Patterns of diversity and adaptation in Glomeromycota from three prairie grasslands
Abstract
Arbuscular mycorrhizal (AM) fungi are widespread root symbionts that often improve the fitness of their plant hosts. We tested whether local adaptation in mycorrhizal symbioses would shape the community structure of these root symbionts in a way that maximizes their symbiotic functioning. We grew a native prairie grass (Andropogon gerardii) with all possible combinations of soils and AM fungal inocula from three different prairies that varied in soil characteristics and disturbance history (two native prairie remnants and one recently restored). We identified the AM fungi colonizing A. gerardii roots using PCR amplification and cloning of the small subunit rRNA gene. We observed 13 operational taxonomic units (OTUs) belonging to six genera in three families. Taxonomic richness was higher in the restored than the native prairies with one member of the Gigaspora dominating the roots of plants grown with inocula from native prairies. Inoculum source and the soil environment influenced the composition of AM fungi that colonized plant roots. Correspondingly, host plants and AM fungi responded significantly to the soil–inoculum combinations such that home fungi often had the highest fitness and provided the greatest benefit to A. gerardii. Similar patterns were observed within the soil–inoculum combinations originating from two native prairies, where five sequence types of a single Gigaspora OTU were virtually the only root colonizers. Our results indicate that indigenous assemblages of AM fungi were adapted to the local soil environment and that this process occurred both at a community scale and at the scale of fungal sequence types within a dominant OTU.
Introduction
Arbuscular mycorrhizas are ancient symbioses between plants and Glomeromycotan fungi that arose simultaneously with the evolution of land plants (Pirozynski & Malloch 1975; Redecker et al. 2000; Brundrett 2002). An estimated 80% of all plant species form these symbioses (Smith & Read 2008), and they are particularly common in grasslands (Miller 1987; Hartnett & Wilson 2002). The Glomeromycota often appear to have little host specificity because the root systems of individual plants are generally colonized by multiple fungal species, and likewise, individual fungal clones may colonize multiple plant species across space and time (Fitter 2005; Rosendahl 2008; Helgason & Fitter 2009). Nevertheless, biogeographical studies suggest that the distribution of Glomeromycota is related to the distribution of plant communities and soil properties (Chaudhary et al. 2008; Murray et al. 2010; Öpik et al. 2010). Within grasslands, the species composition of arbuscular mycorrhizal (AM) fungal communities has been related to precipitation, soil properties, plant community composition and time since disturbance (Johnson et al. 1992; Egerton-Warburton et al. 2007; Fitzsimons et al. 2008). Although the structure of Glomeromycotan communities is often related to plant community properties, there is not always a strong relationship between plant and fungal diversity – highly diverse plant communities may have seemingly depauperate AM fungal communities and vice versa (Johnson & Wedin 1997; Rosendahl & Stukenbrock 2004; Johnson et al. 2010a). This emphasizes the scale dependency of ecological patterns involving AM symbioses (Fitzsimons et al. 2008) and suggests that mycorrhizal diversity needs to be considered at appropriate taxonomic scales. Studies demonstrate that functionally important genetic diversity occurs among isolates of a single species of Glomeromycotan fungi (Munkvold et al. 2004; Koch et al. 2006) and even among nucleotypes within an individual fungal clone (Angelard et al. 2010; Angelard & Sanders 2011). Hence, just as McMillan (1959) demonstrated over 50 years ago that considerable ecotypic diversity exists within species of North American prairie grasses, such variation may also exist for Glomeromycotan fungi. More research is needed to link the functional variation in Glomeromycotan taxa with selection pressures in the environment.
Soil fertility is likely to be a strong selection pressure because mycorrhizas are nutritional symbioses in which photosynthate is traded for soil minerals. The relative availability of essential soil nutrients, particularly phosphorus (P), often determines the degree to which plants may benefit from mycorrhizas; when P and other soil nutrients are in luxury supply, plants have little to gain from AM symbioses (Mosse 1973; Bethlenfalvay et al. 1982). In contrast, AM fungi are obligate biotrophs whose existence requires association with living plants regardless of soil fertility levels. This asymmetry generates a continuum of mycorrhizal functioning from mutualism to parasitism (Johnson et al. 1997). Geographic isolates of Glomeromycota have been shown to contribute more positively to host performance in the environment in which they evolved when they are subjected to water stress (Stahl & Smith 1984; Bethlenfalvay et al. 1989) or nutrient limitation (Johnson et al. 2010b); however, there is a great deal of variation in plant responses to native and exotic fungi (Lambert et al. 1980; Klironomos 2003; Ji et al. 2010). Functional differences among geographic isolates of AM fungi probably occur because of fine-scale local adaptation to both the soil environment and to the plant genotypes in the landscape. Klironomos (2003) showed that extreme responses of mutualism and parasitism were more common in local plant–fungal pairs compared with foreign pairs. This suggests that both positive and negative feedbacks between plants and Glomeromycota may be important forces in structuring communities (Bever 2002; Bever et al. 2002). More studies are needed to determine the mechanisms that generate the range in symbiotic performance among ecotypes of AM fungi and also determine whether there are dominant AM fungi within the community that are responsible for the functional differences among communities.
