SNP rs688 within the low-density lipoprotein receptor (LDL-R) gene associates with HCV susceptibility
[The copyright line for this article was changed on 27th November after original online publication]
Funding information
The research leading to these results has received funding from the European Community's Seventh Framework Programme [FP7-2007-2013] under grant agreement no. HEALTH-F3-2012-305578.
Abstract
Background & Aims
Despite high-risk behaviour, 10%-20% of HCV multiple exposed individuals remain uninfected (MEU), whilst the remainder become infected (MEI). We hypothesize that host factors play a role in HCV susceptibility. We aimed to identify polymorphisms in host genes that encode for proteins involved in viral entry: CD81, Scavenger receptor 1 (SR-1), Low-density lipoprotein receptor (LDL-R), Claudin-1 (CLDN1), Occludin (OCLN) and Niemann-Pick C1–like 1 (NPC1L1).
Methods
Multiple exposed infected and MEU from two observational cohorts were selected. From the MSM study of acute infection with HCV (MOSAIC), HIV-1 infected MEU cases (n = 30) and HIV-1 infected MEI controls (n = 32) were selected based on reported high-risk behaviour. From the Amsterdam Cohorts Studies (ACS) injecting drug users (IDU) cohort, MEU cases (n = 40) and MEI controls (n = 22) were selected who injected drugs for ≥2 years, in the nineties, when HCV incidence was high. Selected single nucleotide polymorphisms (SNPs) were determined by sequencing or SNP assays.
Results
No associations were found for SNPs within genes coding for CD81, SR-1, Claudin-1 or Occludin between the MEU and MEI individuals from either cohort. We did observe a significant association for rs688 within the LDL-R gene with HCV infection (OR: 0.41 P = 0.001), however, LDL cholesterol levels did not vary between individuals carrying the differential SNPs. Additionally, a marginal significant effect was found for rs217434 and rs2072183 (OR: 2.07 P = 0.032 and OR: 1.76 P = 0.039, respectively) within NPC1L1.
Conclusions
Our results demonstrate that the rs688 SNP within the LDL-R gene associates with HCV susceptibility through mucosal as well as intravenous exposure.
Abbreviations
-
- ACS
-
- Amsterdam Cohort Studies
-
- CLDN-1
-
- Claudin-1
-
- HCV
-
- Hepatitis C virus
-
- IDU
-
- Injecting drug use
-
- LDL-R
-
- LDL-receptor
-
- MEI
-
- Multiple exposed infected
-
- MEU
-
- Multiple exposed uninfected
-
- MOSAIC
-
- MSM Observational Study of Acute Infection with Hepatitis C
-
- MSM
-
- Men having sex with men
-
- NMD
-
- Non-sense mediated decay
-
- NPC1L1
-
- Niemann-Pick C1 like 1
-
- OCLN
-
- Occludin
-
- SNP
-
- Single nucleotide polymorphism
-
- SR-B1
-
- Scavenger receptor class B
Key points
- Despite high-risk behaviour, a proportion of HCV multiple exposed individuals remain uninfected and where host factors may play a role in HCV susceptibility. We aimed to identify whether genetic modifications in genes encoding for proteins that are known to be involved with viral entry associate with infection risk. We identified alterations in the gene coding for the Low-density lipoprotein receptor (LDL-R) associated with risk of infection. This was found in individuals exposed sexually or through intravenous drug use. Genetic alterations within the LDL-R gene can therefore provide individuals with a modest protection against HCV infection.
1 INTRODUCTION
With 71 million people chronically infected and 399 000 deaths from HCV-related liver disease, hepatitis C virus (HCV) is a major global health burden.1 Risk factors for acquiring an HCV infection in developed countries are injecting drug use (IDU) and high-risk sexual behaviour among men having sex with men (MSM).2 However, some individuals remain uninfected despite high-risk behaviour. Several studies have shown that from the high-risk IDU ultimately 10%-20% do not become infected with HCV, suggesting a biological reason why those multiple exposed uninfected (MEU) individuals are less prone to acquire HCV.3-5 A number of immune specific genotypes have been associated with risk of HCV transmission, how an individual spontaneously resolves their HCV infection or response to treatment6-9 It has been described that three single nucleotide polymorphisms (SNPs) within the promoter region of the C-type lectin receptor DC-SIGN have associated with the risk of HCV transmission, but only through sexual and not intravenous exposure.10 The SNPs identified were shown to result in down-regulation of receptor expression and the link provides an indication that the interaction of HCV with dendritic cells is important when considering mechanisms of viral transmission.
