Adaptive genomic divergence under high gene flow between freshwater and brackish-water ecotypes of prickly sculpin (Cottus asper) revealed by Pool-Seq
Abstract
Understanding the genomic basis of adaptive divergence in the presence of gene flow remains a major challenge in evolutionary biology. In prickly sculpin (Cottus asper), an abundant euryhaline fish in northwestern North America, high genetic connectivity among brackish-water (estuarine) and freshwater (tributary) habitats of coastal rivers does not preclude the build-up of neutral genetic differentiation and emergence of different life history strategies. Because these two habitats present different osmotic niches, we predicted high genetic differentiation at known teleost candidate genes underlying salinity tolerance and osmoregulation. We applied whole-genome sequencing of pooled DNA samples (Pool-Seq) to explore adaptive divergence between two estuarine and two tributary habitats. Paired-end sequence reads were mapped against genomic contigs of European Cottus, and the gene content of candidate regions was explored based on comparisons with the threespine stickleback genome. Genes showing signals of repeated differentiation among brackish-water and freshwater habitats included functions such as ion transport and structural permeability in freshwater gills, which suggests that local adaptation to different osmotic niches might contribute to genomic divergence among habitats. Overall, the presence of both repeated and unique signatures of differentiation across many loci scattered throughout the genome is consistent with polygenic adaptation from standing genetic variation and locally variable selection pressures in the early stages of life history divergence.
Introduction
The build-up of genomic divergence and reproductive isolation during early stages of speciation has often been attributed to allopatric adaptation to ecologically contrasting habitats (e.g. Mayr 1963; Coyne & Orr 2004; Schluter 2009; Langerhans & Riesch 2013). Accordingly, recent reviews have emphasized the importance of adaptation-driven mechanisms (Shafer & Wolf 2013; Sexton et al. 2014). However, understanding the molecular basis of adaptive divergence and build-up of reproductive isolation remains challenging, particularly in the presence of homogenizing gene flow without a period of allopatric separation (e.g. Feder et al. 2012; Smadja et al. 2012; Tigano & Friesen 2016). Recent ‘speciation with gene flow’ models suggest that genomic divergence is restricted to few genomic regions during the early stage of divergence, while allowing a free exchange of alleles in other genomic regions (e.g. Feder et al. 2012; Nosil & Feder 2012; Smadja et al. 2012). Subsequently, divergence hitchhiking of loci in close linkage to genomic regions under selection can increase differentiation and create so-called genomic islands of speciation (Wu 2001; Turner et al. 2005). Initially, such genomic islands have been proposed to lead to widespread genomic divergence and reproductive isolation among locally adapted populations (e.g. Nosil & Feder 2012). Meanwhile, an increasing number of empirical studies revealed highly variable patterns of genomic differentiation, including pronounced divergence of a few and large ‘genomic island’ regions (e.g. Atlantic cod, Hemmer-Hansen et al. 2013; Berg et al. 2015), the presence of many small divergent regions scattered throughout the genome (e.g. threespine stickleback, Rösti et al. 2012; Guo et al. 2015; Helianthus sunflower, Renaut et al. 2013) or some intermediate pattern with a few dozen genomic island of moderate differentiation (e.g. African crater lake cichlids, Malinsky et al. 2015). Such observations have questioned the importance of divergence hitchhiking and instead promoted a significant role of genomic architecture, reduced recombination rates and reduced genetic diversity for genomic island formation (e.g. Renaut et al. 2013; Yeaman 2013; Cruickshank & Hahn 2014). Consequently, the relative importance of ecologically driven genomic divergence through natural selection remains unclear, because many other possible causes may exist, including sexual selection, reduced recombination rates, mutation, drift or demographic effects (e.g. Nielson 2005; Gagnaire et al. 2013; Nosil & Feder 2013).
Repeated evolution of similar phenotype–environment associations is usually regarded as a strong indicator for natural selection (Endler 1986; Rundle et al. 2000; Schluter 2000; Butlin et al. 2013). Accordingly, the inclusion of replicate ecotype pairs has strengthened inferences on adaptive genomic divergence (e.g. Rösti et al. 2012; Gagnaire et al. 2013; Ravinet et al. 2016). Because the genomic basis underlying repeated phenotypic divergence can vary considerably (e.g. Prunier et al. 2012; Holliday et al. 2015), it becomes increasingly important to identify the targets of selection (‘candidate genes’) that can be functionally linked to ecological variables that likely affect the fitness of individuals. A promising approach is to focus on a priori determined candidate genes or gene families that are suspected to underlie important ecotypic traits, as has been demonstrated by Smadja et al. (2012) for the role of chemosensory gene families for host specialization in aphids. This allows a more straightforward interpretation of candidate genes underlying well-defined selection pressures, as opposed to post hoc selection of putatively interesting genes from a large list of candidates (Haasl & Payseur 2016). When combined with whole-genome scans, ecologically relevant genes under selection as well as new potential candidate genes can be detected while controlling for genomewide patterns of differentiation.
In prickly sculpin (Cottus asper), two life history ecotypes can be found in coastal river systems of western North America. Brackish-water groups occupy estuarine or nearby mainstream habitats and follow an amphidromous life cycle, which is characterized by spawning migrations towards the estuary, an extended planktonic larval stage that may allow wide dispersal along the coast to adjacent river systems, and upstream migrations of the juveniles. In contrast, purely freshwater spawners complete their whole life cycle in landlocked freshwater lakes and streams, or in tributaries to coastal rivers (McAllister & Lindsey 1961; McPhail 2007). Gene flow among amphidromous and freshwater ecotypes in coastal rivers and their tributaries can be high due to downstream drift of numerous planktonic larvae and upstream migration of juveniles and adults (Dennenmoser et al. 2014). Despite ongoing gene flow, genetic differentiation at 14 neutral microsatellite markers has been found independent of geographic distance among estuarine and tributary habitats within the same river system, which could indicate the presence of strong divergent selection (Dennenmoser et al. 2014). The assumption of divergent adaptive phenotypes is further supported by physiological differences in osmoregulatory capability between estuarine and tributary freshwater populations, with freshwater populations being more efficient in retaining iodides and more sensitive to salinity changes compared to estuarine populations of C. asper (Bohn & Hoar 1965). Similarly, differentiation among freshwater and brackish-water groups in the presence of gene flow is known from other euryhaline teleosts such as Atlantic killifish (Fundulus heteroclitus), European whitefish (Coregonus lavaretus) and threespine stickleback (Gasterosteus aculeatus), in which numerous adaptive candidate genes for salinity tolerance have been characterized (Whitehead et al. 2011, 2013; Jones et al. 2012a, b; Papakostas et al. 2012). In C. asper, divergent life history ecotypes occupying brackish-water and freshwater niches were presumably established following the colonization of coastal river systems after the last glacial maximum around 14 000 years BP (Dennenmoser et al. 2015), thus presenting a suitable study system for investigating the early stage of genomic divergence in the presence of gene flow.