The tallgrass prairie is one of the most critically endangered ecosystems in North America; more than 99% of the tallgrass prairie once existing east and north of the Missouri River has been destroyed (Samson & Knopf 1994). Of the original Illinois prairie (over 85 000 km2), <1 km2 of high-quality remnant prairie remains (Robertson et al. 1997). The undesired consequences of converting diverse prairies into crop monocultures have inspired efforts to restore tallgrass prairie communities on decommissioned agricultural lands. Native tallgrass prairie plants are generally highly dependent upon AM symbioses (Wilson & Hartnett 1998), and there is evidence that conversion of grasslands to cropland reduces the diversity of Glomeromycota and alters their species composition (Helgason et al. 1998; Fitzsimons et al. 2008; Gao & Guo 2010; Verbruggen et al. 2010). Furthermore, re-introduction of AM fungal communities from native prairie may increase the establishment of native plant species during restoration efforts (Bever et al. 2003). Little is known about whether the geographic origin of introduced fungi is important to the success of prairie restorations, but there is concern that introduction of exotic AM fungal inoculum may have unexpected detrimental effects (Schwartz et al. 2006).
Andropogon gerardii, a long-lived dominant grass of native prairie, may be expected to select for rhizosphere communities of AM fungi, which form symbioses that maximize mutualistic benefits in nutrient-limited soils and minimize parasitism in nutrient-rich soils. This hypothesis was supported in a reciprocal inoculation study that compared the mycorrhizal functioning of native and exotic combinations of plant ecotypes, fungal communities and soils across three prairies, two that were never tilled and one that had been restored from agriculture nearly 30 years ago (Johnson et al. 2010b). Soils of the three sites varied in texture, pH and nutrient availability (Table 1). The 2010 study manipulated whole communities of AM fungi but it did not identify the individual species of fungi involved in the symbioses. This study identifies AM fungi inside the plant roots from that experiment and links the species composition of these intraradical fungal communities with their symbiotic functioning both at a community scale and at the scale of fungal sequence types within a dominant species. Our current study tested the following hypotheses:
| Site | Coordinates | Soil texture | pH | OM (%) | PO4-P (μg/g) | NH4-N (μg/g) | NO3-N (μg/g) | Soil a(N:P)a |
|---|---|---|---|---|---|---|---|---|
| Cedar Creek | 45°24′N | Sandy loam | 5.3 | 1.4 | 46.5 | 5.1 | 4.9 | 0.2 |
| 93°12′W | ||||||||
| Konza | 39°05′N | Silty clay loam | 6.2 | 5.7 | 18.5 | 13.4 | 8.2 | 1.2 |
| 96°18′W | ||||||||
| Fermi | 41°50′N | Silt loam | 7.6 | 3.3 | 11.5 | 7.2 | 5.8 | 1.1 |
| 88°15′W |
- a Soil a(N:P) ratio is the ratio of mineral N (NH4-N+NO3-N) divided by available soil P.
- Soil pH was measured from a 1:1 soil/water paste; soil organic matter was determined by direct combustion; available P was measured using the Bray-1 method; inorganic N was extracted using 2 M KCl and analysed using cadmium reduction/colorimetry. Kansas State University Soil Testing Laboratory (Manhattan, KS, USA) conducted all of the soil analyses.
H1. The community composition of AM fungi in the roots of our experimental A. gerardii plants is influenced by both inoculum source and the background soil environment in which the plant is grown;
H2. Glomeromycotan diversity is lower in the roots of A. gerardii plants grown in restored prairie compared to undisturbed prairie;
H3. Glomeromycotan fungi have the highest fitness and provide the greatest benefit to A. gerardii when they are grown in the soils where they evolved. We expect this pattern to exist at both a community scale and at the scale of geographic sequence types within an individual fungal species.
Materials and methods
Study sites
Soils and AM fungal inocula were collected from three sites: two native prairie remnants, Cedar Creek Ecosystem Science Reserve in Minnesota and Konza Prairie Biological Station in Kansas, and a restored prairie at Fermi National Laboratory in Illinois (Table 1). All available evidence suggests that the prairies at Cedar Creek and Konza have remained untilled since the last glaciation. The Fermi prairie was cultivated and managed using conventional agricultural methods from the late 1840s through 1975; and in 1975, a reconstructed prairie was established using locally collected seeds of prairie plants growing on similar soil types. Soil fertility at the three sites is very different; Cedar Creek soil has the highest available P and lowest available soil nitrogen (N), Konza has the highest available N, and both plant-available N and P are very low in the Fermi soil used in this study (Table 1).
Experimental design
Unlike the full factorial design used in our previous study (Johnson et al. 2010b) which reciprocally crossed soil source, AM fungal inoculum source and Andropogon gerardii ecotype across the three prairies, this study held plant ecotype constant and only investigated soil source and inoculum source effects on the AM fungal communities inside the roots of the Fermi ecotype of A. gerardii. Our previous study showed that although interactions between soil and plant ecotype, and inoculum and plant ecotype influenced the formation of arbuscules and mycorrhizal growth response (MGR), plant ecotype did not have a strong influence on overall plant–fungal interactions. Therefore, only one of the plant ecotypes was examined in this study. We chose the Fermi ecotype because it was more successful in biomass production than the other two ecotypes and thus would provide the most abundant roots for our extensive molecular analysis. In this study, the two-way factorial design resulted in a total of nine treatment combinations (three soil sources by three inoculum sources), each being replicated three times.
