature. However, these long torpor bouts are intermittently interrupted by short interbout arousals during which an individual returns to euthermic body temperatures in almost all hibernating mammal species (van Breukelen & Martin, 2015). The frequency of interbout arousals and their duration varies with the unique ecology and physiology of mammal species and populations, partly due to differences in diet, metabolic rates, and the environment of the hibernaculum (Ruf & Geiser, 2015). Bats in temperate climates use seasonal hibernation to conserve energy and survive prolonged periods of low temperatures and poor food availability, among other benefits (Nowack, Stawski, & Geiser, 2017). Hibernating bats tend to have interbout arousals that are among the shortest of all hibernating mammals. For example, little brown myotis (Myotis lucifugus) arouse for 2–3 hr (Jonasson & Willis, 2012; Reeder et al., 2012), compared to thirteen-lined ground squirrels (Ictidomys tridecemlineatus) that arouse for an average of 18 hours (Kisser & Goodwin, 2012). Despite their short duration in bats, raising body temperature to euthermy is energetically expensive. Even the short interbout arousals of hibernating bats still cost them approximately 84% of their winter energy budget (Thomas, Dorais, & Bergeron, 1990), providing strong evidence for the importance of periods of euthermy.
It is not clear what combination of factors makes interbout arousals nearly universal in mammalian hibernators (van Breukelen & Martin, 2015). Understanding the energetic trade-offs between arousal frequency, arousal duration, length of the hibernation season and other thermoregulatory variables requires a better understanding of the benefits of interbout arousals. Metabolic imbalances that accumulate during torpor have long been proposed to induce interbout arousals (Twente & Twente, 1965), but the metabolites that are accumulated or depleted remain incompletely understood (van Breukelen & Martin, 2015; Ruf & Geiser, 2015). Bats are vulnerable to dehydration due to evaporative water loss during hibernation (Thomas & Cloutier, 1992), but this does not influence torpor bout length in other hibernating mammals (Thomas & Geiser, 1997). Other possible explanations for the necessity of interbout arousals in mammalian hibernators include renewed synthesis of macromolecules (Carey, Andrews, & Martin, 2003), neural regeneration (van Breukelen & Martin, 2015; von der Ohe, Darian-Smith, Garner, & Heller, 2006) and the need for immune surveillance of pathogens (Bouma, Carey, & Kroese, 2010; Burton & Reichman, 1999; Luis & Hudson, 2006; Prendergast, Freeman, Zucker, & Nelson, 2002). This last factor may be particularly important for the majority of mammalian hibernators that utilize cold hibernacula in which they may be exposed to psychrophilic pathogens.
Hibernating bats infected with a psychrophilic pathogen provide an opportunity to determine the importance of the increase in immune competence that may accompany euthermy during interbout arousals. White-nose syndrome (WNS), an emerging infectious disease of hibernating bats, is caused by an invasive fungal pathogen, Pseudogymnoascus destructans (Lorch et al., 2011). In several bat species in North America, outside of the native Palearctic range of the pathogen, WNS has recently led to severe, regional-scale population declines (Ingersoll, Sewall, & Amelon, 2016; Langwig et al., 2012; Thogmartin et al., 2013). This pathogen infects bats during hibernation, and in many North American species, it disrupts torpor patterns and presumably increases energy depletion (Reeder et al., 2012; Warnecke et al., 2012) and causes several other pathological disturbances (Verant et al., 2014; Warnecke et al., 2013). Host responses and mortality are not uniform (Langwig et al., 2016); interbout arousals occur with increased frequency in highly WNS-susceptible populations of M. lucifugus infected with P. destructans (Reeder et al., 2012), but with less frequency during infection in the big brown bat (Eptesicus fuscus) that is resistant to WNS mortality (Frank et al., 2014; Moore et al., 2018). The presence of changes in arousal frequency in both species and the contrasting directions of change, together, raise the question of whether arousals are needed to initiate protective responses to this pathogen during hibernation. More generally, do hibernating mammals use interbout arousals to compensate for decreased immune competence during torpor? It has been hypothesized that interbout arousals are necessary to initiate immune responses to pathogens or to augment responses that occur at low levels during torpor.
Past work has provided some support for this hypothesis. Previous studies examining the ability of antigens to provoke various immune responses in hibernating mammals (Bouma, Henning, Kroese, & Carey, 2013; Bouma, Strijkstra, et al., 2010; Burton & Reichman, 1999; Kurtz & Carey, 2007; Lilley et al., 2017; Maniero, 2002; Moore et al., 2013; Prendergast et al., 2002) have shown that only some pathways remain active throughout hibernation (Bouma, Carey, & Kroese, 2010; Prendergast et al., 2002). For example, leucocytes are sequestered during hibernation (Bouma, Strijkstra, et al., 2010; Bouma et al., 2011; Kurtz & Carey, 2007), preventing some immune responses in peripheral tissues. Other studies have compared transcriptome-wide gene expression changes between torpid and aroused mammals for different species and tissues (Bogren, Grabek, Barsh, & Martin, 2017; Cooper et al., 2016; Grabek, Diniz Behn, Barsh, Hesselberth, & Martin, 2015; Hampton, Melvin, & Andrews, 2013; Hampton et al., 2011; Schwartz, Hampton, & Andrews, 2013), including in the brain tissue of bats (Lei, Dong, Mu, Pan, & Zhang, 2014). These studies have found large changes in metabolic pathways that accompany the torpor–arousal cycle and may regulate thermoregulatory behaviour. Expression of immune genes in the bone marrow of I. tridecemlineatus (thirteen-lined ground squirrels) showed pronounced changes during torpor (Cooper et al., 2016). Lower transcript levels were found during torpor for most immune genes, consistent with our hypothesis. However, this study also showed elevated levels of certain innate immune genes, including complement genes (Cooper et al., 2016), suggesting that some pathways may be enhanced during hibernation. These prior studies have not examined host gene expression responses in a tissue facing an active pathogenic infection during hibernation, nor have any studies compared pathogen gene expression changes between torpor and euthermy. Thus, we sought here to more directly evaluate the role of interbout arousals in immune responses of hibernators. Our objective was to compare local host physiological responses and host–pathogen interactions during torpor to those during euthermy in tissues of a hibernating bat infected with a psychrophilic pathogen. We predicted that in hibernating M. lucifugus infected with P. destructans, local transcriptomic responses during hibernation would primarily occur only after arousal to euthermy and would largely be dormant during torpor. We also predicted that host–pathogen interactions would regulate P. destructans gene expression differently during torpor than during an interbout arousal.
