Microevolution in time and space: SNP analysis of historical DNA reveals dynamic signatures of selection in Atlantic cod
Abstract
Little is known about how quickly natural populations adapt to changes in their environment and how temporal and spatial variation in selection pressures interact to shape patterns of genetic diversity. We here address these issues with a series of genome scans in four overfished populations of Atlantic cod (Gadus morhua) studied over an 80-year period. Screening of >1000 gene-associated single-nucleotide polymorphisms (SNPs) identified 77 loci that showed highly elevated levels of differentiation, likely as an effect of directional selection, in either time, space or both. Exploratory analysis suggested that temporal allele frequency shifts at certain loci may correlate with local temperature variation and with life history changes suggested to be fisheries induced. Interestingly, however, largely nonoverlapping sets of loci were temporal outliers in the different populations and outliers from the 1928 to 1960 period showed almost complete stability during later decades. The contrasting microevolutionary trajectories among populations resulted in sequential shifts in spatial outliers, with no locus maintaining elevated spatial differentiation throughout the study period. Simulations of migration coupled with observations of temporally stable spatial structure at neutral loci suggest that population replacement or gene flow alone could not explain all the observed allele frequency variation. Thus, the genetic changes are likely to at least partly be driven by highly dynamic temporally and spatially varying selection. These findings have important implications for our understanding of local adaptation and evolutionary potential in high gene flow organisms and underscore the need to carefully consider all dimensions of biocomplexity for evolutionarily sustainable management.
Introduction
In face of accelerated rates of climate change and other growing anthropogenic pressure, it is important to get a better understanding of how quickly natural populations can adapt to altered conditions. The literature contains many examples of rapid evolution in wild populations over contemporary timescales (e.g. Kinnison & Hendry 2001; Palumbi 2001; Stockwell et al. 2003). However, it is still unclear how widespread such short-term adaptive changes are and under what conditions they occur at rates fast enough to track environmental and human-induced changes (Hendry et al. 2008; Hoffmann & Sgrò 2011). Progress in elucidating these important questions has been hampered by the notorious difficulty in demonstrating a genetic basis for apparent local adaptations in natural populations (Gienapp et al. 2008; Hansen et al. 2012). There are multiple strategies for disentangling the effects of phenotypic plasticity from genetic differences (recently reviewed by Hoffmann & Sgrò 2011; Hansen et al. 2012). Most approaches involve either laboratory experiments such as common garden setups or quantitative genetic analysis that requires knowledge of family relationships—both undertakings that can be logistically prohibitive with large, long-lived and highly abundant organisms. For such systems, molecular genetic methods often offer more accessible opportunities for directly observing the underlying genomic signature of selection and adaptive divergence (Nielsen 2005; Storz 2005). Yet, because patterns of genetic diversity integrate effects accumulated over millennia, it remains challenging to distinguish historical selection predating colonization of current habitats from ongoing selection. Hence, snapshot observations of the current distribution of genetic variation often tell us little about how stable these patterns are over time or how quickly they may change in response to human activities.
Temporally spaced DNA samples offer a unique opportunity for studying genetic change directly. By comparing the genetic composition of a population before and after a change in environmental conditions, it is possible to track changes in allele frequencies for retrospective ‘real time’ assessment of genetic impacts. Previously, retrospective studies using presumably neutral markers have offered important insights about demographic processes including estimates of effective population sizes, loss of diversity and stability of population structure and migration rates [see reviews by Wandeler et al. (2007); Leonard (2008); and Nielsen & Hansen (2008)]. Also, studies targeting specific candidate genes expected to be under selection have begun to elucidate the temporal dynamics of adaptive variation (e.g. Umina et al. 2005; Marsden et al. 2012).
Now, with advances in molecular techniques, efforts to study temporal adaptive genetic variation are no longer limited to genes a priori expected to be under selection. While neutral evolutionary forces such as drift and migration are expected to leave genome-wide signatures, selection is expected to act only on specific loci and closely linked genomic regions (Cavalli-Sforza 1966; Lewontin & Krakauer 1973). Therefore, comparisons of locus-specific levels of differentiation among large panels of genetic markers potentially allow for disentangling the effects of neutral processes from the effects of selection. Such ‘genome scan’ approaches are often applied to identify loci affected by selection in space (Storz 2005; Stinchcombe & Hoekstra 2007), but have only in a few cases been utilized to identify signatures of selection and ongoing adaptation over time in wild populations [notable examples include Hansen et al. (2010); Bourret et al. (2011); and Orsini et al. 2012)], most often due to technical constraints and limited sample availability. Yet, where such challenges can be overcome, simultaneous assessment of both temporal and spatial scales over which different evolutionary forces are acting offers extraordinary prospects for gaining more comprehensive insights about the potential for rapid adaptation.
Like many marine fish species, the Atlantic cod (Gadus morhua) is characterized by high dispersal ability and a wide distribution with few obvious barriers to migration. Previously, local adaptation was expected to be rare or absent for such species as the homogenizing effects of presumed high levels of gene flow would swamp the diversifying effects of local selection. However, recent studies based both on genomic signatures of selection on specific loci (e.g. Nielsen et al. 2009; Bradbury et al. 2010) and on common garden experiments (e.g. Marcil et al. 2006; Grabowski et al. 2009) have provided strong evidence in support of adaptive divergence in cod. Signatures of divergent selection have been observed even over surprisingly small spatial scales where neutral genetic markers have typically revealed very limited levels of population structure (Hutchings et al. 2007; Poulsen et al. 2011).
Recent research has also indicated that cod may possess very high potential for rapid adaptation in response to human impacts. Being one of the historically most important commercial fish species in the North Atlantic, it has been subjected to substantial fishing pressure throughout its range. Theory and modelling work predict that the selection and high mortality imposed by such exploitation can cause large and rapid adaptive changes in the targeted populations (e.g. Ernande et al. 2004; Law 2007). Time series of phenotypic data for many cod populations do indeed demonstrate marked changes in life history traits such as growth and timing of maturation over recent decades (e.g. Trippel 1995; Olsen et al. 2005; Swain et al. 2007). Statistical analysis has indicated that these changes represent an evolutionary response to fishing (reviewed by Jørgensen et al. 2007), although the degree to which such results reflect genetic as opposed to environmentally induced effects remains somewhat controversial (e.g. Kuparinen & Merilä 2007; Hilborn & Minte-Vera 2008). Further, as pointed out by Andersen & Brander (2009), the geographical variation in the affected traits among different populations is often as large as the observed changes over time within single areas (see e.g. Olsen et al. 2004, 2005). This overlap between spatial and temporal patterns of variation raises questions about the role of distributional shifts or altered migration patterns (as opposed to local fisheries selection on stationary populations) as a cause of the observed temporal trait changes. At the same time, the recent trait changes within single populations may impact the stability of apparent signatures of spatially varying local adaptation in this high gene flow species.