Greenhouse methods
Seeds of A. gerardii ecotypes from Fermi were pregerminated, and 14-day-old seedlings were transplanted into square plastic pots (14 cm tall by 11 cm wide), one seedling per pot. Each pot was filled with 1 kg of sterilized soils from the three prairies. Soils were steam pasteurized at 80 °C for 2 h to eliminate biotic communities but retain abiotic soil traits. Communities of AM fungi and other soil organisms were added back in a controlled manner by inoculating pots with 20 g of living soil (including small root fragments) from each of the sites in a reciprocal design. These soils contained diverse communities of AM fungal spores (Table S1, Supporting information) as well as fragments of hyphae and mycorrhizal roots, all of which function as fungal propagules. Inoculum soils were collected fresh from each of the three sites and added directly below the seedling roots during transplantation. Differences in the fertility of the three types of soil inocula were eliminated by adding 20 g of additional sterilized soil from the two sites that were not used in the inoculum treatment. Additional pots were established using only sterile soil to create nonmycorrhizal (NM) controls for each plant and soil combination. Each of the nine treatment combinations was replicated three times for a total of 27 pots. All pots were amended with 60-ml nonsterile soil sievate, 20 mL from each of the three sites. The sievates were prepared by blending soil/water in a 1:2 ratio and passing the slurry through a 38-μm sieve. The relatively large AM fungal spores and hyphae were trapped on the sieve, while smaller organisms pass through, allowing for the addition of the majority of soil microbes while excluding AM fungi.
Plants were grown for 14 weeks with full sunlight during the summer in a glasshouse (20–25 °C) at Kansas State University, Manhattan, Kansas, USA. Plants were watered daily to field capacity and received no fertilizer throughout the study. At harvest, roots were removed from the soil and gently washed. A subset of the roots were stained with trypan blue and scored for AM colonization using the magnified gridline intersect method (McGonigle et al. 1990). This method uses a compound microscope (200–400X) to quantify cortical root length colonized by fungal structures (hyphae, vesicles, arbuscules and coils). An additional subset of roots were placed in plastic bags and stored at −20 °C for subsequent molecular analyses. The remaining roots were used to measure total plant dry weight and correct for the fresh root subsamples that were analysed for mycorrhizas. External AM hyphal lengths were determined by extracting the hyphae from 5 g of soil, followed by collecting the hyphae on a membrane filter, staining with trypan blue and quantifying hyphal length by the gridline intercept method (Miller et al. 1995). Total oven-dried plant weight was measured and the MGR of plants was calculated as ln (AM/NM) where AM is the total dry weight of mycorrhizal plants and NM is the mean total dry weight of the plants receiving no live inoculum and grown in the same background soil as the AM plants. Positive MGR values indicate mutualistic relationships, and negative values indicate parasitic relationships. Shoot tissue N and P concentrations were determined after wet digestion with sulphuric acid and hydrogen peroxide of dried plant material was ground through a 20-μm mesh. P concentrations were determined on the digest using the molybdate-blue method (Murphy & Riley 1962), and nitrogen concentrations were determined by a Kjeldahl method with a rapid-flow autoanalyser. All chemical analyses were conducted at the Kansas State University Soil Testing Laboratory (Manhattan, KS, USA).
Molecular methods
The AM fungal communities colonizing the roots of A. gerardii were determined through PCR amplification and sequencing of a portion of the small subunit ribosomal gene (SSU rDNA). A 25 mg (fresh weight) sample of the fine roots of each plant was obtained by pooling small pieces of roots (2–3 cm) from throughout the root system. Roots were cleaned using distilled water, and DNA was extracted from them using the UltraClean Soil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, California, USA) following the manufacturer's instructions. Roots of the NM plants inoculated with sterile soil were not colonized by AM fungi (data not shown) and were thus not included in our DNA analyses.
Partial SSU rDNA was amplified using the nested PCR protocol and primers developed by Lee et al. (2008). These primers (AML1 and AML2) were chosen because they have better specificity and coverage of AM fungi than the more widely used NS31-AM1 primer pair (Helgason et al. 1998), but they amplify a region that overlaps with that amplified by NS31-AM1, thereby allowing us to utilize the large number of sequences currently found in the GenBank database for identification. Despite the improved selectivity of these primers, our initial testing routinely revealed non-AM fungal amplicons in PCR products, and these contaminant sequences were not specific to any particular soil–inoculum treatment group. To reduce the amplification of non-AM fungal DNA, we searched for a restriction enzyme that cuts the AML1-AML2 region of common contaminants found in our samples but not of AM fungi. Of hundreds of commercially available enzymes tested, BsmI was selected because it was able to effectively eliminate most of the fungal contaminants. Our intensive testing found that no AM fungal sequence available in GenBank was cut by this enzyme.
A 10-fold dilution of extracted DNA was first amplified using the general eukaryotic primers NS1 and NS4 (White et al. 1990). The PCR product was then digested with the restriction enzyme BsmI. The second-step PCR was performed using a 10-fold dilution of the digest and the primers AML1 and AML2. Second-step PCR products were visualized on an agarose gel, and bands of ~800 bp, indicating amplification by AML1 and AML2, were cut out and purified using a QIAquick gel extraction kit (QIAGEN, Valencia, CA, USA) following the manufacturer's protocol. Purified DNA was sequenced using an ABI 3130 DNA analyzer to verify the presence of AM fungi prior to further cloning and sequencing. This step was included because of problems with amplification of non-AM fungi from some root samples. Purified DNA was cloned into the pDrive cloning vector and transformed into QIAGEN EZ competent cells using QIAGEN PCR cloning kits (QIAGEN, Valencia, CA, USA). The DNA of a minimum of ten positive transformants per sample was sequenced using sequencing primers SP6 and T7 on an ABI 3730 DNA analyzer. The resulting forward and reverse DNA sequences were assembled in BioEdit version 7.0.5.3 to create a consensus sequence for data analysis (Hall 2011).