2 MATERIALS AND METHODS
2.1 Ethics statement
This study was carried out on bats from a nonendangered species in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All methods were approved by the Institutional Animal Care and Use Committee at Bucknell University (protocol DMR-17). Animals were collected and studied with Wisconsin Department of Natural Resources permits 1044 and 1047 and Pennsylvania Game Commission Special Use Permit 33085.
2.2 Sample collection
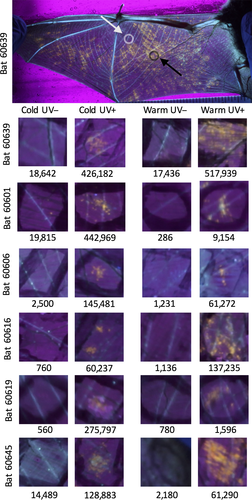
We obtained 23 adult M. lucifugus males in November 2015 from a hibernaculum in Pierce County, Wisconsin (Supporting Information Table S5) that had no known history of WNS. The bats were confirmed negative for P. destructans by quantitative PCR and transported to Lewisburg, Pennsylvania while torpid. The bats were infected with 50,000 conidia of P. destructans (strain 20631-21) in 50 μl phosphate-buffered saline with 0.05% Tween-20 as previously described (Johnson et al., 2014), then allowed to hibernate in climate-controlled environmental chambers under similar conditions that we have used for previous studies (Johnson et al., 2014, 2015), between 5 and 6°C and >90% relative humidity. The bats were removed from hibernation after 20 weeks, and long-wavelength UV transillumination was used to identify putative P. destructans-infected and uninfected areas of the bat wing by the characteristic fluorescence of the fungus (Turner et al., 2014). The fluorescence is thought to be caused by metabolites produced by the fungus during invasion of bat tissue (Flieger et al., 2016; Mascuch et al., 2015). Four 5-mm wing biopsies from each bat were collected and preserved in RNA-Later: UV-negative and UV-positive tissues were sampled, although the bats were still torpid (4–6°C) and again, from the opposite wing, after the bats were allowed to warm to euthermic temperature for 60–90 min (Supporting Information Table S5). Based on our previous studies (Moore et al., 2018), we expect the rate of warming to be similar to a natural arousal from torpor. Six bats of 17 surviving at the termination of the study were chosen for the RNA-Seq study because photographs available most clearly showed UV-positive and UV-negative areas on both wings. Power analysis (Busby, Stewart, Miller, Grzeda, & Marth, 2013) determined that six replicates with a read depth of 40 million per sample would be sufficient to detect 75% of genes differentially expressed at a minimum of twofold change with an adjusted p-value cut-off of 0.05.
2.3 RNA sequencing
Library preparation (Clontech SMARTer Stranded Total RNA-seq) and quality control were performed at the UCLA Technology Center for Genomics & Bioinformatics. RIN values for all samples were between 6.8 and 7.9 prior to library preparation. RNA sequencing was performed on three lanes of an Illumina HiSeq 3000 with single-end 50-bp stranded sequences obtained (Supporting Information Table S6). Prior to analysis, all data sets were quality trimmed using trimmomatic v.0.35 (Bolger, Lohse, & Usadel, 2014) with the parameters SLIDINGWINDOW:4:5 LEADING:5 TRAILING:5 MINLEN:25.
2.4 Differential expression analysis
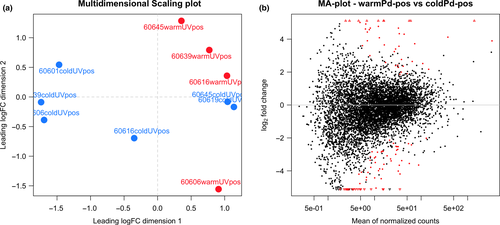
The quality trimmed reads were aligned using star v.2.5.2a (Dobin et al., 2013) to the concatenated genomes of M. lucifugus and P. destructans. For M. lucifugus, we used genome assembly Myoluc2.0 and gene models from Ensembl release 84 (Yates et al., 2016). For P. destructans, we used the genome assembly and gene models from Drees et al. (2016). rsem v1.2.29 (Li & Dewey, 2011) was then used to apply an expectation maximization algorithm to predict gene expression counts for each transcript. The expected count matrix for all samples is available as Supporting Information Table S7. To confirm the infection status of each sample, we totalled the number of reads mapped to P. destructans transcripts after median normalization across samples by sartools v.1.4.0 (Varet et al., 2016) (Supporting Information Table S7 and Figure 1). The RNA-Seq data showed that the 12 UV-negative biopsies contained 6,650 ± 8,190 P. destructans transcripts. The reads that map to the P. destructans transcriptome represent 0.10% ± 0.12% of the total mapped reads in the UV-negative samples, similar to what we find in tissue samples from bats with no exposure to P. destructans (Field, 2018). The 12 UV-positive biopsies contained significantly more transcripts (190,000 ± 181,000, p = 0.004, paired t test). We used a threshold of 39,000 transcripts (mean of the UV-negative samples plus 3 standard deviations) to determine that two of the UV-positive samples were outliers that contained insignificant levels of viable P. destructans. These samples presumably represented areas of the wing that were fluorescent due to lesions from infection earlier in the hibernation period but that no longer contained viable P. destructans. Because our focus was on host response to ongoing infection and on host–pathogen interactions, we excluded the two outliers from further analysis.