Multiple host molecules are involved in the multi-step entry process of HCV into target cells. Hepatocyte surface receptors and co-receptors including Tetraspanin CD81, human scavenger receptor class B (SR-BI), Claudin-1 (CLDN-1) and Occludin (OCLN) are essential for HCV cell entry. In addition, LDL-receptor (LDL-R) and Niemann-Pick C1-like 1 (NPC1L1) are associated with capturing HCV and cell attachment. More recently, the NPC1L1 cholesterol absorption receptor was identified as an HCV entry factor.11 The aim of our study was to identify polymorphisms in host genes and their promoters that encode for proteins that modulate virus entry into cells; CD81, SR-BI, LDL-R, CLDN-1, OCLN and NPC1L1.
2 STUDY POPULATION AND METHODS
2.1 Study populations
2.1.1 HIV-1 infected MSM with high-risk sexual behaviour
Serum samples were collected from HIV-1 infected, Western European MSM (n = 62) from the MSM Observational Study of Acute Infection with Hepatitis C (MOSAIC) cohort recruited from either the Academic Medical Center or the OLVG Oost hospital in Amsterdam, the Netherlands. Serum samples were studied from MEU individuals (n = 30) and multiple exposed infected (MEI; n = 32). Risk behaviour data were available from behavioural questionnaires collected at 6-month intervals, and MOSAIC Risk scores were subsequently calculated.12 Participants were categorized as MEU or MEI, based on reported behavioural risk factors at inclusion or any of the follow-up visits which we previously found to be associated with sexual transmission of HCV in this cohort.13 Behaviour with increased risk of acquiring HCV infection was defined as having reported at either inclusion or any of the follow-up visits any of the following: no or inconsistent condom use and anal intercourse with an HCV-infected sex partner; fisting; use of sex toys; rectal bleeding during or after sex; group sex.14-18 The MOSAIC study was approved by the Institutional Review Board of the Academic Medical Center under assigned study numbers NL26485.018.09 and NL48572.018.14. Informed written consent was obtained from all study participants.
2.1.2 IDU
Serum samples of Western European individuals from the Amsterdam Cohort Studies (ACS) among IDU were selected. A total of 62 individuals were selected (40 MEU and 22 MEI), who started injecting before 1990, which was a period with high HCV incidence among drug users (up to 27.5/100 person years in the 1980s). Participants injected for ≥2 years and either seroconverted for HCV (MEI) or remained HCV seronegative (MEU) during follow-up.
The ACS is an open prospective cohort study that started in 1995 and investigates the epidemiology, the natural history and pathogenesis of HIV-1 infection and other blood-borne and/or sexually transmitted diseases, as well as the effects of interventions. ACS participants complete a standardized questionnaire about their health, risk behaviour and socio-demographical situation every 4-6 months.19, 20 Blood is drawn for laboratory testing and storage. The ACS study was approved by the Institutional Review Board of the Academic Medical Center at the University of Amsterdam and ethical committees/board of directors of each institute recruiting participants. The assigned study numbers are MEC 07/182 and MEC 09/040.
2.1.3 DNA isolation and genotyping
Single nucleotide polymorphisms were selected covering the array of HCV entry genes, with only those that had a minor allele frequency (MAF) score >0.05 being included. DNA was isolated from 200 µL serum with the QIAamp DNA blood mini kit according to the manufacturers protocol (Qiagen, Venlo, the Netherlands). The SNPs were determined for each sample by the appropriate SNP assay or by sequencing. SNP assay: The genotypes were assessed using the Ready-to-use hot start reaction mix for High Resolution Melting (HRM) curve analysis using the LightCycler® 480 (Roche, Almere, the Netherlands) in a volume of 20 µL as follows: 10 µL HRM 2x master mix, 2 µL MgCl2 (25 mmol/L), 0.4 µL α-casein, 0.18 µL Fwd primer (50 µmol/L, Biolegio), 0.18 µL Rev primer (50 µmol/L, Biolegio, Nijmegen, the Netherlands), 5.24 µL Baker water, 2 µL DNA template. Amplification was performed under the following conditions: 50°C for 2 minute, denaturation at 95°C for 10 minute, followed by 45 cycles at 95°C for 15 seconds and 60°C for 15 seconds, 72°C for 20 seconds, followed by a HRM protocol of 95°C for 1 minutes, 40°C for 1 minutes and an acquisition step of 60°C for 45 seconds. Sequencing: PCR amplification was performed with a touchdown-PCR protocol with the following cycling conditions: denaturation at 95°C for 5 minutes, followed by 5 cycles at 94°C for 30 seconds, 61°C for 30 seconds (−0.5°C every cycle) and 72°C for 45 seconds followed by 32 cycles at 94°C for 30 seconds, 60°C for 30 seconds and 72°C for 45 seconds and a final extension step at 72°C for 10 minutes. The amplicons were sequenced in both directions with the same primers using Big dye terminator according to manufacturer's instructions (Applied Biosystems, Inc, Norwalk, CT, USA).