Sequence analysis
To identify the clone DNA sequences, we compared them with the GenBank database using a blastn search on the NCBI website (http://www.ncbi.nlm.nih.gov/). Per cent query coverage, per cent maximum identity and bit score data were used to identify the closest match of our sequences to those in GenBank. Only sequences that were unambiguously identified as AM fungi were included in the subsequent analysis.
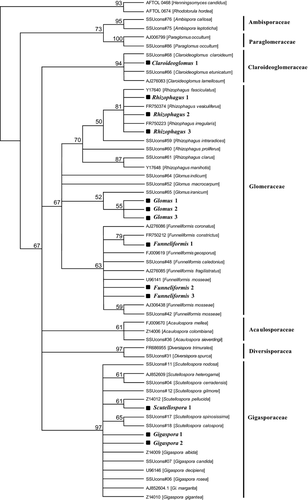
To determine operational taxonomic units (OTUs), we pooled all individual AM fungal sequences together and performed a standard OTU analysis using the software Mothur (Schloss et al. 2009) and a 97% similarity to group sequences into OTUs (Helgason et al. 1998). We then selected one sequence to represent each OTU and aligned these representative sequences together with reference sequences of all major Glomeromycota clades using the ClustalX algorithm in BioEdit. All reference sequences were chosen from Krüger et al. (2012) with one exception. The sequences representing Gigaspora margarita were much shorter than other sequences and therefore were replaced by more appropriate sequences from GenBank (accession numbers AJ852603.1 and AJ852604.1). Maximum-likelihood (ML) phylogenetic analysis was computed in MEGA version 5 (Tamura et al. 2011) with 1000 bootstrap replicates to evaluate support of the tree and with Rhodotorula hordea and Henningsomyces candidus as outgroups to root the tree. We chose R. hordea and H. candidus as outgroups because they were used by Krüger et al. (2012) for the same purpose. Parameters for the ML analysis were estimated using the jModelTest program (Posada 2008). The best model inferred for our data was GTR+I+G. Branches corresponding to partitions reproduced in <50% bootstrap replicates were collapsed. Representative sequences for each OTU used in the phylogenetic analysis were deposited in GenBank under accession numbers JQ837420, JQ837422, JQ837424, JQ837428, JQ837429, JQ837454 and JX970622-JX970628.
For sequences identified as the OTU Gigaspora 1, our initial phylogenetic analysis discovered greater genetic diversity than grouping at 97% similarity could reveal, so we conducted an additional analysis on these sequences focusing on a 94-bp hypervariable region of the SSU rDNA, located between 274 and 368 bp from the 5′ end of the primer AML1. Hypervariable regions of the SSU have been used to distinguish among members of the Glomeromycota in the past although these studies emphasized regions outside of the area amplified by the AML1 and AML2 primers that we used (Bago et al. 1998; de Souza et al. 2004). We constructed a maximum-likelihood phylogeny with the model GTR+I+G calculated as described above. The phylogenetic tree was created using 103 sequences of Gigaspora 1 and five additional members of the genus Gigaspora. The out-group was composed of Scutellospora spinosissima, Scutellospora gilmorei and Racocetra fulgida.
Statistical analysis
Two-way analysis of variance (anova) was performed to test for differences in plant and fungal responses and OTU richness for each soil–inoculum combination, followed by Tukey's HSD tests (P < 0.05) when significant differences were observed. These analyses were performed using JMP 9.0.2 (SAS Institute Inc. Cary, NC, USA). To determine whether sampling intensity was sufficient, species richness was estimated using a first-order jackknife procedure in PC-ORD 5.10 (McCune & Mefford 2006), a measure that accounts for species that occur in only one sampling unit. In addition, an experiment-wide species accumulation curve was generated using the same program. Data on the community composition of AM fungi in the roots of A. gerardii were visualized using a nonmetric multidimensional scaling ordination with a Bray–Curtis distance measure in PC-ORD 5.10 (McCune & Mefford 2006). The influence of soil source and inoculum source on AM fungal community composition was tested with a permutation-based nonparametric multivariate analysis of variance (Permanova) using relative abundance data, also in PC-ORD. The main effects of soil source and inoculum source were analysed together as a two-way factorial design, followed by pairwise tests of soil or inoculum combinations when the main effects were significant (P < 0.05). If significant differences were detected, an indicator species analysis was conducted in PC-ORD to determine whether particular species made a greater contribution to the community differences. Qualitatively, similar patterns were observed with presence/absence data, but only the relative abundance data are presented. To determine whether sequence types of Gigaspora 1 differed significantly in A. gerardii roots inoculated with Cedar Creek versus Konza soil, a multiresponse permutation procedure (MRPP) was conducted using relative abundance data in PC-ORD, followed by an indicator species analysis. The ‘A’ statistic generated by the MRPP is a descriptor of within-group similarity compared to random expectation. An A value >0.1 is a strong indicator of a difference among groups (McCune & Grace 2002).