Differential expression was analysed separately for M. lucifugus transcripts and P. destructans transcripts. False discovery rate (FDR) was used to control for multiple comparisons in all models using the Benjamini–Hochberg procedure (Benjamini & Hochberg, 1995). Differential expression between conditions was determined using SARTools (Varet et al., 2016) and edgeR v.3.16.5 (Robinson, McCarthy, & Smyth, 2010) after normalizing across samples using the trimmed mean of M-values (TMM) method (Robinson & Oshlack, 2010) and a minimum expression level of two transcripts per million mapped reads (TPM) combined across all samples. All generalized linear models used to fit the data incorporated individual as a batch effect, as determined using a likelihood ratio test. Initially, a generalized linear model that incorporated an interaction effect between temperature (warm/aroused or cold/torpid) and infection (~individual + temperature * infection) did not find any genes with significant interaction coefficients (FDR < 0.05). Therefore, the interaction effect was dropped and the following model was used for all subsequent analyses: ~individual + group. We also tested a model that used normalized P. destructans transcript counts (log transformed) as a quantitative variable to measure pathogen load. This model performed almost identically to the model using the categorical variable. Similar results were obtained using transcripts instead of genes or using DESeq2 (Love, Huber, & Anders, 2014) instead of edgeR.
This study utilized a novel design that allowed a paired approach to identifying differential gene expression within individuals before and after arousal from torpor (Figure 1), enabling us to control for interindividual variation. However, because paired samples are from the same individual, it is possible that some of the gene expression we observed in uninfected tissue is due to regional or systemic effects of P. destructans infection. The bats in this captive study showed extensive P. destructans infection (Supporting Information Figure S1), and this could prompt a systemic response. Further, it is likely that some regional changes in gene expression were occurring, as we have found for IL6 and IL17A cytokine gene expression in the locoregional lymph nodes of aroused M. lucifugus with P. destructans infections (Lilley et al., 2017). We assume, however, that these regional and systemic effects would be expressed similarly within the four samples from each individual, and therefore, the differences that we observe between uninfected and infected tissues represent the local effects of infection.
For M. lucifugus, gene ontology annotations were extracted from Ensembl release 84 (Yates et al., 2016) and gene ontology enrichment analysis was performed using g:Profiler (Reimand et al., 2016) v.r1730_e88_eg35 with enrichment measured by Fisher's one-tailed test and an FDR threshold of 0.05. Enrichment was measured using ranked lists (by FDR) against the background of all 16,761 annotated M. lucifugus genes. Similar results were obtained using a background of the 12,012 annotated genes expressed in this data set.
3 RESULTS
We used fluorescence induced by long-wavelength ultraviolet (UV) light to identify likely areas of P. destructans infection (Turner et al., 2014) on transilluminated wings of M. lucifugus 20 weeks after inoculation and hibernation in captivity. We found that 10 of 12 (83%) wing biopsy samples positive for UV fluorescence contained significant P. destructans transcripts (>39,000 counts per million mapped reads (CPM)) and that 12 of 12 (100%) of UV-negative biopsies did not (Figure 1 and Supporting Information Figure S1). We removed the two UV-positive samples negative for P. destructans transcripts from further analysis as described in Materials and Methods.
Of the 25,849 M. lucifugus genes (Yates et al., 2016), 22,880 genes were expressed in at least one of the 22 samples and 14,632 were expressed by at least one CPM in four or more samples (Supporting Information Table S1). The latter set of genes was used for differential expression analysis with edgeR using a generalized linear model that included individual as a batch effect. We identified a total of 1412 genes that were differentially expressed in pairwise comparisons among torpid and euthermic tissue samples that were either negative or positive for P. destructans at a false discovery rate (FDR) of < 0.05 (Table 1, Figure 2). A much greater number of genes were differentially expressed in tissue locally infected with P. destructans during euthermy (Figure 2a, 1098 genes) than during torpor (Figure 2b, 49 genes), and 94% of these transcripts showed greater levels in locally infected than in uninfected tissue (Table 1, Figure 2f). In the comparison of infected and uninfected tissues, the majority (36 of 49, 73%) of the genes differentially expressed in the torpid tissue were also differentially expressed after arousal (Figure 2f).
| Test Group | Reference Group | Down | Up | Total |
|---|---|---|---|---|
| Cold Pd-pos | Cold Pd-neg | 1 | 48 | 49 |
| Warm Pd-pos | Warm Pd-neg | 65 | 1033 | 1098 |
| Warm Pd-neg | Cold Pd-neg | 31 | 180 | 211 |
| Warm Pd-pos | Cold Pd-pos | 46 | 417 | 463 |
Notes
- The number of differentially expressed genes is shown for each pairwise comparison. Down genes are more highly expressed in the reference group, and Up genes are more highly expressed in the test group. Row colours correspond to the Venn diagram in Figure 2f.
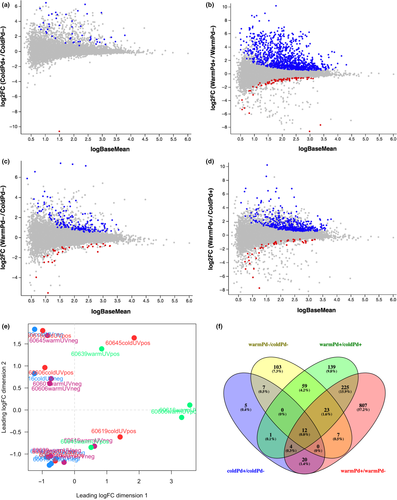
We also determined differential gene expression due to arousal in both uninfected tissue (Figure 2c) and tissue locally infected with P. destructans (Figure 2d). More genes were differentially expressed due to arousal in infected tissue (463 genes) than uninfected tissue (211), and 44% of the genes differentially expressed in uninfected tissue were also differentially expressed in infected tissue (Figure 2f).