2.1.4 Measurement of LDL cholesterol level
The determination of the LDL serum levels was calculated using the Friedewald formula; LDL cholesterol = total cholesterol level- HDL cholesterol level- (0.45 × triglyceride level) mmol/L. Total cholesterol and HDL cholesterol determination were performed using the Cobas c702 (Roche Diagnostics).
2.2 Statistical analysis
Univariable logistic regression analysis was used to estimate the odds ratio (OR) and corresponding 95% confidence interval and P-values between MEU and MEI using R software version 3.2.2.21 An additive genetic model was used with the minor allele as the coded allele. This model assumes a linear increase (b) in the ln-odds ratio for each additional coded allele an individual carries. The resulting odds ratio are thus eb and e2b for an individual carrying one or two copies of the coded allele vs a person carrying null copies of the coded allele (ie, homozygous for the non-coded allele). A (conservative) Bonferroni correction for multiple testing was applied by dividing the nominal significance threshold by the number of SNPs in the study. Thus, a P value of <0.00185 (0.05/27) was considered significant. Markers were removed when not in Hardy-Weinberg equilibrium in the controls (P < 0.0001) or when more than 10% of the samples were missing.
3 RESULTS
3.1 Association between HCV susceptibility and host SNP genotypes
Participant characteristics are shown in Table 1. Individuals from the MSM exposed MOSAIC (n = 62) and IDU exposed ACS (n = 62) cohorts) were genotyped for 27 SNPs in four different genes known to be involved in HCV infection (No SNPs were selected in CD81 and OCLN). We investigated whether associations could be found between the selected SNPs and HCV infection susceptibility. SNP genotype frequencies were compared between cases (HCV infected or ever HCV infected, n = 54) and high-risk controls (HCV uninfected n = 70). A marginal significant effect was found for rs9869236 in CLDN1 (OR: 0.46 P = 0.016) and in NPC1L1 rs217434 (in combined cohort but also in MOSAIC alone) and rs2072183 (OR: 2.07 P = 0.032 and OR: 1.76 P = 0.039, respectively) but did not pass the Bonferroni test for multiple testing. We found the rs688 T variant in the LDL-R gene to be significantly associated with decreased HCV susceptibility (OR: 0.41 P = 0.0001). When the cohorts were analyzed separately the same trend was observed. A significant effect was found in the MOSAIC cohort but not when corrected with Bonferroni (OR: 0.37 P = 0.0176) and the same trend, although not significant, was found in the ACS cohort (OR: 0.22 P = 0.09; Table 2).