Results
All 27 Andropogon gerardii root samples yielded positive PCR product of the expected size (c. 800 bp) after the second-step PCR. A total of 282 clones were sequenced, and 231 of them were identified as AM fungi. Our phylogenetic analysis of these AM fungal sequences revealed a total of 13 OTUs (Fig. 1) from three families of AM fungi, the Gigasporaceae, the Glomeraceae and the Claroideoglomeraceae. Approximately 63% of the root samples contained a single OTU, 26% contained two OTUs, while the remaining samples (11%) contained three OTUs. An experiment-wide species accumulation curve (Fig. S1, Supporting information) and the first-order jackknife estimates of OTU richness (15.6) suggested that our sampling captured most of the community. Ten of the identified OTUs were affiliated with genera within the families Claroideoglomeraceae: Claroideoglomus (one OTU) or Glomeraceae: Funneliformis (three OTUs), Glomus (three OTUs) and Rhizophagus (three OTUs). The other three OTUs belonged to the family Gigasporaceae: Gigaspora (two OTUs) and Scutellospora (one OTU) (Fig. 1). Overall, Gigaspora was the most abundant AM fungal genus in this study, with 57% of clones identified as Gigaspora. The AM fungal spore community in the soil from each site was more species rich than the AM fungal community observed in A. gerardi roots inoculated with that soil. All of the genera observed in roots were also observed as spores, but eight genera were observed as spores but not in roots (Acaulospora, Ambispora, Archaeospora, Diversispora, Entrophospora, Paraglomus, Racocetra and Sclerocystis) (Table S1, Supporting information).
The richness of OTUs per A. gerardii plant was significantly influenced by both soil origin (F = 3.85, P = 0.040) and inoculum source (F = 4.65, P = 0.023), but there was no significant interaction (F = 2.00, P = 0.138). The AM fungal communities of A. gerardii were more species rich when grown in Fermi soil [mean (SE) = 1.89 (0.18)] or with Fermi inoculum [mean (SE) = 1.89 (0.18)] than when grown with Cedar Creek soil [mean (SE) = 1.22 (0.18)] or inoculum [mean (SE) = 1.11 (0.18)]. Values for Konza soil [mean (SE) = 1.33 (0.17)] or inoculum [mean (SE) = 1.44 (0.18)] were intermediate and not significantly different from either of the other prairies.
The community composition of AM fungi inside A. gerardii roots was significantly influenced by both inoculum source (F = 17.821, P = 0.0002) and the soil type in which plants were grown (F = 4.631, P = 0.003) (Fig. 2). There was also a significant interaction between soil type and inoculum source (F = 4.513, P = 0.0002). Fermi soil was driving the differences in community composition (Fermi vs. Cedar Creek, t = 2.769 P = 0.0004; Fermi vs. Konza, t = 1.901, P = 0.022; Cedar Creek vs. Konza, t = 1.249, P = 0.203). This can be seen in Fig. 2 with the communities generated from Fermi soil forming a line in the centre of the figure away from the other communities. There was no significant indicator OTU for soil source.
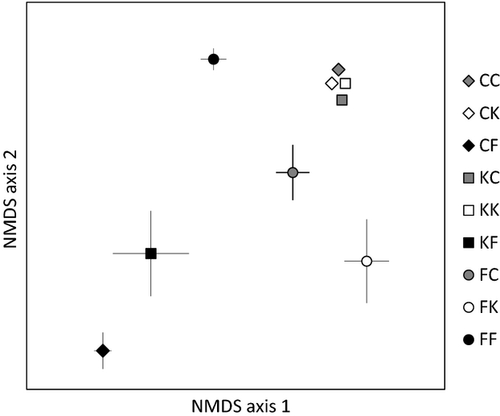
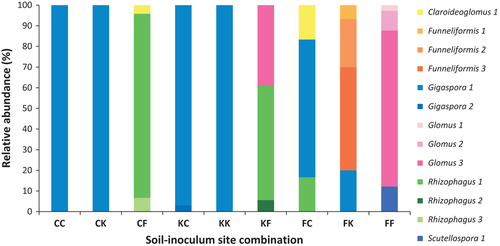
The three inoculum sources resulted in statistically distinct AM fungal communities on A. gerardii (Fermi vs. Cedar Creek, t = 4.819, P = 0.0002; Fermi vs. Konza, t = 5.105, P = 0.0002; Cedar Creek vs. Konza, t = 1.526, P = 0.1230; Fig. 2). Glomeromycotan communities in plants inoculated with AM fungi from Fermi were dominated by members of the Glomeraceae, except for small amounts of Scutellospora and Claroideoglomus. Glomus 3 [indicator value (IV) = 55.6, P = 0.0042] and Rhizophagus 1 (IV = 43.1, P = .0228) were significant indicators of Fermi inoculum. In contrast, soils inoculated with AM fungi from Konza or Cedar Creek were dominated by members of the genus Gigaspora, particularly Gigaspora 1, except for the Fermi soil–Konza inoculum combination where Gigaspora 1 was present, but members of the genus Funneliformis dominated (Fig. 3).