Capitalizing on recently developed genomic resources and invaluable archived specimen collections (see Nielsen & Bekkevold 2012), we here address these issues with the—to our knowledge—most extensive spatiotemporal genome scan study on wild populations to date. We focus on a complex of Canadian cod populations that over recent decades have suffered major collapses due to overexploitation and experienced large ecosystem changes in their habitats (Morissette et al. 2009). By screening temporal and spatial variation in allele frequencies at up to >1000 gene-associated single-nucleotide polymorphisms (SNPs), some of which appear to be under selection over larger spatial scales (Nielsen et al. 2009; Bradbury et al. 2010), we search for loci that show elevated levels of differentiation, indicative of selection, over the past 80 years. Specifically, we ask (i) whether there are any genomic signatures of selection over time within individual populations, (ii) whether we observe parallel temporal patterns across space and (iii) whether patterns of spatial divergence appear stable over time. We also explore whether temporal allele frequency shifts correlate with life history changes or fluctuations in potential drivers of selection. Overall, our study illustrates the advantages of conducting simultaneous spatial and temporal analysis for revealing the genetic basis of microevolutionary change.
Materials and methods
The study populations and samples
The study was centred on an 80-year time series of samples from a cod population in the southern Gulf of St Lawrence (management division 4T), Canada, but to assess relationships between temporal and spatial patterns of variation, it also included samples from three nearby management areas (divisions 3NO, 3Ps, 4VsW; Fig. 1). The cod populations in these four areas have exhibited variable demographic trends over the years, but none of them have fully recovered from the severe overexploitation that led to major collapses in virtually all Canadian cod populations in the 1990s (Hutchings & Reynolds 2004). Exemplifying the pattern of apparent adaptive divergence in both time and space described above, common garden experiments have indicated clear, genetically based functional differences among these particular populations for a number of traits (Marcil et al. 2006; Hutchings et al. 2007), and more or less parallel reductions in growth rate and/or age and size at maturation have been observed within all of them over recent decades. These temporal changes potentially reflect fisheries-induced evolution (Hutchings 2005; Olsen et al. 2005; Swain et al. 2007; Swain 2011), but over the same time period, the populations have also been exposed to fluctuations in a number of other possible drivers of selection, for example temperature (Swain et al. 2007).
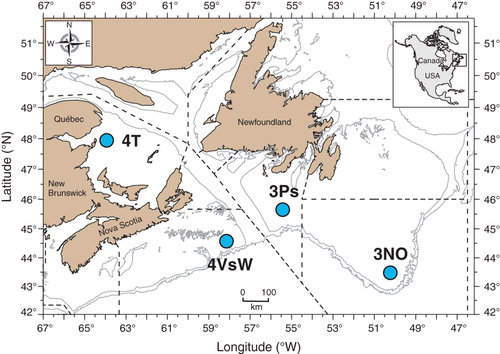
Contemporary samples of gill tissue were collected from all populations on research cruises during 2008–2010. Historical samples consisted of archived otoliths that had been stored individually in paper envelopes at room temperature since collection. The oldest available set of otoliths was from 1928 and was obtained from DTU Aqua, Denmark [previously analysed by Therkildsen et al. (2010a)]. All other otoliths were obtained from Fisheries and Oceans Canada, where the archived collections extend back to 1960. Here, we selected sets of at least 30 otoliths for single years between 1960 and 2010 based on availability (see Table 1 for final sample sizes and years). All individuals, with the possible exception of the 1928 sample, for which the sampling time is unknown, were collected during the spawning season and were of reproductive age.
| Population | Year | n a | # loci | % variable | H obs | H e |
|---|---|---|---|---|---|---|
| 4T | 1928 | 29 | 1047 | 0.87 | 0.27 | 0.28 |
| 1960 | 37 | 160 | 0.84 | 0.25 | 0.26 | |
| 1968 | 31 | 160 | 0.84 | 0.28 | 0.27 | |
| 1974b | 14 | 160 | 0.78 | 0.27 | 0.26 | |
| 1976 | 36 | 160 | 0.85 | 0.28 | 0.27 | |
| 1983 | 37 | 160 | 0.86 | 0.28 | 0.28 | |
| 2002 | 29 | 160 | 0.84 | 0.28 | 0.26 | |
| 2008 | 39 | 1047 | 0.86 | 0.28 | 0.28 | |
| 3NO | 1960 | 28 | 160 | 0.82 | 0.25 | 0.26 |
| 1973 | 37 | 160 | 0.85 | 0.26 | 0.26 | |
| 1990 | 37 | 160 | 0.86 | 0.25 | 0.25 | |
| 2010 | 32 | 160 | 0.87 | 0.27 | 0.26 | |
| 3Ps | 1964 | 33 | 160 | 0.82 | 0.25 | 0.25 |
| 1993 | 25 | 160 | 0.84 | 0.26 | 0.25 | |
| 2010 | 26 | 160 | 0.86 | 0.28 | 0.27 | |
| 4VsW | 1961 | 16 | 160 | 0.86 | 0.27 | 0.26 |
| 2010 | 22 | 160 | 0.92 | 0.28 | 0.28 |
- a Sample sizes represent the number of individuals included in the analysis (i.e. excluding samples that did not pass the quality filtering criteria).
- b Due to the small n, this sample was pooled with the 1976 sample for analysis (there was no significant difference in allele frequencies between 1974 and 1976).