| Characteristics | MOSAIC | ACS | ||
|---|---|---|---|---|
| MEI | MEU | MEI | MEU | |
| n (total = 124) | 32 | 30 | 22 | 40 |
| Mean age ± SD | 43.0 ± 6.9 | 48.5 ± 7.9 | 52.0 ± 6.9 | 52.8 ± 7.2 |
| % Male gender | 32 (100%) | 30 (100%) | 11 (50%) | 29 (72.5%) |
| % Dutch Nationality | 28 (87.5%) | 29 (96.7%) | 19 (86.4%) | 37 (92.5%) |
| % HIV positive at entry | 32 (100%) | 30 (100%) | 0 (0%) | 0 (0%) |
| HIV seroconversion during follow-up | n.a | n.a | 3 (13.6%) | 0 (0%) |
| Median start date of Follow-up (IQR) | 22/2/2011 (4/2/2010-2/8/2011) | 14/2/2011 (19/5/2010-20/12/2011) | 23/2/1988 (15/1/1987-08/02/1992) | 20/10/1992 (12/09/1988-22/04/1998) |
| Median time of follow-up ± SD | 4.01 ± 1.80 | 3.78 ± 1.30 | 14.96 ± 5.65 | 14.31 ± 5.62 |
| Mean duration IDU in years | 4 IDU in last 6 mo (no duration) | n.a | 7.21 ± 3.42 | 8.45 ± 4.83 |
| % Reported sharing of needlesa | 0 (0%) | 0 (0%) | 15 (75%) | 22 (55%) |
| Having an HCV-infected sex partner* | 7 (21.9%) | 1 (3.3%) | n.a | n.a |
| Fistinga | ||||
| With steady partner | 9 (28.1%) | 5 (16.7%) | n/a | n/a |
| With casual partner(s) | 10 (31.3%) | 8 (26.7%) | n/a | n/a |
| Use of sex toysa | ||||
| With steady partner | 13 (40.6%) | 12 (40%) | n/a | n/a |
| With casual partner(s) | 15 (46.9%) | 4 (13.3%) | n/a | n/a |
| Rectal bleeding during or after sexa | ||||
| With steady partner | 10 (33.3%) | n/a | n/a | |
| With casual partner(s) | 15 (46.9%) | 8 (26.7%) | n/a | n/a |
| Group sexa | 24 (75%) | 23 76.7%) | n/a | n/a |
| Rectal bleeding during or after sex* | 17 (53.1%) | 5 (16.7%) | ||
| CD4 count last negative moment(cases)/last visit (controls)* | 523 ± 138 | 621 ± 222 | n.a | n.a |
| CD4 count nadir | 277 ± 160 | 269 ± 179 | n.a | n.a |
| Baseline Mosaic Risk score (medium)10,* | 2.9 | 1.1 | ||
- na, not applicable.
- a Reported at least once.
- *P < 0.05.
| Gene | SNP | Major allele | Minor allele | Combined cohorts | MOSAIC | ACS | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF MEU | MAF MEI | OR | 95% CI | P-value | MAF MEU | MAF MEI | OR | 95% CI | P-value | MAF MEU | MAF MEI | OR | 95% CI | P-value | ||||
| CLDN1 | rs8298 | C | T | 0.343 | 0.439 | 1.41 | 0.86-2.32 | 0.176 | 0.362 | 0.452 | 1.32 | 0.7-2.49 | 0.3914 | 0.329 | 0.417 | 1.47 | 0.64-3.37 | 0.3659 |
| rs8798 | G | A | 0.358 | 0.449 | 1.37 | 0.84-2.23 | 0.2059 | 0.362 | 0.452 | 1.32 | 0.7-2.49 | 0.3914 | 0.355 | 0.444 | 1.43 | 0.65-3.15 | 0.3805 | |
| rs1060679 | C | T | 0.152 | 0.083 | 0.55 | 0.25-1.21 | 0.1368 | 0.183 | 0.063 | 0.43 | 0.15-1.21 | 0.1089 | 0.128 | 0.114 | 0.85 | 0.25-2.92 | 0.7998 | |
| rs17500920 | T | A | 0.123 | 0.111 | 0.9 | 0.42-1.9 | 0.7813 | 0.117 | 0.141 | 1.18 | 0.46-3.01 | 0.726 | 0.128 | 0.068 | 0.46 | 0.11-1.88 | 0.2789 | |
| rs9842576 | G | A | 0.383 | 0.480 | 1.34 | 0.85-2.13 | 0.2107 | 0.308 | 0.450 | 1.57 | 0.8-3.1 | 0.1893 | 0.434 | 0.526 | 1.32 | 0.67-2.61 | 0.4219 | |