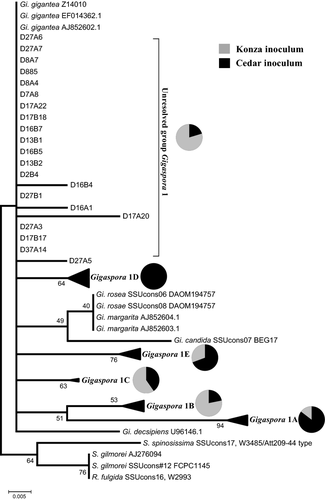
Plants inoculated with Cedar Creek and Konza inocula and grown in Cedar Creek and Konza soil were colonized almost entirely by Gigaspora 1, but the phylogenetic composition of this OTU differed with inoculum source (Fig. 4). Although the genus Gigaspora had strong phylogenetic support (bootstrap value of 70), the internal resolution at the species level was conflictive as some described species could not be distinguished (Fig. 4). This analysis grouped most (>80%) Gigaspora 1 sequences into five sequence types, with bootstrap values between 53 and 94, although some sequences were not well resolved (see Fig. S2, Supporting information for a phylogeny including all sequences and Fig. 4 for a phylogeny with sequences of each of the five sequence types compressed into a single node). The relative abundance of the five sequence types of Gigaspora 1 varied significantly among A. gerardii inoculated with Cedar Creek versus Konza AM fungi [(A) = 0.1198, P = 0.029], with Gigaspora 1B a significant indicator of Konza inoculum (IV = 55.3, P = 0.0480) and Gigaspora 1D an indicator of Cedar Creek inoculum (IV = 87.9, P = 0.002) (Fig. 4).
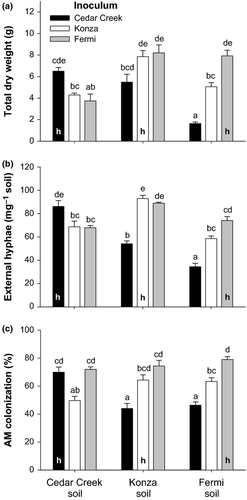
Both plants and AM fungi responded significantly to the soil–inoculum combinations (plant total dry weight, F = 20.578, P < 0.0001; external hyphae, F = 32.404, P < 0.0001; AM colonization, F = 13.582, P < 0.0001). With the exception of the Konza soil and Fermi inoculum combination, plants tended to grow largest and AM fungi produced the most external hyphae in the ‘home’ soil–inoculum combinations (indicated by ‘h’ in Fig. 5a, b). The Glomeraceae-dominated Fermi inoculum produced as much fungal colonization and external hyphae and was equally beneficial to plants grown in Konza soil as the Gigaspora-dominated ‘home’ inoculum from Konza. Even though root colonization was uniformly high with Fermi inoculum, it was not significantly higher than the home soil–inoculum combinations in the other soil types (Fig. 5c).
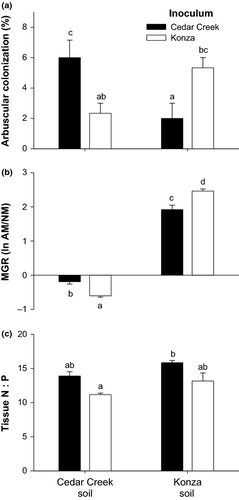
We were able to compare the symbiotic functioning of sequence types of Gigaspora 1 because this OTU was the only taxon identified from roots grown with inoculum from Cedar Creek and Konza, the two native prairie sites. As with the full-scale analysis involving multiple genera of Glomeromycota, this analysis of Gigaspora 1 isolates indicates that the home soil–inoculum combination generates the most efficient symbioses. Home soil–inoculum combinations produced the most arbuscules, the site of symbiotic exchange between plants and fungi (Fig. 6a). Furthermore, the MGR of plants was significantly higher in the home combination (Fig. 6b). Shoot tissue N:P ratios tended to be greater in plants colonized by Cedar Creek Gigaspora (Fig. 6c).
Discussion
In support of our first hypothesis, we discovered that the composition of Glomeromycotan communities inside our experimental Andropogon gerardii roots is determined by both the source of the fungal inoculum and the soil environment in which the plant was grown (Fig. 2). It is not surprising that the community composition of AM fungi differs among three geographically separated prairies with different soil properties and climatic conditions. The important insight from this reciprocal inoculation experiment is that the same fungal inoculum generated three different communities inside A. gerardii roots grown in three soils with very different properties (Table 1, Fig. 2). Uibopuu et al. (2009) found that the same fungal inoculum developed different intraradical AM fungal communities when grown with different plant species. These findings support the hypothesis that plants are not passive participants in the colonization process, but rather, they select AM fungal partners in response to different soil conditions and plant requirements (Reinhardt 2007). Alternatively, fungal taxa with the highest growth rates in the environment may come to dominate roots. It is likely that both plant and fungal factors control the community composition of AM fungi that we observed in the roots of A. gerardii grown for 3 months in our greenhouse experiment.
Our finding of 13 OTUs from just over 230 clones isolated from a single plant species compares well with the six studies reviewed by Dumbrell et al. (2010a) that examined 150–400 clones from one to two species of plants and observed an average OTU richness of 14. Likewise, at the Konza site, six to nine Glomeromycota OTUs were observed per sampling period in field collected roots of A. gerardii sampled four times during the year (Jumpponen 2011). Our species accumulation curve was levelling (Fig. S1, Supporting information), and our jackknife estimates of species richness were close to our observed values. Nevertheless, it is possible that we missed some OTUs in our analysis due to problems intrinsic to all PCR-based molecular techniques and limitations specific to our PCR primers. PCR is known to bias towards certain template DNA (Polz & Cavanaugh 1998), and as a result, taxa with low DNA abundance in roots may be completely missing from PCR products. Roots grown with Cedar Creek and Konza inocula contained some intraradical vesicles (data not shown), a mycorrhizal structure not produced by Gigasporaceae but commonly formed by other Glomeromycota (Gerdemann & Trappe 1974). This indicates that AM fungi other than Gigaspora were present in these samples even though our molecular analysis only detected a single Gigaspora OTU. Although the primers we used amplify most genera of AM fungi, they do not amplify the genus Archaeospora (Lee et al. 2008), which was present in the spore communities of all three inoculum soils (Table S1, Supporting information). Studies that analyse the composition of both AM fungal spores in soil and the fungi inside roots grown in that soil regularly show that the two communities are different (e.g. Clapp et al. 1995; Liu et al. 2012). There are many possible reasons for a lower diversity of fungi inside plant roots compared to the diversity of spores in field soil, particularly in greenhouse studies. Sýkorová et al. (2007) compared the AM fungal community composition between field and greenhouse conditions and found short-term greenhouse cultivation (3 months) favoured a subset of mycorrhizal taxa that were rapid colonizers that could competitively exclude other fungi. Greenhouse conditions may have introduced a similar bias in our study. Also, it is well known that there is selective association between certain plant and AM fungal species (e.g. Johnson et al. 1992; Eom et al. 2000; Uibopuu et al. 2009; Murray et al. 2010), and this would generate root communities that are a subset of the spore community presents at a site.