DNA extraction and genotyping
DNA was extracted with Omega EZNA Tissue DNA kits (Omega Bio-Tek, USA) following the manufacturer's instructions for fresh tissue and the procedure described by Therkildsen et al. (2010b) for otoliths. To prescreen DNA extracts, we amplified four highly polymorphic microsatellites (mean number of alleles = 19) in all samples using a PCR multiplex kit (Qiagen, Germany) and analysed the fragments on an ABI 3130 Genetic Analyzer (Applied Biosystems, USA). We removed individuals that showed evidence of cross-sample contamination (amplification of >2 alleles for any locus) or that failed to produce reliable amplification within 2–3 attempts. For the historical samples, both DNA extraction and PCR preparation were conducted in an ancient DNA laboratory or a separate facility where no contemporary fish samples had been processed.
Samples that passed the prescreening were genotyped for a set of gene-associated SNPs, primarily developed by the Canadian Cod Genomics and Broodstock Development Project (Hubert et al. 2010; Bowman et al. 2011). We used an initial panel of 1536 SNPs, 1474 of which could be anchored on the cod linkage map (Hubert et al. 2010 see Appendix S1, Note 1, Supporting information). In a trade-off between the number of samples and the number of SNPs to analyse, we applied a two-step approach: initially, we only scanned the end points of the longest available time span, that is, the 1928 and the contemporary sample from 4T, with the full 1536 SNP panel. As the majority of SNPs showed no temporal variation in allele frequencies among these samples (see below), all other samples were analysed with only a subset of SNPs, including the 50 loci that showed the largest temporal changes in the initial scan, 29 candidate genes for life history traits (Hemmer-Hansen et al. 2011), 23 loci that have been shown to be under selection in this species on broader geographical scales (Nielsen et al. 2009; Bradbury et al. 2010), and a random selection of 80 among the remaining loci, for a total of 182 SNPs.
All SNP genotyping was performed by the Roslin Institute at the University of Edinburgh, Scotland, using the Illumina GoldenGate platform following the manufacturer's protocol. This array-based technology relies on hybridization of short (<60 bp) locus- and allele-specific probes to the template DNA and should therefore be well suited for historical DNA that typically is fragmented. To minimize the risk of cross-sample contamination, historical and contemporary samples were kept separate during all steps. The SNP data were visualized and analysed with the GenomeStudio Data Analysis Software package (llumina Inc.).
Data quality control
To ensure the reliability of the SNP data despite the degraded nature of DNA from historical samples, we implemented several data control procedures and applied a conservative quality filtering. First, genotypes were called based on manual editing of all SNP cluster positions. Second, 29 DNA extracts were re-genotyped in independent assays to assess the reproducibility of results, and we excluded SNPs yielding <0.7 reproducibility rate between the genotypes in replicate samples. Third, we only included data points with a GenCall score of >0.4 and excluded SNPs and samples that following this strict filtering yielded call rates (percentage of successful genotype calls) <0.5.
We computed expected (He) and observed heterozygosity (Hobs) and tested for Hardy–Weinberg equilibrium (HWE) in all samples using 105 permutations with the Monte Carlo procedure implemented in the R-package adegenet (Jombart 2008). The degree of linkage disequilibrium (LD) between all pairs of loci within each sample was evaluated with the genetics package in R v2.14 (Warnes 2003). Here, and where appropriate throughout the analysis, we corrected for multiple testing by computing the expected false discovery rate (FDR), or q-value, for each test based on the distribution of P-values using the R-package qvalue (Storey & Tibshirani 2003). We considered tests significant when the FDR was <5% (q < 0.05).
Temporal outlier detection
Identifying loci that show divergent patterns of differentiation among samples collected over time from a single population is conceptually similar to searching for outliers in samples collected at a single time point from different populations. Recent evaluations have suggested that BayeScan (Foll & Gaggiotti 2008) is the most reliable of available methods for spatial genome scans (e.g. Narum & Hess 2011; Vilas et al. 2012), so we used this program (v2.01) to search for temporal outliers within each of the four populations. We used the default MCMC parameters, varied the prior odds in favour of a model excluding selection between 3 and 10, and considered loci with a Bayes factor >3 in three independent runs outliers.
Although BayeScan has several advantages over alternative outlier detection methods, little is known about its performance on temporal data. The method is based on the multinomial Dirichlet likelihood to estimate locus- and sample/population-specific effects from the observed variation in allele frequencies (Foll & Gaggiotti 2008), and while this likelihood function should apply to a range of demographic equilibrium models (Beaumont & Balding 2004), it has not been demonstrated to be valid for differentiation over time within a single population. The clear violation of a basic model assumption thus could complicate interpretation of the results from temporal data.
We therefore supplemented the BayeScan analysis with an outlier test based on an explicitly temporal null model, implemented under a modified version of the commonly used Fdist approach of Beaumont & Nichols (1996). We adapted the original method to fit our scenario by generating the expected neutral distribution through simulations of drift within a single isolated population, rather than as drift–migration equilibrium between multiple demes. Our null model was based on multigenerational sampling of a Wright–Fisher population and to quantify how much temporal variation we would expect from drift and sampling error alone, the simulations were parameterized for each population with the number of generations between sampling points, the harmonic mean of sample sizes (number of individuals per sampling point) and an estimate of the effective population size (Ne; see Appendix S1, Note 2, Supporting information for details). Following the procedure in Beaumont & Nichols (1996), the simulated distribution was then used to identify outlier loci that varied more over time than expected under neutrality (we call the method Ftemp, see Appendix S1, Note 2, Supporting information). All simulations and computations were completed with custom R-scripts (available upon request).
For both BayeScan and Ftemp analyses, we conducted separate temporal genome scans for the initial SNP panel genotyped in the 4T 1928 and 2008 samples (1047 SNPs) and for the subsequent samples (1960–2010) from each of the four populations (160 SNPs).
Spatial outlier detection
In addition to testing for temporal outliers within each population, we also looked for spatial outliers among the populations at three periods in time: the 1960s, the 1980s–1990s (here we had no sample from 4VsW) and among contemporary samples. For this analysis, we applied both BayeScan (with settings as above) and the standard spatial Fdist2 model (Beaumont & Nichols 1996) as implemented in the software Lositan (Antao et al. 2008). In Lositan, we based simulations on 105 iterations, the infinite alleles mutation model and assumed 30 demes.