In this study, we aimed to characterize the patterns of genomic divergence between two brackish-water and two freshwater groups of prickly sculpin. These populations represent two distinct life history ecotypes (amphidromous and purely freshwater life cycles) and have been previously shown to belong to the same mitochondrial lineage and to be genetically more differentiated between brackish-water and freshwater habitats than within habitats (Dennenmoser et al. 2014, 2015). To address the possibility of adaptive genomic divergence underlying the osmoregulatory phenotypes described by Bohn & Hoar (1965), we used whole-genome sequencing of pooled samples (‘Pool-Seq’, Futschik & Schlötterer 2010), in combination with a targeted candidate gene search for known teleost salinity tolerance genes. We asked (i) whether a priori selected teleost salinity candidate genes show the congruent patterns of divergence among replicate habitat-type pairs, (ii) whether genomewide differentiation resembles more a pattern of either many small or few large divergent regions and (iii) whether highly divergent FST outlier regions are enriched with gene categories that might be functionally associated with adaptation to different osmotic niches.
Materials and methods
Sampling, DNA preparation and sequencing
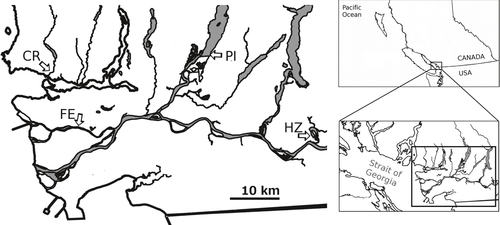
We sampled two estuarine and two tributary groups of C. asper that have been previously shown to represent genetically distinct, potentially locally adapted life history ecotypes in the Lower Fraser River system in southern British Columbia (Canada; Dennenmoser et al. 2014). Brackish-water sampling sites were located in the Capilano River Estuary (49°19.416′N, 123°8.163′W) and the Fraser River Estuary (49°12.317′N, 123°2.679′W). Estuarine groups of Fraser River and the nearby (~15 km) Capilano River have been shown to be highly genetically similar at 14 microsatellite loci, presumably due to extensive gene flow facilitated by along-shore dispersal of a salt-tolerant, planktonic larval stage typical for an amphidromous life cycle (Dennenmoser et al. 2014). Freshwater sampling sites were two tributary systems (Pitt Lake, 49°20.973′N, 122°36.522′W; Hatzic tributary) connected to the Lower Fraser River (Fig. 1). Samples from the Hatzic tributary included 12 samples from Hatzic Lake (49°9.935′N, 122°13.641′W) and 32 samples from a nearby (~3.5 km) inlet site in Hatzic Slough (49°11.352′N, 122°14.852′W). The potential for gene flow among all sampling sites is high due to downstream drift of planktonic larvae, spawning migrations of adults and upstream migrations of juveniles. Previous migration rate estimates indicated a reduced migration from estuarine (Fraser Estuary) into tributary (Pitt and Hatzic Lake) sites compared to migration rates among freshwater tributaries or migration from tributary into estuarine sites (Dennenmoser et al. 2014). Small fin-clip samples (~50 mg) of 44 individuals per group were collected during May 2012 using baited minnow traps, and subsequently preserved in 95% ethanol.
DNA extraction from tissues followed a standard phenol–chloroform technique (Sambrook et al. 2001). DNA quality and quantity was assessed on a 1% agarose gel and a qubit 2.0 fluorometer (dsDNA BR Array Kit, Invitrogen). For each sampling site, equal amounts of DNA (~100 ng) from each of 44 individuals were pooled and subsequently adjusted to a final concentration of approximately 20 ng/μL for each pool. Although biased representation of individuals in a pool can be problematic in the cases of low read depths and small pool sizes (Anderson et al. 2014), we chose a pooling approach because it provides a cost-effective alternative to individual barcoding and has been repeatedly shown to provide reliable estimates of allele frequencies (e.g. Gautier et al. 2013; Rellstab et al. 2013; Schlötterer et al. 2014; Fracassetti et al. 2015; Barrio et al. 2016). To minimize possible artefacts, we used ‘best practice’ guidelines for sequencing of pooled samples (Schlötterer et al. 2014), which recommends pool sizes of at least 40 individuals, a depth of coverage above 50×, use of paired-end reads with read lengths of at least 75 bp, the use of a maximum coverage to avoid copy number variants, trimming of error prone 3′-ends and filtering for mapping quality above 20. Pooled samples were sent to a sequencing provider (Génome Québec, Canada) for the preparation of population-specific libraries and subsequent Illumina HiSeq 2000 whole-genome sequencing (paired-end 100-bp reads).
Data processing and single nucleotide polymorphism (SNP) calling
We trimmed the 100-bp paired-end reads for a minimum average base quality score of 20 and retained reads with a minimum length of 50 bp using the software PoPoolation (Kofler et al. 2011a). All trimmed and filtered reads were archived at the Dryad Digital Repository (doi: 10.5061/dryad.2qg01). The software nextgenmap (version 0.4.8, Sedlazeck et al. 2013) was used to map the trimmed reads against a repeat-masked contig assembly of C. rhenanus (Smolka et al. 2015), which comprised larger contigs (N50 = 6900 bp) compared to a possible assembly of C. asper Pool-Seq data. This mapping step maximized the amount of assembled sequence that could be anchored to the reference genome, thereby optimizing the transfer of positional genomic information from the stickleback genome. Single nucleotide polymorphisms (SNPs) were called using samtools (Li et al. 2009) to first convert files to binary alignment/map (BAM) format, while at the same time filtering for a minimum mapping quality score of 20, and to subsequently generate a mpileup file that combined all four pools. A synchronized mpileup file for downstream analyses was created in PoPoolation2 (Kofler et al. 2011b). From this mpileup file, allele frequencies for every SNP were estimated in PoPoolation2 (Kofler et al. 2011b). To minimize false SNP calls due to sequencing errors, we used only biallelic SNPs with a minimum count of three reads of the minor allele for each population, a minimum of 15 reads covering that genomic position and a maximum coverage of 200 reads to avoid erroneous SNP calling due to copy number variation.
Alignment with threespine stickleback and salinity candidate genes
Gene annotations of Cottus SNPs were explored through the transfer of positional genomic information from the assembled 21 chromosomes of the threespine stickleback genome (version broads1.56). This approach has been successfully used for European Cottus in previous studies (Stemshorn et al. 2011; Cheng et al. 2013, 2015). To determine the positions of Cottus SNPs relative to the genome of threespine stickleback, we applied a custom perl script (available upon request from the authors) that worked as follows. First, the blast software (blastall 2.2.22; Altschul et al. 1990) was used to anchor the Cottus scaffolds in the stickleback genome, using parameters for distant homology search (parameter settings: -e 0.0000000001 -m 9 -F T -G 2 -E 2 -r 2 -q -3 -W 9). These blast analyses omitted scaffolds shorter than 500 bp. Longer scaffolds were split into 500-mers, from which a maximum of 25 randomly chosen sequences per scaffold were retained for subsequent blast searches against the stickleback genome. Significant blast hits obtained for 500-mers from each scaffold were evaluated for the presence of unambiguous hits (e-value threshold = 1e−008, score threshold = 80). Such hits suggest homology with a genomic region in the stickleback genome. If more than 10% of the scaffold hits mapped to genomic regions that were separated in the stickleback genome by more than five times the length (in bp) of the Cottus scaffold, the scaffold was discarded. Finally, the orientation of the Cottus scaffold with respect to the stickleback genome was determined. All Cottus SNPs with tentatively identified genomic map positions were used for downstream analyses.