Our second hypothesis that Glomeromycotan diversity should be lowest in the restored prairie was not supported by our results. In fact, we observed the opposite pattern (Fig. 3). Fermi inoculum resulted in significantly higher species richness including three genera within the Glomeraceae and one genus within the Claroideoglomeracae. In contrast, Konza and Cedar Creek inocula were dominated by a single OTU of Gigaspora. Overdominance by one taxon is not uncommon in Glomeromycotan communities (Dumbrell et al. 2010a). Data synthesized from over 30 studies showed that, on average, a single taxon represented 40% of the abundance of AM fungal communities (Dumbrell et al. 2010a).
Two factors may explain the pattern of Gigaspora dominance in A. gerardii grown in association with Cedar Creek and Konza inocula. First, it is possible that Gigaspora has come to dominate the native prairies at Konza and Cedar Creek over time, but that this genus is not yet well established in the earlier successional restored prairie at Fermi. Members of the Gigasporaceae have extremely large spores that may be slower to recolonize following disturbance compared to members of the Glomeraceae with smaller spores. Relative to members of the Glomeraceae, AM fungi in the Gigasporaceae lack the ability to form extensive hyphal networks (de la Providencia et al. 2005; Voets et al. 2006), rely more on soilborne spores to regenerate and require more plant resources to establish and grow (Klironomos & Hart 2002; Thonar et al. 2011). The reduced abundance of Gigasporaceae at Fermi could therefore suggest that members of this family are more vulnerable to soil disturbance than other families of AM fungi and may need a longer time to recover.
An alternative explanation for the dominance of Gigaspora in A. gerardii grown in soil from Konza and Cedar Creek is that the neutral soil pH at Fermi is more favourable for the Glomeraceae, while the more acidic soil at Konza and Cedar Creek is better for Gigasporaceae. It is well known that soil pH is an important determinant of microbial community structure (Lauber et al. 2009), including AM fungal community composition (Dumbrell et al. 2010b; Lekberg et al. 2011). For example, across a pH range similar to that of our study sites, differences in pH accounted for more than 40% of the variation in AM fungal community composition in a combined habitat of grassland, woodland and heathland (Dumbrell et al. 2010b). An early field survey by Porter et al. (1987) suggests that Gigaspora species might be more adapted to low soil pH than high pH, which may help explain why members of the Gigasporaceae were dominant at Konza and Cedar Creek. However, our findings do not suggest that the higher pH of Fermi soil prevents colonization by Gigaspora because A. gerardii formed associations with Gigaspora when grown in sterilized Fermi soil and inoculated with soil organisms from Konza or Cedar Creek suggesting that Gigaspora is less able to competitively exclude Glomeraceae in Fermi soil over the short term.
Our phylogenetic analyses employing a hypervariable region of the widely used SSU region of the rRNA gene revealed distinct sequence types within a single Gigaspora OTU, Gigaspora 1. Based on the trees we constructed (Fig. 4), the majority of the Gigaspora 1 sequences we obtained were grouped into one of five sequence types that were distinct from one another and from the OTUs used by Krüger et al. (2012) in their recent phylogenetic analysis of the Glomeromycota. While Krüger et al. (2012) found strong support for a monophyletic origin of the Gigaspora using a 2700-bp data set including multiple regions of rDNA, individual species of Gigaspora were poorly resolved with just the SSU (Krüger et al. 2012). Likewise, de Souza et al. (2004) found that a hypervariable region of the SSU could distinguish among species of Gigaspora, but the most informative hypervariable region (V9) was found outside the primers we used. Therefore, it is unclear whether the five sequence types we identified are different species of Gigaspora or different genotypes of a single Gigaspora species. Sequencing another hypervariable region such as V9 (Bago et al. 1998) or using alternative molecular methods such as the PCR-DGGE approach (de Souza et al. 2004) might help resolve our Gigaspora 1 sequence types. Members of the Gigaspora produce large, multinucleate spores that can be genetically distinct (Zézé et al. 1997), adding to the challenges of understanding the relationships between Gigaspora OTUs and sequence types.