Correlation to environmental variation
The moderate number of samples in this study precludes rigorous statistical testing of how allele frequency shifts may correlate with environmental or phenotypic variables. However, to qualitatively investigate what factors could be associated with temporal shifts within the 4T population (the only population sampled at >4 time points), we computed Pearson's correlation coefficients (r) between allele frequencies at temporal outlier loci and data on a suite of environmental and demographic factors for the sampled years. The factors included fishing mortality, temperature, biomass, and indices of growth rate and length at maturation (see Table S1, Supporting information, for a full list of variables and data sources). Based on the obtained coefficients, we compared the relative degree of correlation between outlier loci and explanatory variables and further examined the strongest observed patterns.
Overall differentiation among samples
We used a hierarchical amova with time points nested within populations to assess how the overall genetic variation was distributed in space and time. This was done in Arlequin v3.5 (Excoffier & Lischer 2010), and the significance of contributions from the different levels was tested with 10000 permutations. Pairwise FST between all samples was computed with the fstat function from the Geneland package in R (Guillot et al. 2005), and we tested for pairwise differences in allele frequencies among all samples using chi-square tests, as implemented in the software Chifish (Ryman 2006). To obtain estimates reflecting signatures of neutral evolutionary forces only, we repeated all these analyses on a reduced set of loci (n = 101), excluding all spatial and temporal outlier loci (see below). We also used the program Powsim v4.1 (Ryman & Palm 2006) to evaluate our power to detect genetic heterogeneity in the different comparisons. As a supplement, we attempted to apply various individual clustering methods, but the level of differentiation was too low to obtain meaningful results (not shown).
Contemporary migration
The low levels of differentiation also precluded estimation of contemporary migration rates (Wilson & Rannala 2003; Faubet et al. 2007). However, to evaluate whether migration rather than selection could explain our observations, we constructed simulations to elucidate how much migration from nearby populations would be needed if gene flow alone had caused the observed temporal variation at outlier loci (see below). Assuming that the 1960s samples reflected baseline allele frequencies for the four populations, we simulated various levels of exchange (migration rate m ranging from 0 to 1) between populations over the sampling period and analysed these simulated data with our Ftemp method (see Appendix S1 Notes 2 and 3 for details, Supporting information). For each population, we evaluated the number of significant temporal outliers under different combinations of m, local Ne and source population of migrants, as well as how many of these outliers were identical to the temporal outliers in the observed data.
Results
Data quality and genetic diversity
A total of 508 samples could be used for analysis, while 137 were discarded due to poor amplification, contamination or low-quality SNP genotyping. For the initial scan, 1047 SNPs (of 1536) were successfully genotyped, passed the quality criteria and were polymorphic in at least one sample. For the follow-up panel, 160 SNPs (of 182) could be used for analysis. The error rate among replicate samples was <5% for all historical samples and <1% for contemporary samples, and the mean SNP call rate was >90% for all included loci (except from in the 1928 sample where the mean call rate was 76%).
On average 85% of the loci were polymorphic in each sample (Table 1). The average He within samples was 0.26, and there was no clear relationship between He or the proportion of polymorphic loci and the sampling year (Table 1). In the 1047 SNP data set, 31 tests involving 25 SNPs showed significant departures from HWE proportions after FDR correction, and 6 loci that exhibited departure in >1 sample were excluded from analysis. In the 160 loci set, we observed only a single significant departure from HWE after FDR control among samples.
In the 1047 SNP data, between 1.7 and 2.7% of pairwise tests for the LD were significant (q < 0.05). In the 160 SNP data, between 2.1 and 5.8% of pairwise tests for LD between loci within each sample were significant (q < 0.05). In this panel, almost all SNPs that showed significant LD in multiple samples originated from one of two blocks on different linkage groups and were outliers either in time or space (see below).
Temporal outliers
In the initial comparison between 1928 and 2008 in 4T, the temporal differentiation at 50 of the 1047 loci exceeded the 95% confidence limit for neutral expectations in the Ftemp analysis. Nine of these loci remained significant after FDR correction (q < 0.05), and all but one of these loci were also identified as outliers in the BayeScan analysis (Fig. 2a). Eight of the strong outliers (q < 0.05) and in total half of the 50 Ftemp outliers were successfully genotyped in the remaining samples (1960–2010). Surprisingly, however, the vast majority of the initial outlier loci did not show increased temporal differentiation among post-1928 samples. Although we observed allele frequency changes of >50% among the outliers between 1928 and 2008 in 4T, none of these loci showed outlier patterns in the intermediate time period from 1960 to 2002. Examination of the temporal data showed that the changes had primarily occurred between 1928 and 1960 and that allele frequencies at these particular loci had remained stable from 1960 to 2008 (Fig. 3a). Similar patterns of stability were observed in the other populations, although one locus from the initial comparison was also a significant Ftemp outlier in 3NO (Fig. 3b–d).
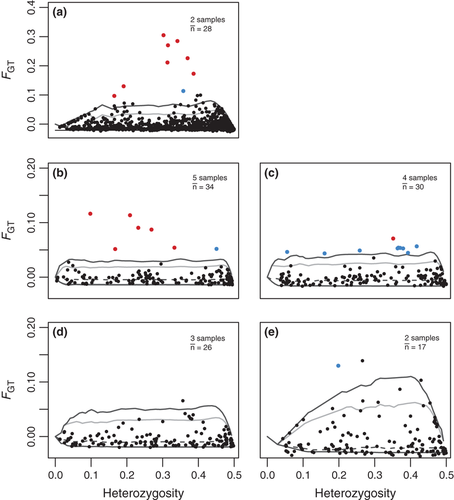
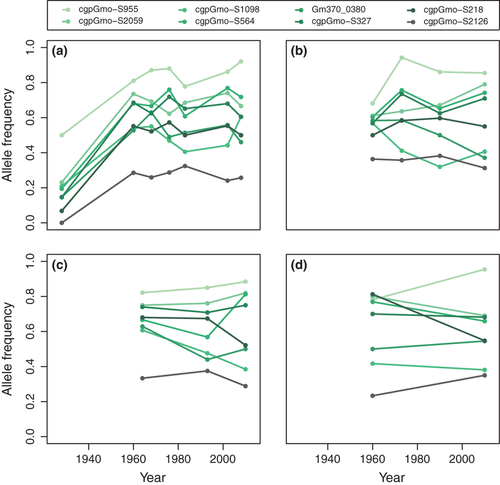
Entirely different sets of loci were identified as temporal outliers among the samples collected between the 1960s and 2000s, however. Between 7 and 14 of the 160 loci genotyped here fell above the 95% confidence envelope for Ftemp null expectations in the different populations, but only in 3NO and 4T were >1 locus significant after FDR correction (Fig. 2b–e, Table 2). BayeScan identified fewer outliers, but was qualitatively consistent, with a single significant outlier in 3NO and six outliers in 4T 1960–2002 (in both cases subsets of the Ftemp outliers), but none in the other two populations (Fig. 2b–e, Table 2). A subset of the significant 3NO Ftemp outliers also showed increased differentiation in 3Ps, but interestingly, there was basically no overlap between the outliers of 3NO and 4T (Table 2). 4VsW showed an intermediate pattern where subsets of both 3NO and 4T outliers as well as an additional group of loci showed increased differentiation (Table 2). Examination of outlier allele frequencies revealed that the 3NO Ftemp outliers in fact also showed large differentiation within 4T, but mostly between 1928 and 1960 (and therefore were not detected in the 1960–2002 genome scan; Fig. 4b). The 4T outliers, however, remained stable in 3NO and 3Ps throughout the study period, indicating clear nonparallel trajectories for these loci among the populations (Fig. S1, Supporting information).