Analyses
To distinguish between repeated and nonrepeated patterns of genomic divergence among two brackish-water and two freshwater habitats, we used all four pairwise comparisons among habitats as replicate population pairs (Fig. 1). Because we first aimed to test for repeated differentiation of teleost salinity genes, we retrieved 48 putative candidate genes for salinity tolerance and osmoregulation from the teleost literature. To facilitate mapping of SNPs to annotated genes, we restricted our selection to 36 candidate genes that could be assigned to chromosomal positions at the Gasterosteus genome (Table 1). SNPs were assigned gene names by matching their mapping position with gene start and end positions from a stickleback gene annotation file (Gasterosteus_aculeatus.BROADS1.75.gtf) retrieved from Ensembl (http://www.ensembl.org). To identify candidate salinity genes with consistent allele frequency differences among the two habitats, we applied a Cochran–Mantel–Haenszel (CMH) chi-squared test implemented in PoPoolation2, using SNPs with a minimum coverage of 15 reads, a minimum count of eight reads for the minor allele (i.e. minimum count of two per population on average) and a maximum coverage of 200 reads. All CMH test P-values were corrected for multiple testing (FDR, <0.05, Benjamini & Hochberg 1995). Finally, SNPs with significant allele frequency differences were positioned to stickleback genes as before, using gene start and end positions retrieved from the Ensembl stickleback gene annotation file.
| Gene name | Ensembl gene ID (Gasterosteus aculeatus) | Gene notation | References |
|---|---|---|---|
| ANXA4 | ENSGACG00000017070 (chrXIV:6770049-6777904) | Annexin A4 | Papakostas et al. (2012) |
| ANXA6 | ENSGACG00000018367 (chrIV:13375189-13385899) | Annexin A6 | Papakostas et al. (2012) |
| AQP3 | ENSGACG00000010315 (chrXIII:10684451-10690114) | Aquaporin 3a | Whitehead et al. (2011), Shimada et al. (2011), Yan et al. (2013) |
| ATP1A1 | ENSGACG00000014324 (chrI:21699051-21730778) | ATPase Na+/K+ transporting alpha 1 (sodium/potassium ATPase) | Papakostas et al. (2012), Shimada et al. (2011) |
| ATP2C1 | ENSGACG00000002968 (chrXX:138187-148751) | ATPase, Ca++ transporting, type 2C, member 1 | Shimada et al. (2011) |
| CFTR | ENSGACG00000009039 (chrXIX:10185359-10208352) | Cystic fibrosis transmembrane conductance regulator | Yan et al. (2013), Norman et al. (2011) |
| CLCN4 | ENSGACG00000015506 (chrI:27635716-27641746) | Chloride channel, voltage-sensitive 4 | Shimada et al. (2011) |
| DCT | ENSGACG00000004101 (chrXVI:9223846-9229163) | Dopachrome tautomerase | Shimada et al. (2011) |
| DIO1 | ENSGACG00000012104 (chrVIII:15377362-15380410) | Deiodinase, iodothyronine, I | Whitehead et al. (2011) |
| GHRA | ENSGACG00000006842 (chrXIII:5674429-5680840) | Growth hormone receptor a | Yan et al. (2013) |
| GHRB | ENSGACG00000017924 (chrXIV:10387950-10398780) | Growth hormone receptor b | Aykanat et al. (2011) |
| GTF2B | ENSGACG00000015696 (chrIII:8929813-8936195) | General transcription factor IIB | Shimada et al. (2011) |
| HNRNPM | ENSGACG00000010265 (chrVIII:12982618-12991104) | Heterogeneous nuclear ribonucleoprotein | Papakostas et al. (2012) |
| HSPA14 | ENSGACG00000019293 (groupIV:23830489:23838903) | Heat shock protein 14 | Papakostas et al. (2012), Jones et al. (2012a), Larsen et al. (2008) |
| IGF1 | ENSGACG00000020042 (chrIV:32097861-32108728) | Insulin-like growth factor 1 | Yan et al. (2013) |
| IGF2b | ENSGACG00000011125 (chrXIX:13286563-13292182) | Insulin-like growth factor 2b | Norman et al. (2011) |
| MAP3K3 | ENSGACG00000009795 (chrXI:7412764-7421428) | Mitogen-activated protein kinase kinase kinase 3 | Papakostas et al. (2012) |
| MAP3K7 | ENSGACG00000006251 (chrXVIII:3565010-3578113) | Mitogen-activated protein kinase kinase kinase 7 | Papakostas et al. (2012) |
| MAPK1 | ENSGACG00000014421 (chrXIII:18962727-18976830) | Mitogen-activated protein kinase 1 | Shimada et al. (2011) |
| Muc5AC | ENSGACG00000014095 (chrII:394200-413477) | Mucin-5AC | Jones et al. (2012a) |
| Muc5B | ENSGACG00000014109 (chrII:472443-479508) | Mucin-5B | Jones et al. (2012a) |
| NBCE1 (SLC4A4A) | ENSGACG00000014471 (chrXIII:19003120-19035911) | Electrogenic sodium bicarbonate cotransporter 1 (solute carrier family 4) | Kozak et al. (2013) |
| NCC (SLC12a1) | ENSGACG00000016765 (chrII:18335882-18347375) | Na+/Cl− cotransporter, solute carrier family 12, member 1 | Shimada et al. (2011), Yan et al. (2013) |
| NCC (SLC12A2, 1 of 2) | ENSGACG00000014721 (chrXIII:19856460-19871357) | Na+/Cl− cotransporter (solute carrier family 12, member 2, 1 of 2) | Shimada et al. (2011), Yan et al. (2013) |
| NCC (SLC12a2, 2 of 2) | ENSGACG00000018343 (chrXIV:13968407-14004955) | Na+/Cl− cotransporter, solute carrier family 12, member 2, 2 of 2 | Shimada et al. (2011), Yan et al. (2013) |
| NCC (SLC12a10.1) (Danio rerio) | ENSGACG00000018955 (chrVII:1832511-1841285) | Na+/Cl− cotransporter, solute carrier family 12, member 10 | Wang et al. (2009) |
| NHE3 (SLC9A3.2) | ENSGACG00000002446 (chrX:2084882-2101301) | Solute carrier family 9 (sodium/hydrogen exchanger) | Shimada et al. (2011) |
| ODC1 | ENSGACG00000012974 (chrXV:14293863-14298917) | Ornithine decarboxylase 1 | Whitehead et al. (2011) |
| PLECA | ENSGACG00000002566 (groupX:2359321:2399237) | Plectin a | Papakostas et al. (2012) |
| PRL | ENSGACG00000006561 (chrXI:3125864-3130591) | Prolactin | Yan et al. (2013) |
| PRLRA | ENSGACG00000016473 (groupXIV:3992906:3998872) | Prolactin receptor a | Yan et al. (2013), Jones et al. (2012a) |
| Rhogtp (arhgap4a) | ENSGACG00000008645 (chrXII:10779457-10784569) | Rho GTPase activating protein 4a | Shimada et al. (2011) |
| STIP1 | ENSGACG00000020272 (chrVII:15340112-15348214) | Stress-induced phosphoprotein | Papakostas et al. (2012) |
| TGM1 | ENSGACG00000014362 (chrVIII:18925268-18928490) | Transglutaminase 1 | Whitehead et al. (2011) |
| TGM 1|1 | ENSGACG00000014435 (chrIII:4385971-4397309) | Transglutaminase 1-like 1 | Papakostas et al. (2012) |
| VCAN | ENSGACG00000015542 (chrXIV:271307-289240) | Versican | Whitehead et al. (2011) |
For a genomewide screen of highly divergent genes that could be under natural selection, we performed FST outlier analyses for all between-habitat comparisons (Fraser Estuary vs. Pitt Lake, Fraser Estuary vs. Hatzic Lake, Capilano Estuary vs. Pitt Lake, Capilano Estuary vs. Hatzic Lake) using the software bayescan 2.5 (Foll & Gaggiotti 2008). This method is based on a finite-island model with migration between populations and mutation–drift equilibrium (Beaumont & Nichols 1996). It uses reversible-jump Markov chain Monte Carlo sampling to estimate posterior odds for a model with selection against a model without selection. For all outlier analyses, we set the prior odds to 1000 and performed 40 short pilot runs of 5000 MCMC steps, followed by a long run of 100 000 MCMC steps (using a burn-in of 100 000 and a thinning interval of 10). SNPs were only considered as candidates if they emerged in all four between-habitat comparisons as outliers using a false discovery rate (FDR) threshold of 0.05, and were positioned to stickleback genes using gene start and end positions retrieved from the Ensembl stickleback gene annotation file.