Our third hypothesis that Glomeromycotan fungi have the highest fitness and provide the greatest benefit to A. gerardii when they are grown in the soils where they evolved was partially supported. The differences in the composition of fungal communities from the different sites corresponded with significant differences in AM colonization, external hyphae and plant biomass, and for the most part, matched pairs of soil and inoculum performed the best (Fig. 5). This exciting result supports the hypothesis that local adaptation is an important force structuring Glomeromycotan communities (Dumbrell et al. 2010b; Ji et al. 2010; Johnson et al. 2010b, 2012). However, there was one exception that is noteworthy; plants grown in P-limited Konza soil and inoculated with soil organisms from the restored prairie at Fermi had a similar total biomass to plants grown in the home soil–inoculum combination (Fig. 5). Hyphal lengths also did not differ between the two groups. These results suggest that the Gigasporaceae-dominated Konza community and the Glomeraceae-dominated Fermi community had similar mycorrhizal benefits and costs in Konza soil. This result is surprising given previous work suggesting that compared to Glomeraceae, members of the Gigasporaceae tend to produce more mycelium in soil, a presumed advantage in low P soils (Hart & Reader 2002; Thonar et al. 2011).
Comparisons of the performance and symbiotic function of the Gigaspora in the Konza and Cedar Creek inocula revealed a tremendous degree of intra-OTU variation and strongly support the hypothesis that AM fungi have the highest fitness and provide the greatest benefit to plants in the soils where they evolved. In Cedar Creek soil, the local sequence types of Gigaspora 1 formed significantly more external hyphae (Fig. 5b) and arbuscules (Fig. 6a) than Konza sequence types of Gigaspora 1; the reverse was equally true – Konza Gigaspora 1 sequence types performed best in Konza soil. Perhaps most importantly, in the P-rich Cedar Creek soil, local Gigaspora 1 sequence types generated a commensal symbiosis, while Konza Gigaspora 1 sequence types generated a parasitic symbiosis (Fig. 6b). Both sequence types generated mutualistic symbioses in Konza soil, but the benefits were significantly greater when roots were colonized with fungi that evolved at Konza.
The properties of Cedar Creek and Konza soils are extremely different (Table 1), and it is likely that N limitation at Cedar Creek and P limitation at Konza are important selection pressures on plants and Glomeromycota. Fungal OTUs that best facilitate plant acquisition of these limited resources should be favoured because Glomeromycota are obligate biotrophs so that fungal fitness is tightly linked to plant carbon allocation, and it is known that plants preferentially allocate carbon to fungi that provide them the greatest benefit (Bever et al. 2009; Hammer et al. 2011; Kiers et al. 2011). If preferential allocation occurs in our system, the selection of different geographic sequence types in Cedar Creek and Konza soils can be directly linked to the difference in plant nutritional demands. In both soil types, tissue N:P ratio tends to be higher in plants inoculated with Cedar Creek soil organisms (Fig. 6c), indicating that Cedar Creek inoculum enhances N uptake, while Konza inoculum enhances P uptake. The statistical significance of these comparisons would likely increase if the sample size was larger (N = 3).
A uniform microbial wash composed of non-AM microorganisms from all three sites was added to each pot in an attempt to create a common community of bacteria and non-AM fungi across all treatments. Nevertheless, it is possible that differences in the microbial communities remained, and consequently, some of the plant responses to the different inocula may have been generated by soil organisms other than AM fungi. Although we cannot rule out this possibility, it is unlikely that the observed patterns in plant growth are independent of the AM symbioses in their roots because of the close correspondence between plant dry weight and the formation of internal and external structures of AM fungi (Fig. 5).
In conclusion, our study supports the idea that Glomeromycota are ‘more diverse than meets the eye’ (Bever et al. 2001) and suggests that ecological studies should focus on variation within geographic isolates of fungi. Although Glomeromycota are asexual, their unusual genetic structure maintains a high degree of genetic diversity within individual fungal clones because of diverse populations of nuclei within heterokaryotic, coenocytic mycelium (Sanders 2002; Reinhardt 2007; Rosendahl 2008). These fungi exhibit high rates of segregation and recombination, which generate functionally distinct nucleotypes that have the potential for rapid adaptation to the environment (Angelard et al. 2010; Angelard & Sanders 2011). Consequently, intraspecific diversity of Glomeromycota may be very important (Johnson et al. 2012), and thus, application of non-native AM fungi in commercial inoculants may not always have the expected outcomes because of a mismatch between the ambient soil conditions and the edaphic adaptations within the inoculant fungi (Schwartz et al. 2006).
Acknowledgements
This work was funded by the National Science Foundation (DEB-03116136, DEB 0842327 and DEB 0816675) and the Fulbright Commission of the Czech Republic. RMM's participation was in part funded by the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. We thank Jacqueline Wilson for her help with the greenhouse study, Carlyn van Camp and Carolyn Myren for their assistance with the molecular analysis and Anita Antoninka for analysing the spore communities. Five anonymous reviewers and the laboratories of C. Gehring, N. Johnson and J. Bever provided valuable comments on the manuscript.
References
N.C.J., C.A.G. and B.J. initiated this research project. N.C.J., G.W.T.W. and R.M.M. designed and conducted the greenhouse experiment, BJ designed and conducted the molecular analysis with assistance from L.R.-F. and C.A.G., L.R.-F. and B.J. conducted the phylogenetic analyses, and C.A.G, N.C.J and B.J. conducted the statistical analyses. N.C.J., C.A.G., L.R.-F. and B.J. drafted the manuscript with editorial input from G.W.T.W. and R.M.M.
Data accessibility
DNA sequences: GenBank accession numbers JQ837420, JQ837422, JQ837424, JQ837428, JQ837429, JQ837454 and JX970622-JX970628.
Phylogenetic data: TreeBASE study accession number: #13814.
Full sequence data, sequence description file, plant and fungal trait measurements: Dryad entry doi:10.5061/dryad.k4m41.