| SNP Name | LG | Temporal outliers | Spatial outliers | |||||
|---|---|---|---|---|---|---|---|---|
| 3NO | 3Ps | 4T | 4VsW | 1960s | 1980–1990a | 2000s | ||
| Rhod_1_1 | 1 | 99b, BS3b | 95 | 99 | 99b | 95, BS3 | ||
| cgpGmo-S1874 | 1 | 99b | 95 | 95 | 99 | 95, BS3 | ||
| cgpGmo-S1955 | 1 | 99b | 95 | 95 | 99 | 95, BS3 | ||
| cgpGmo-S1166 | 1 | 99b | 95 | 99 | 99, BS3 | |||
| cgpGmo-S985 | 1 | 99b | 95 | 99 | 95 | |||
| Pan1 | 1 | 99b | 99 | 95 | 95 | |||
| cgpGmo-S1456 | 2 | 99b | 95 | |||||
| cgpGmo-S1101a | 2 | 95 | 99 | |||||
| cgpGmo-S1068 | 2 | 99 | ||||||
| cgpGmo-S1970 | 5 | 95 | ||||||
| cgpGmo-S1200 | 7 | 99b | 95 | 99b | ||||
| cgpGmo-S1017 | 9 | 95 | ||||||
| LDHB | 9 | 95 | ||||||
| cgpGmo-S1737 | 12 | 99b, BS3b | 99b | 99 | 99b | |||
| cgpGmo-S180b | 12 | 99b, BS10b | 99 | 99b, BS10b | 99b, BS10b | |||
| cgpGmo-S816a | 12 | 99b, BS10b | 95 | 99b, BS10 | 99b, BS3 | |||
| cgpGmo-S866 | 12 | 99b, BS10 | 95 | 99, BS3 | 99, BS3b | |||
| cgpGmo-S57 | 12 | 99b, BS10 | 95 | 95 | ||||
| cgpGmo-S2101 | 12 | 99b | ||||||
| cgpGmo-S1046 | 12 | 99b, BS3 | 95 | 95 | ||||
| cgpGmo-S316 | 12 | 95 | 95 | |||||
| cgpGmo-S142 | 14 | 95 | ||||||
| cgpGmo-S1467 | 14 | 95 | ||||||
| cgpGmo-S955 | 17 | 99b | ||||||
| cgpGmo-S1340 | 18 | 99 | ||||||
| cgpGmo-S442a | 18 | 95 | ||||||
| Gm370_0380 | 22 | 95 | ||||||
| Anti_1 | 22 | 95 | 95 | |||||
| Gm0588_0274 | ? | 99b | ||||||
| cgpGmo-S1406 | ? | 95 | 99b | |||||
| cgpGmo-S1731 | ? | 95 | ||||||
| Gm335_0159 | ? | 99b | ||||||
| Total number of outliers | 14 | 7 | 10 | 15 | 7 | 15 | 8 | |
| Total with FDR control | 10 | 0 | 7 | 1 | 1 | 4 | 5 | |
- a This comparison does not include 4VsW.
- b indicates that the outlier remained significant following FDR control (q < 0.05).
Spatial outliers
The Lositan analysis indicated that the differentiation at 7–15 loci exceeded the 95% confidence limit on neutral expectations in the spatial comparisons for different time periods (Table 2, Fig. S2, Supporting information). BayeScan generally identified fewer outliers (between 0 and 7 in the different comparisons; Table 2), but all BayeScan outliers were also Lositan outliers, and the two programs were qualitatively consistent in identifying the most differentiated loci (Fig. S3, Supporting information).
Comparison of the three snapshots in time revealed a marked sequential shift in which loci exhibited spatial divergence, with no overlap between the 1960s and the contemporary spatial outliers, while the spatial outliers from the intermediate 1980s–90s comparison showed overlap with both the early and the late period (Table 2). No locus remained a spatial outlier in all time periods. Removing the 4VsW sample from the spatial comparisons resulted in much fewer and some different outliers (Fig. S4, Supporting information), suggesting that this population is driving much of the overall outlier pattern observed. However, this population was not sampled in the 1980s–90s period that overall showed the highest number of outliers (in both Lositan and BayeScan), so the locus-specific patterns of spatial divergence are clearly highly dynamic.
The dynamic pattern of spatial divergence is further supported by the match between temporal and spatial outliers. The spatial outliers in the 1960s were a subset of the loci that were temporal outliers within 3NO (and with weaker support in 3Ps and 4VsW; Table 2). As these loci were no longer spatial outliers at later time points, the temporal changes have homogenized allele frequencies among populations. The contemporary spatial outliers, on the other hand, were for a large part the same loci that were temporal outliers in 4T (and with weaker support 4VsW; Table 2). As these loci were not spatial outliers in the 1960s, the temporal changes in outlier loci in 4T and 4VsW caused greater divergence among populations over time.