To further assess whether regions of elevated FST would be small and scattered throughout the genome, or cluster together to form larger ‘genomic islands of divergence’, we evaluated the patterns of genomewide differentiation in nonoverlapping 100- and 50-kb windows along the stickleback chromosomes. We applied permutation tests using the ‘sample’ function in r 3.2.2 to characterize ‘outlier’ windows that are significantly enriched with highly differentiated SNPs falling within the top three percentile of the genomewide FST distribution (e.g. see Renaut et al. 2013). For this purpose, we counted the total number of SNPs and the number of top three percentile SNPs for each window, randomly sampled with replacement the same number of SNPs from the genome and calculated the proportion of top three percentile SNPs per window for each of the 10 000 permutations. Significance was then inferred from the percentage of 10 000 permutations that showed a larger or equally large proportion of highly differentiated SNPs. We corrected significance values for multiple testing using a false discovery rate (FDR) threshold of 0.05.
We further characterized genomewide patterns of Tajima's D, nucleotide diversity π and absolute nucleotide divergence dxy with the aim to distinguish between background selection, divergence hitchhiking and locally reduced gene flow underlying elevated differentiation of FST outlier windows. Locally restricted gene flow (sensu Nachman & Payseur 2012) may act, for example, through divergent selection against maladaptive loci underlying reproductive isolation or ecological adaptation. This can facilitate genomewide differentiation in the presence of gene flow without the need of hitchhiking (e.g. Nachman & Payseur 2012). To account for the possibility of reduced recombination rates causing elevated FST regions (Nachman & Payseur 2012; Cruickshank & Hahn 2014), we also evaluated the patterns of absolute nucleotide divergence dxy (Nei 1987) among population pairs (x,y) estimated from allele frequencies (A,a) as (fA,x fa,y) + (fA,y fa,x) (see also Delmore et al. 2015). For dxy calculations, we adjusted a perl script from Owens (2015; https://github.com/owensgl/pop_gen/blob/master/sync2dxy.pl).
Because dxy may not be a reliable indicator of locally reduced gene flow during the early stages of population differentiation (e.g. Nachman & Payseur 2012; Feulner et al. 2015), we also calculated the estimates of Tajima's D (TD, Tajima 1989) and nucleotide diversity π using PoPoolation2. Tajima's D is sensitive to deviations from the neutral allele frequency spectrum and should be shifted towards positive values under balancing selection and towards negative values (excess of rare frequency allelic variants) under the influence of natural selection as, for example, in a hitchhiking scenario (e.g. Charlesworth et al. 1993). In contrast, FST outlier regions are not expected to deviate from a neutral allele frequency spectrum under the influence of locally restricted gene flow (Nachman & Payseur 2012). Similar to other studies (Feulner et al. 2015; McGaughram et al. 2016), we tested whether FST outlier windows showed decreased TD (defined as lower 5% quantile of the genomewide average) and distinguished between (i) background selection (decreased TD in both habitats), (ii) directional selection with hitchhiking (decreased TD in one of both habitats), (iii) locally restricted gene flow (TD not decreased compared to genomewide average) and (iv) balancing selection (TD increased, π decreased).
Gene ontology enrichment analyses
To test whether differentiated candidate loci common to all pairwise between-habitat comparisons were enriched with potential osmoregulatory molecular functions or other specific functional categories, we performed gene ontology (GO) enrichment tests in blast2go (Conesa et al. 2005), using a Fisher's exact test corrected for multiple testing with a false discovery rate (FDR) of 0.05. As a reference gene set, we retrieved Gasterosteus Ensemble gene identifiers from the biomart database (BROADS1) for 14 871 genes that matched mapping positions of our full set of SNPs aligned to the stickleback genome. As test gene sets, we first used 1138 candidate genes resulting from the Cochran–Mantel–Haenszel (CMH) analysis for overrepresentation of specific gene functions. Second, we tested the set of 25 candidate genes that emerged from the FST outlier analysis. Finally, we tested the sets of genes falling within highly differentiated 100- and 50-kb FST outlier windows for the enrichment of gene functions.
Results
A total of 760 282 616 reads (94.91%) could be mapped against the C. rhenanus genome assembly (Fraser Estuary: 114 323 511 reads; 15.5× coverage; Capilano Estuary: 154 396 036 reads, 21.7× coverage; Hatzic tributary: 355 074 685 reads, 50× coverage; Pitt Lake: 136 488 384 reads; 12.9× coverage), resulting in a total average coverage of 25× and a total of 1 116 630 SNPs after quality filtering. Of those, 761 871 SNPs (68.2%) could be positioned to the 21 chromosomes of the Gasterosteus genome, which were used for calculating average FST values as well as Tajima's D, π and dxy across 100- and 50-kb windows.
The Cochran–Mantel–Haenszel (CMH) test identified 8600 SNPs (after FDR correction for multiple testing) that showed a consistent differentiation in allele frequencies among estuarine and tributary habitats. Of those, 6160 SNPs (71.6%) could be positioned to the Gasterosteus chromosomes, which included 2668 within-gene SNPs (43.3%) distributed across 1138 genes. Significant SNPs that matched the a priori selected salinity candidate genes included 17 SNPs in sodium/potassium ATPase (atp1a1a), six SNPs in a Na+/Cl− cotransporter gene (NCC, slc12a10.1) and one SNP in the heat shock 70-kDa protein 14 (HSPA14).