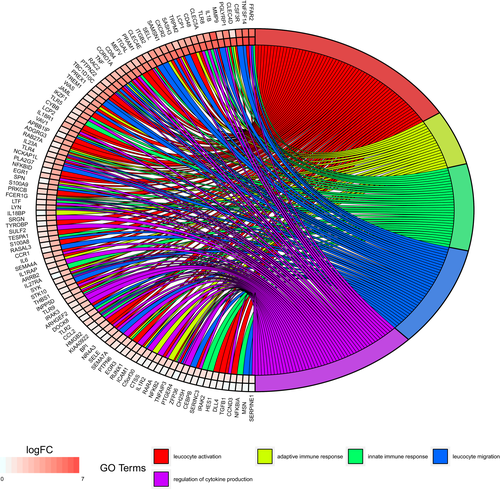
Putative functions for 1335 of 1412 differentially expressed host genes were annotated in the UniProt KnowledgeBase. Using g:Profiler, we examined the biological process pathways that were enriched in each of the pairwise comparisons (Table 2 and Supporting Information Table S3). We found that genes differentially expressed due to P. destructans infection in torpid bats were especially enriched in pathways that involved chemotaxis and fever generation. Expression of host genes for some cytokines and other immune regulatory proteins was upregulated in response to P. destructans infection in tissue from both torpid and aroused bats (Figure 3 and Supporting Information Table S3). For example, during both torpor and arousal, infection caused significant increases in transcript levels of both the Fos and Jun components of the AP-1 transcription factor that regulates immune cell activation (Table 3).
| Biological Process | BP ID | p-value | T | Q | Q&T | Q&T/Q | Q&T/T |
|---|---|---|---|---|---|---|---|
| Cold Pd+ vs. Cold Pd− | |||||||
| Cell chemotaxis | GO:0060326 | 0.000000793 | 178 | 47 | 7 | 0.149 | 0.039 |
| Regulation of fever generation | GO:0031620 | 0.00029 | 5 | 46 | 2 | 0.043 | 0.4 |
| Positive regulation of fever generation | GO:0031622 | 0.00029 | 5 | 46 | 2 | 0.043 | 0.4 |
| Positive regulation of chemokine biosynthetic process | GO:0045080 | 0.000308 | 8 | 26 | 2 | 0.077 | 0.25 |
| Leucocyte migration | GO:0050900 | 0.00032 | 181 | 47 | 5 | 0.106 | 0.028 |
| Warm Pd+ vs. Warm Pd− | |||||||
| Immune system process | GO:0002376 | 8.7E-43 | 1355 | 509 | 137 | 0.269 | 0.101 |
| Cytokine metabolic process | GO:0042107 | 1.44E-09 | 71 | 492 | 15 | 0.03 | 0.211 |
| Regulation of endopeptidase activity | GO:0052548 | 0.000000121 | 242 | 892 | 33 | 0.037 | 0.136 |
| Regulation of protein complex assembly | GO:0043254 | 0.00000029 | 213 | 1072 | 33 | 0.031 | 0.155 |
| Mast cell degranulation | GO:0043303 | 0.000039 | 21 | 481 | 6 | 0.012 | 0.286 |
| Warm Pd− vs. Cold Pd− | |||||||
| Regulation of immune system process | GO:0002682 | 0.000000162 | 676 | 182 | 23 | 0.126 | 0.034 |
| Positive regulation of endocytosis | GO:0045807 | 0.0000331 | 75 | 137 | 6 | 0.044 | 0.08 |
| Cellular extravasation | GO:0045123 | 0.000173 | 28 | 55 | 3 | 0.055 | 0.107 |
| Positive regulation of lipid localization | GO:1905954 | 0.000362 | 49 | 38 | 3 | 0.079 | 0.061 |
| Interleukin-1 beta secretion | GO:0050702 | 0.000405 | 16 | 134 | 3 | 0.022 | 0.188 |
| Warm Pd+ vs. Cold Pd+ | |||||||
| Response to external stimulus | GO:0009605 | 9.88E-18 | 1083 | 460 | 77 | 0.167 | 0.071 |
| Regulation of NF-kappaB import into nucleus | GO:0042345 | 0.00000438 | 29 | 389 | 7 | 0.018 | 0.241 |
| Phagocytosis | GO:0006909 | 0.0000499 | 81 | 184 | 7 | 0.038 | 0.086 |
| Vesicle organization | GO:0016050 | 0.000291 | 135 | 459 | 12 | 0.026 | 0.089 |
| Positive regulation of feeding behaviour | GO:2000253 | 0.000453 | 3 | 131 | 2 | 0.015 | 0.667 |
Notes
- Enrichment of Biological Process gene ontology categories was calculated for each comparison group using g:Profiler. The five categories with the lowest FDR are shown along with their gene ontology ID; the FDR-adjusted p-value of category enrichment; the number of genes annotated to the functional term (T); the number of genes in the input list from the top of the list where the optimal enrichment occurs (Q); the number of genes in input list with functional annotation, corresponding to the enrichment (Q&T); the predictive rate, that is, the proportion of genes in the input list that have the function (Q&T/Q); and the recall rate, that is, the proportion of functionally annotated genes that the query recovers (Q&T/T). See Supporting Information Table S3 for complete results.