In the post-1928 data, a total of 32 loci were outliers either in space, time or both. These were spread over at least 11 linkage groups and generally displayed low and nonsignificant levels of LD between them (Fig. S5, Supporting information). However, 13 of the outliers clustered into two high-LD groups, each spanning 10–14 cM and mapping to 5–6 different scaffolds that combined cover >2 Mb in the Ensembl cod genome assembly (www.ensembl.org; release 65, Dec 2011). Most outlier SNPs were located in the 3′ UTR of gene models (Tables S3 and S4, Supporting information).
Correlation with environmental variation
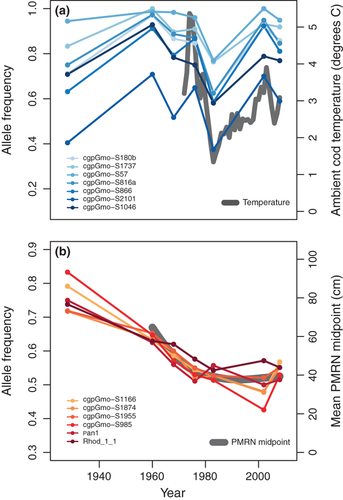
An index of ambient temperature for cod during the feeding season showed the best correlation with allele frequencies at the 1960–2002 temporal outlier loci in 4T (r > 0.8 for most loci), indicating a covarying temporal pattern (Fig 4a). A similar correlation was also evident for two other outlier loci (Fig. S6, Supporting information). Within 4T, a set of loci initially identified as temporal outliers in 3NO also showed a strong correlation (r > 0.9) to temporal shifts in estimated probabilistic maturation reaction norm midpoints (Fig. 4b), a life history change suggested to reflect an evolutionary response to fishing (Swain 2011). These loci also showed correlation with the temporal trend in total mortality rate (r < −0.8; Fig. S6, Supporting information).
Differentiation among samples
The amova based on all loci suggested that there was slightly more variation between time points within populations than overall between populations, with both levels highly significant (P < 0.00001, Table 3). However, for the nonoutlier loci—that presumably reflect neutral population structure—a much smaller proportion of the variance was partitioned among the hierarchical levels. There was low but significant spatial variation (P = 0.0064), but no significant variation among time points within populations (P = 0.87; Table 3). These results of very weak but temporally stable spatial structure were corroborated by the pairwise tests for differences in allele frequencies. With all loci, pairwise FST ranged from −0.007 to 0.086, and 31 tests were significant after FDR correction including comparisons both within and among populations (Table S5, Supporting information). When only considering the ‘neutral’ set of loci, no comparisons were significant after FDR correction (Table S6, Supporting information). The mean pairwise FST among samples for these loci was 0.0015, and simulations indicated that with the applied panel of markers and sample sizes, we would only have a 14% chance of detecting significant differentiation at this level (results not shown). However, by pooling the time points within areas, we should have >75% chance of detecting this level of differentiation among populations, and we did indeed find significant differences in allele frequencies (P < 0.04) between all combinations except those involving 3Ps, which could not be differentiated from 3NO and 4T (Table S7, Supporting information).
| Data | Source of variation | F-index | d.f. | Var components | % variation (95% CI) | P-value |
|---|---|---|---|---|---|---|
| All loci | Among populations | F CT | 3 | 0.10 | 0.44 (0.21 to 0.68) | <0.00001 |
| Among time points within populations | F SC | 12 | 0.12 | 0.54 (0.28 to 0.83) | <0.00001 | |
| Among individuals within time points | F IS | 463 | 0.22 | 1.03 (−0.41 to 2.67) | 0.01059 | |
| Within individuals | 21.35 | 97.99 | ||||
| Non-outlier loci | Among populations | F CT | 3 | 0.02 | 0.15 (0.024 to 0.27) | 0.00639 |
| Among time points within populations | F SC | 12 | −0.01 | −0.09 (−0.24 to 0.064) | 0.86560 | |
| Among individuals within time points | F IS | 463 | 0.19 | 1.14 (−0.58 to 3.156) | 0.01269 | |
| Within individuals | 16.16 | 98.80 |
Contemporary migration
Simulations indicated that in general, migration rates of 0.2–0.3 would be required to generate a similar number of significant Ftemp outliers as were observed in the different populations (Fig. S7, Supporting information). For 3NO, somewhat higher migration rates of >0.6 could have driven allele frequency changes at the specific loci that were observed as significant temporal outliers in the actual data (Fig. S8, Supporting information). However, this was only if migrants originated from 4VsW—the population most differentiated from 3NO at neutral markers (Tables S5 and S6, Supporting information). In 4T, even complete replacement by any of the other populations could not account for the frequency shifts at the observed outlier loci (on average ≪1 of these loci were identified as outliers in the migration simulations regardless of Ne and m values assumed; Fig. S8, Supporting information).
Discussion
This study revealed highly heterogeneous patterns of differentiation among SNPs from different regions of the cod genome, with certain gene-linked SNPs showing substantially elevated divergence in either time, space or both. Over the short timescale (6–12 generations, Table S2, Supporting information) and relatively small geographical area considered, we observed markedly different microevolutionary trajectories within four adjacent populations and found a temporal shift in the identity of loci showing increased differentiation during different time periods. Because the majority of genomic locations showed almost complete stability in allele frequencies, the significantly higher differentiation at specific loci probably reflects effects of selection either directly at these loci or at closely linked sites.
Patterns of selection
If selection indeed is the primary driver behind the observed allele frequency shifts at outlier loci, then this study indicates highly variable local selection pressures that both target different regions of the genome and work at varying strengths and directions depending on the area and time period considered. Most notably, the majority of temporal outliers in the 1928–2008 comparison in 4T showed little change between 1960 and 2008, but a completely different set of loci exhibited large allele frequency shifts during this period. This indicates that different parts of the genome may have been under selection early (1928–1960) and late (1960–2010) in the study period. Similarly, the identification of nonoverlapping sets of temporal outliers in 4T and 3NO suggests that selection was acting on different genomic regions in these two populations during the 1960–2010 period.
Varying power to detect outliers caused by the nonsymmetric sampling pattern (imposed by limited sample availability) probably explains a portion of the variation in which and how many loci were detected as statistically significant temporal outliers in the different populations. For example, the smallest numbers of significant temporal outliers were found in 3Ps and 4VsW—the populations for which we had the fewest sampling points and the smallest number of individuals per sample (see Table 1). A sensitivity analysis suggested that increased statistical power conferred by larger sample sizes could have resulted in identification of additional temporal outliers in these populations (Appendix S1, Note 2, Supporting information). Thus, the lack of significant temporal outliers here may reflect sampling limitations rather than stable allele frequencies. In any case, the data make it clear that contrasting sets of loci exhibited the largest temporal variation in the different populations, and that patterns of allele frequency change at particular loci were often nonparallel (see Supporting information Fig. S1).