| rs55724972 | C | A | 0.109 | 0.102 | 0.94 | 0.45-1.98 | 0.8786 | 0.115 | 0.133 | 1.13 | 0.43-3 | 0.8053 | 0.105 | 0.053 | 0.56 | 0.14-2.35 | 0.4305 | |
| rs1043747 | A | C | 0.383 | 0.480 | 1.35 | 0.85-2.16 | 0.2046 | 0.269 | 0.450 | 1.83 | 0.9-3.7 | 0.0932 | 0.461 | 0.526 | 1.23 | 0.61-2.47 | 0.5561 | |
| rs3172404 | G | A | 0.185 | 0.200 | 1.08 | 0.61-1.91 | 0.7985 | 0.241 | 0.150 | 0.66 | 0.3-1.47 | 0.3121 | 0.145 | 0.275 | 1.96 | 0.82-4.72 | 0.1327 | |
| rs9869263 | G | A | 0.307 | 0.167 | 0.46 | 0.25-0.87 | 0.0159* | 0.183 | 0.109 | 0.57 | 0.21-1.55 | 0.2704 | 0.400 | 0.250 | 0.48 | 0.2-1.13 | 0.0938 | |
| rs9848283 | A | G | 0.400 | 0.472 | 1.29 | 0.8-2.07 | 0.2921 | 0.350 | 0.453 | 1.39 | 0.74-2.61 | 0.3126 | 0.438 | 0.500 | 1.29 | 0.61-2.71 | 0.5038 | |
| rs17429833 | G | C | 0.107 | 0.065 | 0.58 | 0.23-1.48 | 0.2512 | 0.117 | 0.047 | 0.41 | 0.1-1.6 | 0.198 | 0.100 | 0.091 | 0.89 | 0.23-3.37 | 0.8624 | |
| rs12696600 | A | C | 0.471 | 0.519 | 1.17 | 0.74-1.86 | 0.5021 | 0.417 | 0.484 | 1.22 | 0.66-2.25 | 0.5248 | 0.513 | 0.568 | 1.24 | 0.6-2.59 | 0.5584 | |
| NPC1L1 | rs217434 | T | C | 0.150 | 0.259 | 2.07 | 1.07-4.03 | 0.0315* | 0.083 | 0.281 | 5.51 | 1.72-17.7 | 0.0041* | 0.200 | 0.227 | 1.17 | 0.48-2.83 | 0.7249 |
| rs10264715 | C | T | 0.093 | 0.130 | 1.43 | 0.65-3.15 | 0.3699 | 0.017 | 0.141 | 8.78 | 1.06-72.92 | 0.0443* | 0.150 | 0.114 | 0.72 | 0.23-2.23 | 0.5704 | |
| rs2072183 | C | G | 0.214 | 0.343 | 1.76 | 1.03-3.01 | 0.0392* | 0.183 | 0.344 | 1.95 | 0.92-4.15 | 0.0832 | 0.238 | 0.341 | 1.65 | 0.73-3.71 | 0.2282 | |
| rs41279633 | C | A | 0.157 | 0.250 | 1.59 | 0.9-2.81 | 0.1096 | 0.033 | 0.250 | 6.66 | 1.49-29.81 | 0.0132* | 0.250 | 0.250 | 1 | 0.46-2.18 | 1 | |
| rs17655652 | A | G | 0.321 | 0.231 | 0.66 | 0.38-1.14 | 0.137 | 0.383 | 0.266 | 0.63 | 0.31-1.28 | 0.203 | 0.275 | 0.182 | 0.57 | 0.22-1.46 | 0.2379 | |
| SR-B1 | rs5888 | C | T | 0.443 | 0.426 | 0.94 | 0.57-1.54 | 0.7946 | 0.450 | 0.422 | 0.9 | 0.45-1.78 | 0.7616 | 0.438 | 0.432 | 0.98 | 0.47-2.04 | 0.9517 |
| rs4238001 | C | T | 0.100 | 0.094 | 0.95 | 0.44-2.03 | 0.8951 | 0.067 | 0.097 | 1.44 | 0.41-5.01 | 0.5713 | 0.125 | 0.091 | 0.77 | 0.27-2.22 | 0.624 | |
| LDL-R | rs1799898 | T | C | 0.214 | 0.213 | 0.99 | 0.48-2.03 | 0.9764 | 0.183 | 0.234 | 1.52 | 0.55-4.21 | 0.4166 | 0.238 | 0.182 | 0.63 | 0.22-1.84 | 0.3989 |
| rs688 | C | T | 0.550 | 0.324 | 0.41 | 0.24-0.7 | 0.001** | 0.417 | 0.203 | 0.37 | 0.16-0.84 | 0.0176* | 0.650 | 0.500 | 0.5 | 0.22-1.13 | 0.0972 | |
| rs5927 | G | A | 0.471 | 0.389 | 0.64 | 0.35-1.16 | 0.1374 | 0.533 | 0.422 | 0.59 | 0.27-1.29 | 0.1876 | 0.425 | 0.341 | 0.56 | 0.21-1.5 | 0.2475 | |
| rs2738464 | C | G | 0.129 | 0.130 | 1.01 | 0.53-1.92 | 0.9831 | 0.150 | 0.109 | 0.77 | 0.32-1.89 | 0.5743 | 0.113 | 0.159 | 1.37 | 0.53-3.51 | 0.5161 | |