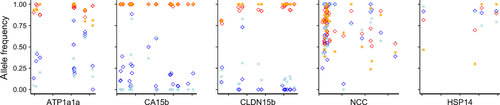
The bayescan FST outlier analyses identified 555 outlier SNPs between Capilano Estuary and Pitt Lake, 443 outlier SNPs between Fraser Estuary and Hatzic Lake, 509 outlier SNPs between Capilano Estuary and Hatzic Lake and 468 outlier SNPs between Fraser Estuary and Pitt Lake (Figure S1, Supporting information). Of those, 158 (>28%) significant outlier SNPs were common to all four pairwise comparisons. Significant outlier SNPs matched 25 common candidate genes and included sodium/potassium ATPase (atp1a1a), as well as two other genes related to osmoregulation and salinity tolerance (CA15b and CLDN15; Table 2). All candidate genes identified by the bayescan outlier analyses were also identified by the CMH test (Table S4, Supporting information). However, allele frequency differences among habitats in candidate salinity genes were much more pronounced in bayescan FST outlier genes compared to genes only identified by the CMH analysis (Fig. 2). The average sequencing depth for all common candidate genes was 34.5× (±19.5 SD; range: 17–96×).
| Gene | CMH test | bayescan FST outlier | Gene product |
|---|---|---|---|
| Candidate salinity genes | |||
| ATP1a1a | * | * | ATPase, Na+/K+ transporting, alpha 1a |
| NCC | * | - | Na+/Cl− cotransporter, solute carrier family 12, member 10 |
| HSP14 | * | - | Heat shock protein 14 |
| CA15b | * | * | Carbonic anhydrase XV b |
| CLDN15b | * | * | Claudin 15b |
| Other candidate genes | |||
| CALB2a | * | * | Calbindin2a (calcium ion binding) |
| CAPN1b | * | * | Calpain 1 large subunit b (calcium ion binding) |
| CEP290 | * | * | Centrosomal protein 290 |
| CSAD | * | * | Cysteine sulfinic acid decarboxylase |
| GUCY2f | * | * | Guanylate cyclase 2f, retinal |
| HMP19 | * | * | Hypothalamus Golgi Apparatus Expressed Protein |
| KIF3a | * | * | Kinesin family member 3A |
| KITLGA | * | * | Kit ligand a |
| LYVE1b | * | * | Lymphatic vessel endothelial hyaluronic acid receptor 1b |
| MME | * | * | Membrane metallo-endopeptidase (neprilysin) |
| MMP25 | * | * | Matrix metallopeptidase 25 |
| PIF1 | * | * | PIF 5′–3′ DNA helicase homolog |
| PNP6 | * | * | Purine nucleoside phosphorylase 6 |
| PRKD3 | * | * | Protein kinase D3 |
| Si:dkey-106n21.1 | * | * | Transmembrane transport? |
| Si:ch211-173d10.4 | * | * | Uncharacterized (carbonic anhydrase IV?) |
| Si:ch211-253 h3.1 | * | * | ABR, active BCR-related |
| SLA1 | * | * | Src-like adaptor 1 |
| SSUH2RS1 | * | * | SSU-2 homolog, related sequence 1 |
| TMTC3 | * | * | Transmembrane and tetratricopeptide repeat containing 3 |
| Zgc:174906 | * | * | Unknown |
| Novel | * | * | Unknown (ENSGACG00000007888) |
Plotting of 3995 100-kb FST windows and permutation tests revealed 299–391 outlier windows in between-habitat comparisons (Table 3). Of those, 111 (34%) outlier windows were common to all between-habitat comparisons, which were scattered along the stickleback chromosomes (Fig. 3). The 111 common outlier windows contained 336 genes (see Table S1, Supporting information).
| Comparisons | 100-kb FST outlier windows | 50-kb FST outlier windows | FST (mean; SD) | dxy (mean; SD) |
|---|---|---|---|---|
| Between-habitat comparisons | ||||
| Capilano Est.–Pitt Lake | 326 (8.2%) | 574 (7.2%) | 0.112 (±0.027) | 0.0011 (±0.0004) |
| Fraser Est.–Pitt Lake | 299 (7.5%) | 570 (7.2%) | 0.115 (±0.023) | 0.0008 (±0.0003) |
| Capilano Est.–Hatzic Lake | 391 (9.8%) | 659 (8.3%) | 0.098 (±0.025) | 0.0024 (±0.0009) |
| Fraser Est.–Hatzic Lake | 336 (8.4%) | 612 (7.7%) | 0.102 (±0.02) | 0.0016 (±0.0006) |
| Within-habitat comparisons | ||||
| Fraser Est.–Capilano Est. | 118 (3%) | 216 (2.7%) | 0.1 (±0.014) | 0.0015 (±0.0006) |
| Hatzic Lake–Pitt Lake | 216 (5.4%) | 439 (5.5%) | 0.096 (±0.028) | 0.001 (±0.0004) |
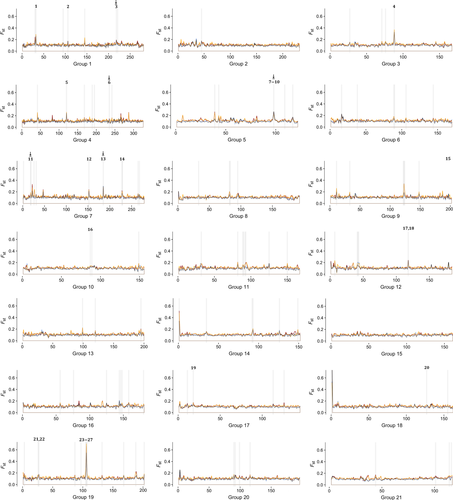
 (1 = Guyc2f, 2 = MME, 3 = atp1a1a, 4 = Pif1, 5 = Kif3a, 6 = Hsp14, 7 = mmp25, 8 = Ca15b, 9 = unknown, 10 = si:ch211-173d10.4, 11 = NCC, 12 = si:ch211-253 h3.1, 13 = Cldn15b, 14 = hmp19, 15 = Capn1b, 16 = sla, 17 = Csad, 18 = Zgc:174906, 19 = ssuh2rs1, 20 = prkd3, 21 = Calb2a, 22 = Pnp6, 23 = Si:dkey-106n21.1, 24 = Kitlga, 25 = Tmtc3, 26 = Cep290, 27 = Lyve1b).
(1 = Guyc2f, 2 = MME, 3 = atp1a1a, 4 = Pif1, 5 = Kif3a, 6 = Hsp14, 7 = mmp25, 8 = Ca15b, 9 = unknown, 10 = si:ch211-173d10.4, 11 = NCC, 12 = si:ch211-253 h3.1, 13 = Cldn15b, 14 = hmp19, 15 = Capn1b, 16 = sla, 17 = Csad, 18 = Zgc:174906, 19 = ssuh2rs1, 20 = prkd3, 21 = Calb2a, 22 = Pnp6, 23 = Si:dkey-106n21.1, 24 = Kitlga, 25 = Tmtc3, 26 = Cep290, 27 = Lyve1b).Within habitats, the number of 100-kb FST outlier windows was 118 between Fraser Estuary and Capilano Estuary and 216 between Hatzic and Pitt Lake (Table 3). Ten outlier windows emerged in both within-habitat comparisons and one outlier window on stickleback chromosome 17 (12.5–12.6 Mb) overlapped with a between-habitat outlier window. Average FST values showed higher differentiation between habitats compared to within habitats (Wilcoxon rank-sum test, two-tailed, P < 0.001; Table 3).
bayescan FST outlier genes were mostly dispersed across multiple 100-kb outlier windows (Fig. 3). Clustering of multiple bayescan outlier genes within a single 100-kb FST window was found on chromosome V (9.7–9.8 Mb: Ca15b, Mmp25, si:ch211-173d10.4 and one novel gene) and chromosome XII (11.7–11.8 Mb: Csad, Zgc:174906). Clustering of multiple bayescan outlier genes within a region of two or more consecutive outlier windows occurred only on stickleback sex chromosome XIX (2.5–2.7 Mb: calb2a, pnp6; 10.4–10.7 Mb: si:dkey-106n21.1, kitlga, tmtc3, cep290, lyve1b; Fig. 3).