| Gene | Name | Description | Torpor | Arousal | ||
|---|---|---|---|---|---|---|
| FC | FDR | FC | FDR | |||
| ENSMLUG00000027823 | CLEC4D | C-type lectin pattern recognition receptor | 2.4 | 1 | 69.6 | 8.47E-05 |
| ENSMLUG00000009941 | IL1B | Inflammatory cytokine | 76.7 | 0.02 | 59.7 | 1.02E-03 |
| ENSMLUG00000027388 | Putative IL4 receptor | 59.1 | 0.1 | 56.5 | 1.30E-02 | |
| ENSMLUG00000012745 | TLR8 | Toll-like pattern recognition receptor | 15.1 | 0.485 | 55.7 | 8.85E-04 |
| ENSMLUG00000017259 | CLEC5A | C-type lectin pattern recognition receptor | 11.2 | 0.136 | 55 | 4.57E-05 |
| ENSMLUG00000014375 | NLRP3 | Inflammasome complex | 7.3 | 0.04 | 40 | 1.40E-02 |
| ENSMLUG00000011581 | CLEC4E | C-type lectin pattern recognition receptor | 4.7 | 0.492 | 27.6 | 1.13E-04 |
| ENSMLUG00000013248 | CLEC12A | C-type lectin pattern recognition receptor | 2.4 | 1 | 23 | 2.82E-04 |
| ENSMLUG00000003969 | TNF | Tumour necrosis factor-a | 6.7 | 0.168 | 22.5 | 5.48E-06 |
| ENSMLUG00000030769 | TLR5 | Toll-like pattern recognition receptor | 2.3 | 0.941 | 14.5 | 2.12E-04 |
| ENSMLUG00000001861 | CYBB | NADPH oxidase | 2.4 | 0.827 | 14.2 | 1.61E-05 |
| ENSMLUG00000006778 | IL23A | Inflammatory cytokine | 2.1 | 1 | 11.5 | 5.90E-04 |
| ENSMLUG00000007417 | TLR4 | Toll-like pattern recognition receptor | 2.6 | 0.844 | 10.7 | 3.65E-04 |
| ENSMLUG00000022258 | S100A9 | Calprotectin | 2.7 | 0.536 | 9.4 | 1.38E-04 |
| ENSMLUG00000003347 | CSF3 | G-CSF cytokine | 3.5 | 0.487 | 7.8 | 4.59E-04 |
| ENSMLUG00000002912 | S100A8 | Calprotectin | 2.6 | 0.592 | 7.3 | 7.85E-04 |
| ENSMLUG00000015855 | IL6 | Inflammatory cytokine | 5.3 | 0.099 | 6.7 | 2.13E-03 |
| ENSMLUG00000022206 | S100A12 | Calprotectin | 2.2 | 0.77 | 6.4 | 8.78E-04 |
| ENSMLUG00000009003 | FOSB | AP-1 transcription factor | 2.3 | 0.017 | 6.1 | 2.57E-11 |
| ENSMLUG00000013014 | IL27RA | IL27 receptor | 1 | 1 | 5.9 | 2.65E-04 |
| ENSMLUG00000008204 | PTGS2 | Cyclooxygenase-2 | 2.8 | 0.05 | 5.7 | 1.76E-06 |
| ENSMLUG00000016412 | THBS1 | Thrombospondin | 1.8 | 0.459 | 5.5 | 1.27E-07 |
| ENSMLUG00000007189 | CLEC7A | C-type lectin pattern recognition receptor | 1.4 | 1 | 5.2 | 4.23E-03 |
| ENSMLUG00000015097 | TLR9 | Toll-like pattern recognition receptor | 3 | 0.067 | 5.2 | 1.43E-04 |
| ENSMLUG00000016718 | CCR2 | Chemokine receptor | 1.9 | 0.99 | 4.8 | 4.60E-02 |
| ENSMLUG00000012813 | TLR2 | Toll-like pattern recognition receptor | 1.5 | 1 | 4.6 | 2.67E-04 |
| ENSMLUG00000011369 | CCL2 | Chemokine | 3.2 | 0.05 | 4.5 | 2.59E-04 |
| ENSMLUG00000003729 | FOS | AP-1 transcription factor | 1.6 | 0.592 | 3.7 | 1.43E-06 |
| ENSMLUG00000008750 | IL1R2 | IL1b receptor | 1.6 | 0.953 | 3.5 | 2.55E-03 |
| ENSMLUG00000010038 | PTGER4 | Prostaglandin E2 receptor | 1.1 | 1 | 2.8 | 1.36E-03 |
| ENSMLUG00000012714 | JUNB | AP-1 transcription factor | 2.5 | 0.01 | 2.7 | 4.64E-04 |
| ENSMLUG00000008863 | CYBA | NADPH oxidase | 1.5 | 0.906 | 2.5 | 6.25E-03 |
| ENSMLUG00000013098 | TGFB1 | Cytokine involved in Th17 responses | 1.1 | 1 | 2.2 | 2.37E-03 |
Notes
- Ensembl gene ID and gene name are listed for selected genes differentially expressed in euthermic bats between uninfected and infected tissues. The fold change (FC) and Benjamini–Hochberg adjusted p-value (FDR) calculated by edgeR are shown for each gene for samples from both torpid and euthermic bats. Bold FDR values indicate ≤ 0.05. See Supporting Information Table S1 for results for all host genes.
A much greater number of immune genes exhibited significant infection-dependent expression increases in tissue from euthermic bats than from torpid bats (Figure 3 and Table 3). Except for IL1B (interleukin-1β), all immune genes that were differentially expressed during torpor showed greater fold changes during interbout arousals (Table 3). After arousal, genes showing differential expression due to infection were more heavily enriched in pathways involving immune responses, protease activity, protein complex assembly and mast cell degranulation (Table 2 and Supporting Information Table S3). Some of these differentially expressed genes due to local infection were also found in our previous study that compared the host transcriptomes of wing tissue in uninfected bats to wing tissue from bats captured in the wild with severe symptoms of WNS (Field et al., 2015).
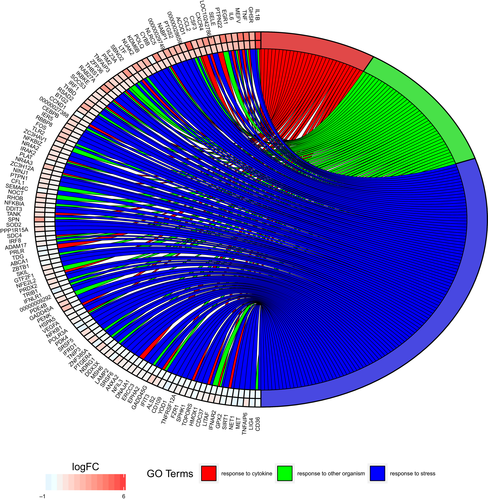
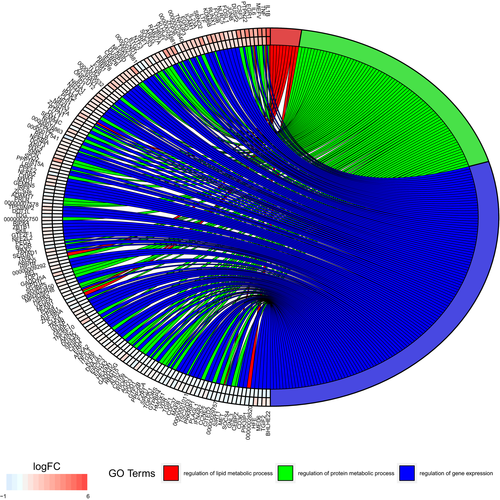
Genes differentially expressed due to arousal include those involved in responses to cytokines, responses to other organisms (such as an infection) and responses to stress (Figure 4). Genes involved in metabolic regulatory pathways are also differentially expressed upon arousal from torpor (Figure 5), with greater changes typically seen in infected tissues (Supporting Information Table S3). Biological processes identified by g:Profiler that showed enrichment upon arousal from torpor in samples without P. destructans infection included regulation of immune responses, as well as endocytosis, extravasation and lipid localization (Table 2). Comparison of the infected tissue before and after arousal from torpor showed enrichment of genes involved in pathways regulating responses to external stimuli, phagocytosis and activation of feeding behaviour. Together, these gene expression changes illustrate complex interactions between hibernation physiology and infection.