A number of outlier loci showed gradual and directional changes over time, but many showed more fluctuating, apparently ephemeral patterns. Such unstable patterns of selection are in line with the combined inference from numerous temporally replicated studies on phenotypic selection in natural populations, which indicate that strong temporal variation in both the direction and strength of selection is the norm rather than the exception over short timescales (recently reviewed by Siepielski et al. 2009; Bell 2010; Kingsolver & Diamond 2011). Although some of the patterns reported in these studies may be caused by sampling error (Morrissey & Hadfield 2012), short-term fluctuations in selection pressures may be particularly common in a highly dynamic and stochastic environment like the ocean, so they would explain our observed patterns well.
Drivers of selection
Our exploratory correlation analysis showed that allele frequencies at temporal outliers in 4T seemed to track variation in ambient fall temperature, indicating that this variable could reflect a possible driver of selection. Although based on few data points, this relationship is supported by a previous finding of strong temperature-associated clines in allele frequencies at these particular loci over large spatial scales (Bradbury et al. 2010). Similarly, allele frequency changes in 4T at a different set of linked loci strongly correlated with temporal shifts in the probabilistic maturation reaction norm that has been suggested to reflect an evolutionary response to fishing (Swain 2011). While more detailed time series data are needed to firmly establish these possible relationships to drivers and traits, our initial synoptic findings here are consistent with both temperature- and fisheries-induced selection pressures acting over the study period.
A central strength of genome scan approaches, as applied here, is that they do not require targets of selection to be known a priori and that observed patterns are intrinsically genetic. The drawback is that, especially for nonmodel organisms, it can be difficult to establish the phenotypic significance and fitness effects of interesting genetic polymorphisms. A first step is to identify the exact targets of selection. Because most of the SNPs studied here were located in 3′ UTR regions and the few that were located in coding sequence were synonymous polymorphisms (results not shown), we are unlikely to have identified the exact causative mutations (although the loci may have regulatory roles). We have, however, narrowed in on important candidate genes and we note that many outlier loci are found in regions with genes related to metabolism (see Table S4, Supporting information). These findings constitute valuable starting points for future more detailed efforts to identify specific targets of selection.
Reliability of results
For robust interpretation, it is important to consider whether factors other than selection could have caused the observed temporal shifts in allele frequencies. First, as the study is based on historical DNA, we must carefully evaluate whether genotyping error or missing data could have caused false impressions of large allele frequency differences. This is particularly true for the 1928 sample that had lower call rate and slightly reduced genotype reproducibility (though still >95%) compared with the other samples. However, while suboptimal DNA quality inevitably has caused a few inaccuracies in our data set, the stringent quality control measures implemented and the resulting high degree of genotype concordance among replicate samples clearly indicate that this effect has been minor. There is no relationship between sample age and observed genetic diversity (Table 1), and outlier loci do not exhibit significantly elevated error rates (Fig. 5) or lower realized sample sizes (after accounting for missing data), neither in 1928 or later samples (Fig. S9, Supporting information). The strongest verification of data reliability, however, comes from the highly consistent patterns of strong LD among outlier loci across all samples (Fig. 4 and Fig. S5, Supporting information).
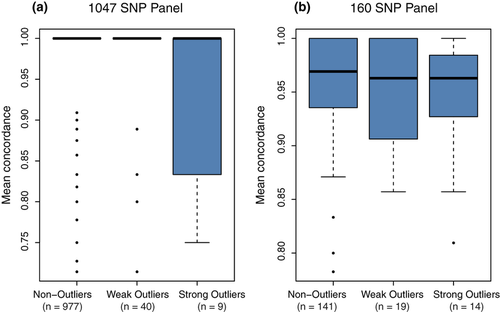
Even with reliable data, violated model assumptions or general methodological limitations could have resulted in false positives among the identified outliers. Evaluations on simulated data have suggested that the Fdist outlier detection method, as we applied here both in its original form and adapted for temporal analysis, in some cases may have higher type I error rates than BayeScan (Narum & Hess 2011). However, in most of our analyses, a subset of Fdist outliers was also identified as outliers by BayeScan. The combined inference from these two methods that are based on completely different underlying models and mathematical approaches provide robust statistical support for the divergent pattern of differentiation at certain loci—especially as the Ftemp method was specifically designed for temporal data. Further, our sensitivity analysis for the Ftemp method suggested that uncertainty in parameter inputs would only cause slight changes in the cut-off for significance. As the differentiation at the majority of outlier loci was substantially elevated compared with the rest of the loci (i.e. not just at a tail of a continuous distribution; Figs 2 and S2), slight shifts in significance cut-offs could change the status of a few weak outliers, but would not affect conclusions about the highly divergent loci. The finding that many outlier loci also show elevated patterns of differentiation across larger spatial scale (Nielsen et al. 2009; Bradbury et al. 2010) also support their affiliation with selection. Hence, overall, it seems unlikely that data errors or statistical artefacts should have strongly impacted our main conclusions.
The role of migration
Another important factor to consider is the possibility of population replacement. If migration patterns are highly dynamic, our sampling over time could potentially reflect various mixtures of populations moving in and out of the study sites rather than changes in which genotypes survive and reproduce within stationary isolated populations. The fact that many temporal outliers were also spatial outliers could suggest that migration between areas may have generated some of the temporal outlier signatures. However, our analysis of nonoutlier loci—which should reflect neutral evolutionary forces—revealed temporally stable spatial differentiation between the sampling locations, indicating that the overall population structure has remained intact over the study period. Previous microsatellite studies have also demonstrated differentiation between several of the studied populations (Ruzzante et al. 1996, 2000, 2001; Beacham et al. 2002), lending further support to the biological relevance of this maintained structure despite the very low levels of differentiation.