Plotting of 7952 50-kb FST windows and permutation tests revealed 574–659 outlier windows in between-habitat comparisons (Table 3). Of those, 178 (31%) outlier windows were common to all between-habitat comparisons, which were scattered along the stickleback chromosomes and overlapped with all common 100-kb outlier windows. The 178 common outlier windows contained 278 genes, of which 175 genes were also matched by the 100-kb Fst outlier windows (listed in Tables S2, S3, Supporting information). Within habitats, the number of 50-kb FST outlier windows was 296 between Fraser Estuary and Capilano Estuary and 439 between Hatzic and Pitt Lake (Table 3). Among all within-habitat outlier windows, 32 outlier windows emerged in both within-habitat comparisons, of which seven outlier windows overlapped with a between-habitat outlier window.
Because the 50-kb window analyses gave qualitatively similar results as the 100-kb window analyses, we focused our subsequent analyses on 100-kb windows to evaluate the patterns of Tajima's D, π and dxy with the aim to better distinguish between background selection, divergence hitchhiking and locally reduced gene flow. Genomewide averages of Tajima's D were negative for all populations (Capilano: −0.33 ± 0.11 SD; Fraser: −0.32 ± 0.11; Pitt: −0.21 ± 0.12; Hatzic: −0.32 ± 0.13), indicating an excess of rare alleles resulting from selection or a recent (postglacial) demographic population expansion. Only a few FST outlier windows showed decreased (lower 5% of genomewide average) Tajima's D values: reduced TD values in freshwater, but not the brackish-water, habitats (indicating directional selection) occurred on chromosome 3 (8.7–8.8 Mb; genes: ptger4c, pif1, nsun4, dmbx1a, gadd45aa, gng12a, wls) and chromosome 11 (8.9–9 Mb; genes: ush1ga, otop2, BAHCC1). Reduced TD values in both brackish-water, but not the freshwater, habitats occurred on chromosome 9 (12.2–12.3 Mb; genes: novel, slc43a3a). Reduced TD values in both habitats (three of four populations) occurred on chromosome 2 (4.4–4.5 Mb; genes: SEMA7A) and chromosome 14 (12.5–12.6 Mb; genes: unknown), which could reflect background selection [see Figure S2 (Supporting information) for some examples of genomic regions putatively under directional or background selection]. The majority of FST outlier windows did not show decreased TD values, suggesting a more plausible role of locally reduced gene flow without apparent effects of divergence hitchhiking underlying elevated FST regions.
Across all 100-kb windows, average dxy values were generally low for all pairwise population comparisons (Table 3). Average dxy values were slightly higher within compared to outside FST outlier windows in all comparisons except Fraser Estuary vs. Capilano Estuary (Wilcoxon rank-sum test, two-tailed, P < 0.05): Fraser Estuary vs. Capilano Estuary: 0.0015 (within) vs. 0.0015 (outside); Fraser Estuary vs. Hatzic Lake: 0.0017 (within) vs. 0.0016 (outside); Fraser Estuary vs. Pitt Lake: 0.00087 (within) vs. 0.0008 (outside); Capilano Estuary vs. Hatzic Lake: 0.0025 (within) vs. 0.0023 (outside); Capilano Estuary vs. Pitt Lake: 0.0012 (within) vs. 0.001 (outside); Hatzic Lake vs. Pitt Lake: 0.0011 (within) vs. 0.001 (outside).
Estimates of nucleotide diversity π (averaged across all 100-kb windows) were similar among populations (mean ± SD): Fraser Estuary: 0.33 ± 0.02; Capilano Estuary: 0.31 ± 0.02; Hatzic Lake: 0.25 ± 0.02; Pitt Lake: 0.37 ± 0.02. Diversity estimates were not lower within than outside FST outlier regions as would be expected under divergence hitchhiking, but instead showed a weak tendency to be increased within the 111 common FST outlier windows (Wilcoxon rank-sum test, two-tailed, P < 0.05; mean ± SD): Fraser Estuary: 0.335 ± 0.03 (within) vs. 0.331 ± 0.02 (outside); Capilano Estuary: 0.316 ± 0.02 (within) vs. 0.308 ± 0.02 (outside); Hatzic Lake: 0.258 ± 0.03 (within) vs. 0.245 ± 0.02 (outside); Pitt Lake: 0.369 ± 0.04 (within) vs. 0.366 ± 0.02 (outside).
Finally, we screened 100-kb FST outlier windows for signatures of elevated (upper 5%) TD values combined with increased levels of nucleotide diversity π (upper 5%) that might suggest balancing selection. We distinguished between elevated TD in all populations and habitat-specific TD peaks in both populations of either brackish-water or freshwater habitats. Increased TD values present in all four populations were evident in four FST outlier windows (chromosome 5, 9.7–9.8 Mb; chromosome 7, 1.8–1.9 Mb; chromosome 17, 12.5–12.6 Mb; and chromosome 21, 2–2.1 Mb). Corresponding elevations in π were found only on chromosome 7 (1.8–1.9 Mb) in Capilano Estuary and Hatzic Lake and on chromosome 17 (12.5–12.6 Mb) in Capilano Estuary, Fraser Estuary and Pitt Lake. Increased TD values unique to brackish-water populations were found in one FST outlier window (chromosome 7, 1–2 Mb), without a corresponding increase in π. Freshwater-specific TD peaks were found in four FST outlier windows (chromosome 5, 4.2–4.3 Mb; chromosome 7, 1.7–1.8 Mb; chromosome 16, 15.7–15.8 Mb; and chromosome 19, 10.4–10.5 Mb), all of which showing elevations in π in the freshwater populations Hatzic Lake and Pitt Lake.
Analyses of GO enrichment for the 1138 genes with significant (FDR-corrected) CMH SNPs revealed ‘protein binding’ as significantly overrepresented (P = 0.0273). In contrast, neither the set of 25 genes associated with FST outlier SNPs nor genes falling within the 100- or 50-kb FST common outlier windows showed any significant enrichment of GO terms.