| rs2738465 | G | A | 0.279 | 0.287 | 1.04 | 0.62-1.74 | 0.8917 | 0.233 | 0.281 | 1.24 | 0.58-2.64 | 0.572 | 0.313 | 0.295 | 0.93 | 0.44-1.96 | 0.8552 | |
| rs1433099 | G | A | 0.224 | 0.250 | 1.13 | 0.65-1.96 | 0.6727 | 0.259 | 0.200 | 0.77 | 0.36-1.66 | 0.5046 | 0.197 | 0.325 | 1.8 | 0.79-4.13 | 0.1623 | |
| rs2738466 | A | G | 0.182 | 0.260 | 1.5 | 0.83-2.72 | 0.1776 | 0.183 | 0.234 | 1.35 | 0.57-3.22 | 0.4928 | 0.181 | 0.300 | 1.73 | 0.75-3.96 | 0.1966 | |
| rs17242683 | G | A | 0.189 | 0.275 | 1.5 | 0.85-2.65 | 0.1619 | 0.200 | 0.258 | 1.33 | 0.6-2.94 | 0.4819 | 0.181 | 0.300 | 1.73 | 0.75-3.96 | 0.1966 | |
- The most common allele was considered the major allele. An additive genetic model was used to determine the Odds ratio (OR) for the SNPs between multiple exposed uninfected (MEU) and multiple exposed infected (MEI) individuals in relation to infection susceptibility. Rs688 appears to be significantly associated with HCV infection and passed the Bonferroni test for multiple testing when the data of the MOSAIC and ACS cohort are combined. The minor rs688 T allele is more abundant in controls, hence the odds ratio of 0.41 suggests a protective effect of the T allele.
- CI, confidence interval; MAF, minor allele frequency; OR, Odds ratio calculated by dividing frequencies major allele over minor allele.
- *Statistically significant (P = <0.05)
- **Passed Bonferroni test for multiple testing (P = <0.00185)
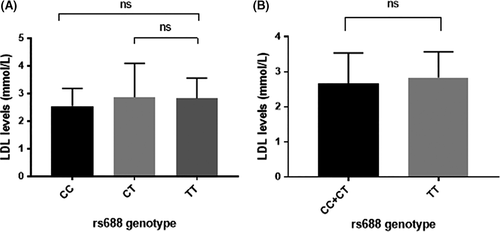
3.2 No association between LDL-R SNP genotypes and cholesterol levels
Since the rs688 genotype within the LDL receptor gene associates with protection from infection, we tested whether serum LDL levels could be associated with HCV susceptibility or with rs688 genotype. We therefore selected participants from the MOSAIC cohort (n = 34) from whom serum was available for measuring LDL levels. No statistical significant differences were found in serum LDL levels for any of the genotypes compared (Figure 1), nor was there a difference in serum LDL levels when HCV MEI were compared with MEU (data not shown).
4 DISCUSSION
We identified a SNP in the LDL-R gene to be associated with susceptibility to HCV infection where the rs688 T allele was found to be associated with decreased susceptibility to HCV infection. The LDL-R plays a role in HCV infection of human hepatocytes. In vitro studies have shown that the receptor is involved in the early stage of HCV infection.22 A study by Petit et al among 68 chronically infected patients found a significant association between HCV viral load and LDL-R expression levels, suggesting that LDL-R is involved in HCV infection and/or replication. We report for the first time an association between LDL-R rs688 and HCV infection susceptibility.