We also examined gene expression in the pathogen and found detectable expression (TPM > 2 in UV+ samples) of 5,610 of 9,575 known P. destructans genes (Supporting Information Table S4). We found a total of 43 P. destructans genes that were more highly expressed in euthermic tissues and 68 pathogen genes more highly expressed in torpid tissues (Figure 6 and Supporting Information Table S4), when the fungus would be actively growing. Pathogen genes more highly expressed after the host aroused from torpor included several heat shock genes and secreted enzymes that may be involved in virulence (O'Donoghue et al., 2015; Pannkuk, Risch, & Savary, 2015; Reeder et al., 2017).
4 DISCUSSION
Interbout arousals are energetically costly but nearly ubiquitous in hibernating mammals. This study determined the role of interbout arousals in responding to local infection during hibernation. We utilized a novel design that allowed a paired approach to identifying local differential gene expression in both infected and uninfected tissues within host individuals just before and after arousal from torpor. Our results demonstrate that in the host, significant changes in gene expression occur in both uninfected and infected tissues after arousal to euthermy. Infection caused widespread changes in transcript levels only after arousal from torpor. Genes that were differentially expressed in infected tissues of the host after arousal included genes involved in host responses to fungal pathogens, in other immune responses, in metabolism, wound healing and in the possible regulation of the torpor–arousal cycle. These results support the hypothesis that torpor is a period of relative immune dormancy in the host but that immune response is enhanced upon arousal to euthermy.
The pattern of host gene expression we observed indicates that inflammatory responses in locally infected tissue are greatly elevated during arousals but decline during torpor. The infection-dependent differences in transcript levels during torpor may indicate that transcripts are being selectively protected from degradation, as transcription is presumably halted during torpor (Grabek, Diniz Behn, et al., 2015). Increased stabilization of some transcripts during torpor has been proposed to allow translation to occur earlier during subsequent arousals (Bogren et al., 2017; Grabek, Diniz Behn, et al., 2015) and would be expected to enhance responses to infection during these arousals. We propose that the host transcripts that show increased levels due to local infection during torpor, such as several transcription factors, are those that are most essential to a rapid response to P. destructans infection during interbout arousals. The ability of some gene expression changes to be preserved into subsequent torpor bouts could suggest a role for interbout arousals in host responses to infectious diseases that extends past the brief arousal period.
4.1 Immune response pathways
During torpor, infection caused increases in transcript levels of multiple transcription factors, including both the Fos and Jun components of the AP-1 transcription factor that regulates immune cell activation (Table 3). The increased transcript levels during torpor would enable the AP-1 transcription factor to more quickly activate immune cells during the brief euthermic arousals in bats. The pro-inflammatory cytokine IL1B (interleukin-1β) and chemokine CCL2 genes also showed increased transcript levels due to infection during torpor. Each of these genes showed similar or greater increases in expression due to infection during arousal as during torpor (Table 3), demonstrating that some responses during torpor and interbout arousals are related.
As expected, euthermic bats showed much greater infection-dependent changes in transcript levels than torpid bats. Many of these genes promote inflammation, such as PTGS2 (the enzyme cyclooxygenase-2 that is the rate-limiting step in the production of inflammatory prostaglandins such prostaglandin E2), calprotectins (S100A9, S100A8, and S100A12 that have antimicrobial and inflammatory activities), IL6 (an inducer of the acute phase response), NLRP3 (a critical component of the inflammasome) and TGFB1 (a potential inducer of a T-helper 17 (Th17) immune response). In addition to causing local inflammation and activation of immune responses in locoregional lymph nodes (Lilley et al., 2017), these inflammatory mediators may have systemic effects. Although the febrile response is inactive during torpor (Prendergast et al., 2002), bats with WNS in some (Lilley et al., 2016; Mayberry, McGuire, & Willis, 2017), but not all (Brownlee-Bouboulis & Reeder, 2013; Moore et al., 2018), previous studies show elevated skin temperature during interbout arousals. Systemic inflammation could also influence the torpor–arousal cycle and contribute to WNS pathology by increasing arousal frequency. It is possible that the increase in arousal frequency seen during WNS in naïve populations of M. lucifugus (Reeder et al., 2012) represents a behavioural response to increase the ability to respond to infection. However, the longer torpor bouts seen in remnant populations of M. lucifugus with long WNS experience (Lilley et al., 2016) suggests that the increased arousal frequency response in naïve populations is maladaptive in this species, which are known to have insufficient energy stores to support an increased number of arousals (Thomas et al., 1990). Thus, it remains unclear which of the infection-dependent responses that we observe are protective and which may be contributing to WNS pathology.
Host genes involved in both innate and adaptive immune responses showed much greater increases in expression levels due to P. destructans infection in tissue from aroused bats than from torpid bats. These included pathways that regulate both activation and migration of leucocytes, as well as expression of pattern recognition receptors in both the Dectin and Toll-like receptor families (Figure 3 and Table 3) that can recognize fungal pathogens (Levitz, 2010). Adaptive immune responses, which are likely needed to control the fungal infection (Romani, 2011), show evidence of activation of both Th1 and Th17 pathways in euthermic tissues but not during torpor. These two cell-mediated pathways control resistance, tolerance and immune pathology in other fungal infections (Collette & Lorenz, 2011; Romani, 2011) and may be critical for understanding WNS susceptibility. The absence of messenger RNAs that would allow an appropriate adaptive response and the limited innate response during torpor likely permit uncontrolled growth of the psychrophilic pathogen. However, it remains unknown whether immune proteins translated during the brief periods of euthermy might persist into torpor, allowing some responses to continue after transcript degradation (Grabek, Martin, & Hindle, 2015).