Discussion
Marine to freshwater transitions have played a major role in the remarkable diversification of the family Cottidae, which underwent extensive adaptive radiations following the colonization of freshwater environments throughout the Northern Hemisphere (Kinziger & Wood 2010; Goto et al. 2014). Hence, uncovering the genetic basis of freshwater adaptation and evolution of subsequent genomic divergence is of major interest to disentangle the relative roles of ecologically driven adaptation from nonselective processes during the early stages of speciation. Cottus asper presents an attractive study system to investigate how natural selection drives adaptive divergence at the genomic level, because it allows testing for repeated patterns of differentiation among estuarine (brackish-water) and tributary (freshwater) habitats. Divergent life history ecotypes have presumably evolved from standing genetic variation of the same genetic lineage following deglaciation around 14 000 years ago (Dennenmoser et al. 2015). Independence of environmental and genetic distance from geographic distance implies that the patterns of differentiation of C. asper are shaped by ongoing gene flow (Dennenmoser et al. 2014). Accordingly, ongoing genetic exchange coupled with large population sizes of C. asper should reduce the influence of genetic drift and enhance signals of natural selection on beneficial allelic variants. In agreement with this, we found candidate genes putatively under selection showing repeated pattern of divergence between brackish-water and freshwater habitats. The presence of both repeated and unique signatures of differentiation across many loci suggests that both polygenic adaptation from standing genetic variation and locally variable selection pressures play a role in the early stages of life history divergence.
We combined a Pool-Seq approach with a targeted search for a priori determined candidate genes, which was facilitated by using threespine stickleback (G. aculeatus) as a reference genome. Our experimental design of sequencing pools of 44 individuals at an average sequencing depth of 25× is comparable to recent studies that have validated the accuracy of allele frequency estimates experimentally (Lamichhaney et al. 2012; Guo et al. 2015; Barrio et al. 2016). Although we did not experimentally validate allele frequency estimates, simulation studies have recommend similar sized pools of 50 haploid individuals sequenced at a read depth of 20–30× (Ferretti et al. 2013) and suggest that robust allele frequency estimates can be reached with lower pool sizes and read depths (Gautier et al. 2013). In general, pools with a larger number of individuals relative to sequencing depth have the advantage of summarizing the mutation frequency spectrum better because it is averaged over more evolutionary histories (Ferretti et al. 2013).
Adaptive divergence of osmoregulatory genes
Adaptation to different osmotic environments includes a variety of physiological mechanisms including ion transport, stress responses, cell signalling or structural permeability of cell membranes (Evans et al. 2005; Evans 2008). Accordingly, a complex interplay among numerous genes affects osmoregulation in fishes (Hwang et al. 2011; Papakostas et al. 2012). This presumably results in polygenic adaptation that may be reflected by weak signatures of selection affecting many loci of small fitness effects. However, osmoregulation can be energetically costly in teleost fishes (>30% of total metabolic cost, Ern et al. 2014), and therefore, divergent selection should be expected to increase the frequency of adaptive alleles. Ion transport genes are one likely target of selection because of their importance in retaining ions in low-salinity environments and removing excess ions in high-salinity environments (Hwang et al. 2011).
Our results support the physiological observations by Bohn & Hoar (1965) that brackish-water and freshwater groups of prickly sculpin are adapted to their respective osmotic environment. The strong signal for divergent selection on the a priori predicted ion transport gene sodium/potassium ATPase (α1 subunit, isoform atp1a1a) suggests a possible role in local adaptation to different osmotic niches. ATPase is crucial for maintaining the ion balance and electrolyte homeostasis in different osmoregulatory epithelia (Evans et al. 2005), with a plethora of studies supporting its adaptive function for ion uptake in freshwater in teleost fishes (e.g. Hohenlohe et al. 2010; Shikano et al. 2010; Norman et al. 2011; Papakostas et al. 2012; Kozak et al. 2013; Urbina et al. 2013; Whitehead et al. 2013; DeFaveri & Merilä 2014; Sutherland et al. 2014; Velotta et al. 2014). Overall, ATPase isozymes appear to be evolutionarily highly conserved (e.g. Mobasheri et al. 2000) and therefore may have repeatedly played an important role during evolutionary transitions from saltwater to freshwater habitats in different euryhaline taxa such as fishes, crabs and copepods (e.g. Nilsen et al. 2007; Tsai & Lin 2007; Lee et al. 2011).
Another interesting candidate gene is the Na+/Cl− cotransporter (NCC), which is involved in chloride uptake in freshwater (Hwang et al. 2011; Zikos et al. 2014) and has also been characterized as osmoregulatory candidate gene under directional selection in threespine stickleback (DeFaveri et al. 2011; Shimada et al. 2011). Our finding of six consistently differentiated SNPs among freshwater- and brackish-water groups in a putative NCC gene could indicate a possibly adaptive function for chloride uptake in C. asper. Osmoregulation in freshwater might be further supported by the carbonic anhydrase gene CA15b, which emerged as a strong FST outlier in the bayescan analyses. Carbonic anhydrases have been shown to assist epithelial Na+ uptake and H+ secretion under freshwater conditions in zebrafish (Ito et al. 2013), and might have similar effects in C. asper. Osmoregulatory capability also depends on the structural permeability of cell membranes (e.g. Evans et al. 2005). Claudin transmembrane proteins appear to be essential for changes in structural permeability in freshwater gills of euryhaline teleosts by supporting the formation of tight junctions that act as a barrier to reduce chloride loss (Hwang et al. 2011; Kozak et al. 2013; Whitehead et al. 2013). Our finding of claudin 15b as an FST outlier could suggest such an adaptive function in C. asper.
Altogether, our finding of various teleost salinity candidate genes that show a consistent differentiation among two brackish-water and two freshwater habitats agrees with previously described osmoregulatory phenotypes of C. asper (Bohn & Hoar 1965) and supports a polygenic basis of osmoregulatory freshwater adaptation. The genetic basis of freshwater adaptation may differ among teleost species, but should affect the same osmoregulatory key functions such as ion transport, stress responses, cell signalling or structural permeability. Although we did not find an enrichment in GO terms related to such functions, we identified a set of highly differentiated candidate loci involved in osmoregulation and various other functions (Table 2). These candidate genes could be part of future resequencing studies to test the link between selection and fitness for a higher number of brackish-water and freshwater habitats repeatedly over time, which would contribute to a better understanding of the genomic basis and temporal stability of freshwater adaptation in euryhaline teleost fishes.
Genomewide differentiation under high gene flow
Genomic divergence can be widespread during the early stages of speciation, but confounding factors such as demography, migration or drift make it challenging to unravel the genomic basis of incipient adaptive divergence in the presence of gene flow (e.g. Seehausen et al. 2014).
Gene flow among brackish-water and freshwater groups of prickly sculpin appears to be reduced enough to allow for genomewide differentiation and adaptive divergence, which could reflect migration–selection balance driven by selection against maladapted immigrants (e.g. Gow et al. 2007; McCairns & Bernatchez 2010; Yeaman & Whitlock 2011). This interpretation is consistent with previous estimates of reduced migration from estuarine into tributary freshwater sites (Dennenmoser et al. 2014). Currently, we do not know whether osmoregulatory divergence alone could be strong enough to reduce gene flow among freshwater and brackish-water habitats. However, fitness costs associated with osmoregulation can be high, and accordingly, adaptation to osmotic niches has been shown to limit gene flow across the salinity boundary in other euryhaline teleost fishes (Bekkevold et al. 2005; McCairns & Bernatchez 2008; Whitehead et al. 2011).