The rs688 LDL-R SNP is located within exon 12 and represents a silent codon mutation. However, this SNP has been shown to affect cholesterol levels and affect splicing of the mRNA resulting in a loss of functional surface expression levels of the receptor. Rs688 has also been associated with total plasma cholesterol levels, together with rs7412 and rs429358 in APOE, rs646776 in CELSR2, rs1367117 in APOB, rs6756629 in ABCG5, rs662799 in APOA5, rs10889353 in DOCK7, rs2304130 in NCAN, rs3846662 in HMGCR, rs2275543 in ABCA1, rs7275 in SMARCA4. All these genes are involved in the cholesterol pathway.23 A study by Gao et al have previously analyzed the mechanism whereby the silent SNP rs688 affects LDL-R expression and demonstrated that rs688 caused an increase in exon 12 alternative splicing, which affects translation of LDL-R resulting in reduced full-length functional LDL-R cell surface expression through introducing a premature stop codon and triggering non-sense mediated decay (NMD). The rs688 TT (minor variant) caused a 6% reduction in splicing efficiency compared to the CT or CC genotype (mRNA not significantly lower, however less for the TT genotype). They also looked at the SNP effect on full-length mRNA production and showed that there was reduced surface expression of LDL-R (21.8%), more LDL-R in the lysosome (25.7%) and reduced uptake of the receptor (24.3%). For these studies, they utilized a plasmid containing the full LDL-R mRNA sequence with and without the SNP introduced. These experiments suggest that besides alternative splicing the rs688 SNP influences LDL-R activity via impaired LDL-R recycling and/or PCSK9 binding.
Since the rs688 LDL-R SNP affects the receptor surface expression level and LDL-R plays a role in HCV cell entry, we hypothesize that in people with the LDL-R rs688 T allele, HCV cell entry is impaired, reducing the risk of acquiring an HCV infection. However, this explanation may be too simplistic. A recent paper by Yamamoto et al24 demonstrated that LDL-R and SR-B1 redundantly participate in HCV entry. When both genes are systematically knocked out, there is a major decrease in HCV entry in comparison to the single knock out phenotypes. Entry in the double knock outs could be rescued by exogenous expression of SR-B1, LDL-R and also VLDLR.
Previously, it has been shown that LDL-R genotype correlates with LDL levels in serum.25-28
As we hypothesized that serum LDL levels could contribute to HCV susceptibility, we compared LDL levels between MEU and MEI, and between the different LDL-R genotypes. However, we did not observe such an effect in the MOSAIC cohort. This could be due to the fact we are only able to test a small subset of individuals. However, this suggests that the protective effect of the LDL-R rs688 T allele in this population is not mediated through serum LDL levels. Further research is warranted to gain better insights into the mechanisms behind the rs688 SNP influencing HCV infection susceptibility and the interaction with other factors.
We observed a marginal effect for rs9869236 in CLDN1 and rs217434 and rs2072183 in NPC1L1 with HCV susceptibility. These SNPs show an association with HCV infection but do not pass the Bonferroni correction. However, the Bonferroni correction is rather strict and some have suggested that Bonferroni correction is not needed unless testing large number of SNPs. Our results suggest that genetic variability in CLDN1 and NPC1L1 may play a role in HCV infection susceptibility but this topic obviously requires further testing in larger cohorts.
Here, we report that the LDL-R rs688 T variant is enriched within MEU. This effect seems to be independent of the transmission route of the virus, which is either through mucosal or IDU exposure. The strong correlation identified using a relatively small number of well characterized individuals is highly supportive of a biological effect, however, this finding needs to be confirmed using larger numbers of individuals.
ACKNOWLEDGEMENTS
We would like to thank all ACS and MOSAIC participants as well as Margreet Bakker and Astrid Newsum for cohort management support. This work was conducted within the framework of the Amsterdam Cohort Studies on HIV infection and AIDS (Website: www.amsterdamcohortstudies.org), a collaboration between the Public Health Service of Amsterdam, the Academic Medical Center of the University of Amsterdam, Sanquin Blood Supply Foundation, the University Medical Center Utrecht and the Dutch HIV Monitoring Foundation. The MOSAIC cohort was supported by the "AIDS Fonds"(grant numbers 2008 026 and 2013 037) and funding was provided from the European Community's Seventh Framework Programme [FP7-2007-2013] under grant agreement no. HEALTH-F3-2012-305578.
CONFLICT OF INTEREST
The authors do not have any disclosures to report.