One of the unique signatures seen in the local response of aroused bats to P. destructans infection that is not present in torpid bats is the increased expression of a set of genes that may be involved in mast cell degranulation (Table 3 and Supporting Information Table S3). Mast cell degranulation could be caused by a hypersensitivity reaction to fungal antigens (allergens). Pseudogymnoascus destructans shows increased transcription of the homolog of the protease, Aspf2, while infecting host tissue compared to growth in culture (Reeder et al., 2017). We also find abundant expression of this P. destructans transcript (VC83_01361) in tissue from both torpid and aroused bats in the current study (Supporting Information Table S4). A homologous protease elicits an immunoglobulin E- and mast cell-mediated allergic response to the fungus Aspergillus fumigatus in humans (Svirshchevskaya, Alekseeva, Andronova, & Kurup, 2001). Although allergic immune responses have not been studied in hibernating animals, if P. destructans produces a similar response in bats, an allergic reaction could generate the release of histamines and other inflammatory compounds. If these small molecules persisted after the return to torpor, they could contribute to WNS pathology and, possibly, disrupt torpor patterns.
Based on our results, regulation of immune responses appears to be important upon arousal from torpor, even in tissue not locally infected by P. destructans. The preponderance of genes involved in cytokine responses and responses to other organisms (i.e., infections) that increase expression upon arousal in uninfected tissue, especially the IL1β pathway, suggests that inflammatory immune responses are part of a systemic response to infection that occurs during arousal. The increased expression of cytokine transcripts that we find in the uninfected tissue does not necessarily lead to inflammation—both translation and inflammasome activation are also needed. However, it remains unclear whether inflammasome activation is occurring in either torpor or euthermy and whether cytokines are secreted and inflammatory lipid metabolites are produced. Bats lack some components of the inflammasome (Ahn, Cui, Irving, & Wang, 2016), and further proteomic and metabolomic studies will be needed to determine which products of inflammasome activation and which inflammatory metabolites are produced in response to infection. The activation of cytokine response pathways during interbout arousals in the absence of a local pathogenic infection may be used to keep psychrophilic microflora in check during hibernation.
4.2 Metabolic pathways
Hibernation requires a shift in whole-body energy utilization that favours the use of fat stored in white adipose tissue over dietary energy sources rich in protein (Ruf & Geiser, 2015). Torpor arousal utilizes energy stores in white and brown adipose tissues for thermogenesis, and previous transcriptomic studies of these tissues provide evidence for these metabolic changes (Hampton et al., 2013; Hao et al., 2015). The arousal-dependent changes in host gene expression that we found included genes that regulate protein and fat metabolism (Figure 5). The increases are typically larger in tissue infected with P. destructans, particularly for genes regulating protein metabolism (Figure 5), possibly reflecting the increased energy demands of an immune response leading to protein catabolism. A large number of genes involved in stress responses that are upregulated upon arousal from torpor also presumably reflect the responses to the oxidative stress generated by arousal (Carey et al., 2003) and that may be increased in infected tissue (Figure 4).
Arousal from torpor affects the expression level of many host genes that, themselves, regulate gene expression (Figure 5). These transcription factors and signalling proteins show even larger changes in tissue infected with P. destructans, indicating that the local tissues are responsive to the presence of pathogens during the brief euthermic arousals. The dramatic differences in gene expression patterns between infected and uninfected tissues demonstrate that arousal from torpor allows physiological responses to pathogens as quickly as 70–80 min after arousals are initiated (Supporting Information Table S5), that is, after <50 min spent at euthermy (Jonasson & Willis, 2012). During an arousal that extends for 12 or more hours, as found in many other mammalian hibernators (Ruf & Geiser, 2015), we would expect even more extensive transcriptional changes in response to local infection. Much fewer infection-dependent differences were found in torpid bats indicating that, for most transcripts, expression returned to similar levels in uninfected and locally infected tissues within 4 to 13 days (Supporting Information Table S5) after the previous torpor bout. Thus, bats with WNS must wait until emergence from hibernation (if they survive with sufficient energy stores) to fully respond to infection and begin the metabolically expensive immune and wound healing responses (Meierhofer et al., 2018).
Of potential interest is the large difference in transcript levels of FFAR2 in aroused bats, with P. destructans-infected tissue expressing 127-fold higher levels than uninfected tissue. This gene encodes a receptor that is activated by short-chain fatty acids and regulates whole-body energy homeostasis and intestinal immunity (Bindels, Dewulf, & Delzenne, 2013; Mohammad, 2015). Previous studies on the effects of hibernation in metabolically active tissues have shown dramatic shifts in the expression of genes that regulate fat metabolism (Hampton et al., 2013; Hao et al., 2015). We speculate that FFAR2 may represent a link between the metabolic changes that occur during torpor and immune activity, both in the intestine of hibernating mammals, which shows large changes in immune function during torpor (Kurtz & Carey, 2007), and by influencing the torpor–arousal cycle in the hypothalamus (Lei et al., 2014; Schwartz et al., 2013; Yan, Barnes, Kohl, & Marr, 2008). Expression of FFAR2 (ENSMLUG00000012387) was 49-fold higher in P. destructans-infected tissue in torpid bats (Supporting Information Table S1), suggesting a possible role in modulating local immune responses. This gene is also more highly expressed in the brains of greater horseshoe bats (Rhinolophus ferrumequinum) during torpor than during the active season (Lei et al., 2014). Activation of this free fatty acid receptor 2 (also known as GPR43) by short-chain fatty acids in the gut can regulate metabolic homeostasis in the intestine and adipose tissues, controlling lipid metabolism and adipokine secretion (Mohammad, 2015). Additional studies will be necessary to determine how the possible increased production of short-chain fatty acids during hibernation might affect local signalling of FFAR2 in infected tissues and whether this can influence systemic torpor–arousal cycles.