The genomewide pattern of differentiation among brackish-water and freshwater groups of C. asper aligns with other studies that report the presence of many small divergent regions scattered throughout the genome (e.g. threespine stickleback, Rösti et al. 2012; Feulner et al. 2015; Guo et al. 2015; Helianthus sunflower, Renaut et al. 2013). Such studies frequently suggest an important role for divergent selection and reduced recombination rates underlying genomic regions of elevated differentiation. Meanwhile, the contributions of divergence hitchhiking and locally restricted gene flow in shaping the patterns of genomic divergence remain difficult to disentangle. For example, similar large proportion for divergence hitchhiking (38–75%) and locally restricted gene flow (25–55%) has been suggested for lake–river stickleback ecotype divergence (Feulner et al. 2015), whereas a predominant role of selection opposed to hitchhiking is thought to maintain highly differentiated haplotype blocks among divergent populations of Atlantic herring (Barrio et al. 2016). Herein, we did not find apparent signatures of divergence hitchhiking such as large clusters of genomic islands, decreased nucleotide diversity within FST outlier windows or decreased values of Tajima's D within regions of elevated FST (e.g. Feulner et al. 2015; Barrio et al. 2016). Instead, nucleotide diversity showed a tendency to be slightly increased within FST outlier windows and the lowest 5% of Tajima's D values rarely overlapped with regions of increased FST values. Such patterns suggest a major role for locally (locus-specific) reduced gene flow and divergent selection acting on many loci throughout the genome. We found some limited evidence for directional selection (in 3 of 111 FST outlier windows) and background selection (in 2 of 111 FST outlier windows). Similarly, only few differentiated regions showed signs of balancing selection, as indicated by increased values of Tajima's D and increased nucleotide diversity. The few genes located within regions showing a signature of directional selection (ptger4c, pif1, nsun4, dmbx1a, gadd45aa, gng12a, wls, ush1ga, otop2, BAHCC1) do not show obvious functions that could be easily related to adaptive divergence among brackish-water and freshwater habitats. This suggests that genomewide patterns of negative-shifted values of Tajima's D observed in all populations may have been shaped more by a recent (postglacial) population expansion rather than by selection.
Recent studies have emphasized that genomic islands are not a prerequisite for adaptive divergence or reproductive isolation in the presence of gene flow, but are rather a by-product of reduced genomic diversity and low recombination rates that can be associated with genomic structures such as centromeric or telomeric regions (e.g. Cruickshank & Hahn 2014; Ruegg et al. 2014; Burri et al. 2015). Although we did not address the possible effects of variable recombination rates on the patterns of genomewide differentiation, the findings of increased rather than decreased nucleotide diversity within FST outlier windows and only weakly increased dxy within such regions do not support a major role of reduced recombination rates as has been suggested in other studies (e.g. Renaut et al. 2013; Burri et al. 2015). However, future studies should test for possible effects of reduced recombination rates and identify the possible mechanisms of recombination suppression that might promote adaptive divergence among ecotypes in the presence of gene flow. Chromosomal inversions are prime candidates of recombination rate suppressors in heterozygotes and have been suggested to promote genomic island formation, as well as to facilitate the ecotypic divergence and speciation with gene flow (e.g. Hoffmann & Rieseberg 2008; Kirkpatrick 2010; Lohse et al. 2015; Wadsworth et al. 2015). For example, in threespine stickleback, marine–freshwater divergence has been related to chromosomal inversions that include divergent adaptive genes such as the ion transporter genes KCNH4 and atp1a1a (Jones et al. 2012b). Interestingly, the most divergent FST outlier region we found in C. asper was located at the centre of the stickleback sex chromosome 19, which seems to be affected by several pericentric inversions in stickleback (Leder et al. 2010). Although speculative at present, this could present a genomic region of low recombination in C. asper, such as a centromere region, an inversion site or a sex chromosome. Remarkably, this extreme divergence peak included the same candidate genes (si:dkey-106n21.1, kitlga, tmtc3, cep290, lyve1b) that have been characterized as FST outliers on the stickleback sex chromosome 19 between benthic and limnetic ecotypes (Jones et al. 2012a), which could indicate a divergence hotspot of reduced recombination.
Overall, our finding of many divergent outlier loci scattered throughout the genome without apparent signatures of divergence hitchhiking is consistent with models emphasizing selection acting on many traits and many loci. Herein, selection is expected to cause many small changes in allele frequencies among many covarying loci (polygenic soft sweeps) rather than few large-effect changes (LeCorre & Kremer 2012; Bourret et al. 2014; Stephan 2016). Consistently, the build-up of genomic divergence and reproductive isolation among evolving ecotypes has been suggested to proceed through selection on multiple unlinked loci (‘genomewide congealing’, Flaxman et al. 2014; Feder et al. 2014). Although we cannot rule out that some outlier loci are caused by drift, large population sizes and the replicate patterns of divergence suggest a role of natural selection.
Besides replicated signatures of selection, we also found unique FST outlier regions that occurred only in one between-habitat comparison. Currently, we do not know whether such locality-specific FST outliers reflect selective processes on adaptive variants or neutral demographic effects, but similar patterns have frequently been observed and attributed to locally variable selection pressures (e.g. DeFaveri et al. 2011; Kaeuffer et al. 2011; Perrier et al. 2013; Feulner et al. 2015; Ravinet et al. 2016). Surprisingly, their evolutionary significance for the strength of selection and the initial build-up of genomic divergence and reproductive isolation remain largely unexplored. For example, MacPherson et al. (2015) developed a model to show that the number of traits under spatially variable selection (‘trait dimensionality’) affects the strength of local adaptation, which they validated using empirical data from 35 reciprocal transplant studies. Thus, pronounced (genomewide) adaptive divergence should be possible even in the presence of high gene flow, if trait dimensionality under selection is high and if the major axis of genetic variation (G-matrix) aligns with the vector of directional selection (MacPherson et al. 2015). Future studies should experimentally evaluate whether barriers to gene flow during the early stages of divergence are supported by a combination of spatially variable selection pressures, increased trait dimensionality and aligned G-matrices. In addition, research programmes aiming to test the ecological importance of suggested candidate genes could (i) begin to characterize the alternate alleles of a candidate gene in vitro (i.e. genotype to biochemical phenotype); (ii) determine candidate gene impacts on cellular function, morphology, behaviour, physiology and performance in vivo (i.e. genotype to performance); and (iii) assess the fitness consequences of genetic variants either directly, with experimental tests in relevant environments, or indirectly by using comparative phylogenetic approaches and analyses looking for molecular signatures of selection (i.e. genotype to fitness; Dalziel et al. 2009).
Acknowledgements
Field assistance by Jonathan Lowey is greatly appreciated. This research was supported by NSERC Discovery Grants (SMR and SMV), an Alberta Innovates Technology Futures New Faculty Award (SMR), an Alberta Innovates Technology Futures Postgraduate scholarship (SD) and an European Research Council starting grant (AN). SMR would like to thank the Bamfield Marine Sciences Centre (BMSC) for resources while working on this study.
References
S.D., S.V., A.N. and S.R. designed the study. S.D. conducted the analyses with input from all authors. S.D., S.V., A.N. and S.R. wrote the manuscript.
Data accessibility
Pool-Seq data were stored at the Dryad Digital Repository (doi: 10.5061/dryad.2qg01).