The 4T-1928 was the only sample not known to be collected during the spawning season and its highly divergent allele frequencies at a subset of loci could suggest that this sample possibly originated from a different spawning population. The very low level of differentiation between populations at ‘neutral’ loci makes it difficult to completely exclude this possibility, although it appears unlikely as no other populations are known to migrate into the 4T area (Robichaud & Rose 2004). However, even if the 1928 sample indeed is of alternative origin, equally large allele frequency changes were observed at different sets of loci in the 1960–2010 period, as between 1928 and the later samples. Hence, the conclusions of strong temporal outliers in this system are by no means solely dependent on the 1928 data point.
For the 1960–2010 data, our simulations suggest that very high migration rates of at least 0.2–0.3 would be needed to cause the strong shifts in allele frequencies we observed within populations (assuming migrants originated from a sampled population). Such high migration rates seem at odds both with the detectable genetic structure (see Waples & Gaggiotti 2006) and with ecological data that generally suggest substantial reproductive isolation of the studied populations: previous analyses have demonstrated consistent phenotypic differences (Swain et al. 2001), and despite extensive seasonal migrations, both traditional tagging and otolith microchemistry studies have indicated that in 4T, almost all fish return to spawn with their local population (Campana et al. 2000; Robichaud & Rose 2004), and this is also true for 4VsW (Robichaud & Rose 2004). Tagging results are more mixed for the two other populations (Robichaud & Rose 2004), but much of the reported population mixing occurs outside the spawning season, and samples from these populations were specifically selected from locations expected to be minimally affected by migration.
Even if migration has caused some of the observed changes, our simulations showed that none of the other populations could have served as a migrant source for divergent allele frequencies at the specific temporal outlier loci in the 4T population between 1960 and 2002. However, as the temporal outlier loci are also spatial outliers on larger geographical scales (Bradbury et al. 2010), we cannot exclude that some unsampled ‘ghost’ population has contributed such migrants. Nevertheless, adult migration into this semi-enclosed basin appears, as mentioned, very limited, and predominant currents out of the Gulf make external larval input unlikely. Oceanographic modelling also predicts a high degree of local retention of larvae here, although a small portion may drift to the other management areas (Chassé 2003). Hence, while we cannot entirely rule out that migration has played an important role in shaping allele frequency variation in some of the other populations, it seems unlikely that it has been the main driver behind the observed patterns at outlier loci in the 4T population, which was the main focus of this study.
Implications and future research
The finding of highly dynamic signatures of selection in this study raises the question of whether our observations reflect a general pattern of microevolution in marine fish and in natural populations in general. Only very few other studies have so far examined temporal variation in genetic markers under selection. In cod, two previous studies have detected temporal shifts in allele frequencies at the well-studied Pan-I locus over decadal timescales (Árnason et al. 2009; Jakobsdottir et al. 2011). We also found temporal variation at this locus but showed that it along with 5 other outliers was part of a tight LD group spanning >2 Mb of the genome, indicating that Pan-I may just be a marker for a genomic region, not the target of selection. Temporal shifts at a number of additional gene-associated SNPs have also been observed for cod populations in the Northeast Atlantic (Poulsen et al. 2011), so there is evidence that ongoing selection over short timescales could be a widespread phenomenon, at least for cod but probably also in other marine fish. Further research using retrospective genetic analysis is needed to gain further insights into its extent.
Even though the phenotypic role and fitness effects of the observed outlier loci remain elusive, the findings here add an important new perspective to recent studies presenting evidence for local adaptation in marine fish (e.g. Marcil et al. 2006; Nielsen et al. 2009; Bradbury et al. 2010). Our study illustrates that where temporal stability of genetic divergence has not been demonstrated, it can clearly not be assumed a priori. If environmental conditions are highly dynamic, local adaptation may also not necessarily imply static differences between populations, but can reflect ongoing changes. Findings of greater temporal than spatial variation at certain loci may be limited to studies that, like here, focus on a relatively small part of a species range over which environmental conditions are somewhat similar. However, future studies over larger spatial scales should provide additional insights about the interactions between temporal and spatial variation in selection pressures. Studies on other organisms will also reveal whether similar patterns are common in different ecological systems. In any case, the large changes observed over just a few generations—in some cases correlating with environmental or life history trait variation—suggest that cod populations potentially can respond rapidly to changes in selection pressures and therefore may be able to quickly adapt to human-induced modifications of their environment.
A better understanding of temporal and spatial scales of adaptation will be crucial both for our fundamental understanding of microevolution in high gene flow organisms and for conservation and fisheries management. The importance of intraspecific diversity in fitness-related traits for ensuring stability and persistence of species and fisheries yields is increasingly being recognized (Hilborn et al. 2003; Hutchinson 2008). Adaptive divergence between populations or between subunits of metapopulations can generate ‘portfolio effects’ that dampen overall fluctuations in abundance because if various population components are adapted to different conditions, they may exhibit independent and complimentary reactions to perturbations (Hilborn et al. 2003; Schindler et al. 2010). Our results here demonstrate that it is critical to consider not only the spatial but also the temporal dimension of this biocomplexity and to evaluate how human activities affect the overall system resilience.
Acknowledgements
We would like to thank Fisheries and Oceans Canada for supplying tissue samples and providing access to archived otoliths. We are also very grateful to Eske Willerslev and M. Thomas P. Gilbert at the Center for GeoGenetics at the Natural History Museum of Denmark for generously providing access to ancient DNA laboratory facilities, and to Rob Ogden, Richard Talbot and David Morrice helpful assistance with the SNP genotyping. Robin Waples, Louis Bernatchez and four anonymous reviewers provided valuable comments on earlier versions of the manuscript. The study received financial support from the European Commission, as part of the Specific Targeted Research Project Fisheries-induced Evolution (FinE, contract number SSP-2006-044276) and from the Danish Agency for Science, Technology and Innovation as part of the Greenland Climate Research Centre.
References
N.O.T. and E.E.N. designed the research with input from D.P.S., M.J.M. and E.A.T. D.P.S., M.J.M. and E.A.T. contributed samples. N.O.T. and D.M. performed the laboratory research. N.O.T. analysed the data with input from E.E.N., S.R.P., T.D.A. and J.H.H. N.O.T. and E.E.N. wrote the paper with input from all other authors.
Data accessibility
A list of accession IDs for the SNPs, individual SNP genotypes for all samples and the analysed environmental data are archived at the Dryad data repository: doi:10.5061/dryad.4v0c5.