4.3 Host–pathogen interactions
Many pathogen genes were differentially expressed between arousal and torpor (Supporting Information Table S4). Many of these genes have putative functions that could contribute to virulence or that could assist pathogen evasion of host immune responses during arousal, such as cell wall remodelling, protease activity and ion transport, consistent with our previous comparison between cultured and tissue-infecting P. destructans (Reeder et al., 2017). This prior study found a number of genes upregulated during infection that are involved in heat shock responses and we speculated that this was a response of the pathogen to host arousal. In the current study, we find four P. destructans putative heat shock genes that are very highly expressed and significantly upregulated in tissue from euthermic bats compared to torpid bats (Supporting Information Table S4), confirming that the expression of heat shock genes is a response of the pathogen to host arousals. The ability of a pathogen to deal with the heat stress of arousals is likely essential for its persistence during interbout arousals when the host temperature extends beyond the viable range of this psychrophilic pathogen (Verant, Boyles, Waldrep, Wibbelt, & Blehert, 2012).
4.4 Implications
During hibernation, we observed dramatic changes in host transcriptomic responses in bat wing tissue locally infected with P. destructans after host arousal from torpor. Infection with the psychrophilic fungal pathogen stimulated host gene expression changes associated with inflammatory immune responses to a limited extent during torpor, and to a much greater extent during arousal. Both innate and adaptive immune responses were similarly dormant during torpor, suggesting that any host immune responses to infections during torpor would have to depend on pre-existing mediators that do not require host transcription or translation. The depressed immune response in bats during torpor represents an important seasonal vulnerability to disease in the host that may be exploited by psychrophilic pathogens like P. destructans as they grow on the bat skin surface and invade skin tissue while host immune response is limited.
Mammals that utilize seasonal hibernation to conserve energy almost universally display repeated interbout arousals that are energetically very costly. Our results suggest that the ability to rapidly and extensively activate host responses to infection may be one of the important functions of these arousals. In species whose ecology and physiology allow extended or frequent interbout arousals (van Breukelen & Martin, 2015; Humphries, Thomas, & Kramer, 2003), individuals are presumably able to mount a robust immune response during euthermy. However, hibernating mammals with a small body size and for which hibernation duration is long may face an important trade-off because their limited energy reserves may preclude long or frequent arousals. For yangochiropteran bats, which may weigh as little as 5 g, hibernate for several months at a time at higher latitudes, and spend 99% of the hibernation season torpid, euthermic arousals are extremely costly but may be essential for a response to infection. The rapid and extensive immune response by the bat host during arousals may be a sufficient response to many pathogens. Nonetheless, the short time bats spend euthermic during hibernation may preclude an effective response to novel pathogens such as P. destructans in its invasive range in North America. The changes we observed in pathogen gene expression during arousals may further complicate host responses and facilitate pathogen persistence during arousals. As the disease progresses during the winter months, additional host arousals occur in M. lucifugus (Reeder et al., 2012). At first glance, these additional arousals may seem beneficial as they could promote a more robust immune response. Indeed, even bats severely affected by WNS clear the infection and heal damage from the disease within 2 weeks of emergence from hibernation (Fuller et al., 2011; Meierhofer et al., 2018; Pikula et al., 2017). In the absence of additional food resources, however, the energetic costs of repeated arousals from torpor are likely maladaptive, as they rapidly deplete host energy reserves and contribute to bat mortality from WNS.
More broadly, given the importance of immune pathways to defence from infectious agents, our results strongly suggest periodic arousals are necessary to respond to infection in any heterothermic animal that undergoes prolonged hibernation (Luis & Hudson, 2006). In our study, arousal from torpor also affected the expression of host genes involved in metabolic pathways, consistent with shifts in energetic demands during hibernation for tissues involved in energetically costly immune responses. Together, these results support a model of interbout arousals in which changes in signalling and gene expression facilitate host responses to pathogens.
Our finding that the psychrophilic fungal pathogen that causes WNS elicits a weak immune response during torpor but a robust response soon after arousal supports the hypothesis that interbout arousals are necessary to initiate immune responses to pathogens or to augment responses that occur at low levels during torpor. This finding also has several other potential implications. It provides an example of a homeostasis-restoring physiological response that can occur during the near-universal interbout arousals observed in hibernating mammals. The limited local immune response of bat hosts during torpor may also help explain why some proposed treatments meant to support bat immune responses during hibernation have thus far had limited effectiveness in reducing the severity of WNS (Johnson et al., 2015). In addition, given the importance of bats as reservoirs of zoonotic disease (Brook & Dobson, 2015; Luis et al., 2013; Mandl et al., 2015), and their common use of heterothermy, the reduced immune competence during torpor that we have observed could contribute to the ability of bats to serve as disease reservoirs.
ACKNOWLEDGEMENTS
We thank Jennifer Redell and Heather Kaarakka for assistance in collecting samples. We acknowledge the generous access to the field site provided by the landowner, Fairmont Sandtrol. Cindy Rhone, Gretchen Long and the rest of the animal care staff at Bucknell University assisted in providing excellent care for the captive animals for this study. We thank Beth Rogers, Naama Kipperman and Cali Wilson for assistance in tissue and data collection.
AUTHOR CONTRIBUTION
K.A.F. designed the study, collected the tissue samples, conducted the bioinformatics analysis, interpreted the results and wrote the manuscript with critical feedback from all of the coauthors; B.J.S. designed the study, collected the animals, collected the tissue samples and helped write the manuscript; J.M.P. collected the tissue samples and isolated the RNA; G.G.T. designed the study, collected the tissue samples and collected animals; MG collected the tissue samples; T.M.L. collected the tissue samples; J.P.W. collected animals; J.S.J. collected the tissue samples; C.H. collected the tissue samples; and D.M.R. designed the study, collected the tissue samples and interpreted the results.
DATA ACCESSIBILITY
RNA sequencing: Sequence Read Archive SRP111376. Interactive plots are available at the following persistent URLs for Figure 2a (https://digitalcommons.bucknell.edu/fac_pubs/130), Figure 2b (https://digitalcommons.bucknell.edu/fac_pubs/129), Figure 2c (https://digitalcommons.bucknell.edu/fac_pubs/128) and Figure 2d (https://digitalcommons.bucknell.edu/fac_pubs/127).




