Weak population-genetic structure of a widely distributed nematode parasite of frogs in the western Palearctic
Contributing authors: Michaela Mešková ([email protected]), Martin Cyprich ([email protected]), Daniel Jablonski ([email protected]), Petr Papežík ([email protected]), Diyar Hamidi ([email protected]), Çiğdem Akın Pekşen ([email protected]), Judit Vörös ([email protected]), David Herczeg ([email protected]), Michal Benovics ([email protected])
Abstract
The genetic structure of parasite populations is affected by various factors such as host–parasite interactions, life-history strategies, and the evolutionary histories of both interacting organisms. In this study, we investigated the distribution, prevalence, and population-genetic structure of Icosiella neglecta (Spirurida, Onchocercidae), a nematode parasite found in Ranidae frogs. We reported this parasite from eight species of water frogs (genus Pelophylax) in Europe, the Middle East, and Central Asia. Its prevalence across investigated localities varied from 3.03% to 95.83%. Based on nucleotide variation in a 28S ribosomal RNA gene, all investigated I. neglecta sequences formed a well-supported phylogenetic clade and were placed in the sister position to the genus Ochoterenella. Despite the substantial genetic variability in a mitochondrial cytochrome c oxidase subunit I (COI) fragment (33 unique haplotypes recognized among 91 sequences), we found only weak population-genetic structure across the study area. There was no obvious association of COI haplotypes with geography, except haplotypes from eastern Turkey, Lebanon, and Iraq which formed a homogeneous, albeit only weakly differentiated group. The historical demographic analyses suggest that the species underwent a sudden and relatively recent population expansion. According to our results, we assume that the population-genetic structure of I. neglecta might be linked to the evolutionary history and dispersal of its dipteran vectors than water frog hosts.
1 INTRODUCTION
Population-genetic structure is a key evolutionary parameter that helps to understand what extent distinct populations have embarked on separate evolutionary trajectories. While the weak genetic structure points to extensive gene flow, strong population differentiation implies divergence and potential incipient allopatric speciation. In organisms that come into close biological interactions, like parasites and their hosts, the population-genetic structure of one species can be affected by the life and evolutionary histories of its ecological counterparts. In host–parasite systems, dispersal capabilities of hosts can ultimately facilitate gene flow among parasite populations (Blouin et al., 1995; Louhi et al., 2010; Prugnolle et al., 2005). Since parasites strongly depend on their hosts, the biology of the hosts, host specificity, and the complexity of life cycles are key factors determining the genetic structure of parasite populations (Barrett et al., 2008; Cole & Viney, 2018). Parasite species with a wide spectrum of hosts, high level of dispersal, and complex life cycles exhibit higher inter-population gene flow than host specialists with limited dispersal, whose populations are spatially more structured because of the stronger genetic drift and local selection (Archie & Ezenwa, 2011; Brant & Orti, 2003; Gustafson et al., 2018).
Besides the contemporary interactions with hosts, the spatial population structure and genetic diversity of parasites might also mirror historical processes like bottlenecks, colonization events, or vicariance. One of the most important historical processes which left deep imprints in the current patterns of intraspecific genetic diversity is range retractions and expansions associated with Pleistocene climatic oscillations (Hewitt, 2000, 2011; Schmitt, 2007). During cold periods (glacials), the ranges of thermophilic species in temperate zones contracted to climatically favorable areas (refugia) and expanded during interglacials. In the western Palearctic, such refugia were mainly located in southern European peninsulas, the Middle East, and the Transcaucasia (Stewart et al., 2010; Taberlet et al., 1998). Long-term persistence of populations in southern refugia during multiple glacial cycles induced genetic differentiation and led to high genetic and taxonomic diversity, the “southern richness” pattern (Hewitt, 2000, 2004; Taberlet & Cheddadi, 2002). By contrast, repeated demographic bottlenecks associated with postglacial expansions from refugia and rapid colonization of the north produced populations with reduced genetic diversity, the “northern purity” pattern. This general scenario has its specific exceptions, like the existence of cryptic northern refugia (Stewart & Lister, 2001) or multiple refugia (Gómez & Lunt, 2007), which make the general biogeographic pattern more complex.
The existence of glacial refugia may explain the unequal distribution of genetic diversity in water frogs from the genus Pelophylax, in Europe. Currently, at least 12 species and three asexual hybrid forms are recognized in the western Palearctic (Frost, 2021; Plötner, 2005). The highest species diversity is observed in the eastern Mediterranean and the Middle East and is probably associated with the complex past geological events and paleoclimatic changes promoting allopatric speciation (Akın et al., 2010; Beerli et al., 1996; Lymberakis et al., 2007; Plötner et al., 2010). Central and Northern Europe is inhabited by only two species and their asexual hybrids whose genetic diversity is declining to the north and west and is putatively connected with the gradual postglacial expansion of frogs from southeastern glacial refugia (Hoffmann et al., 2015; Snell et al., 2005).
Even though the outcomes of the range dynamics on the distribution of genetic diversity during the Quaternary are well-known for many vertebrate species inhabiting temperate zones, little is known about how climatic oscillations influenced the population structure of their parasites. The effect of Quaternary climatic fluctuations on parasitic species distributed in temperate zones is not congruent and depends largely on the host biogeography, host specificity, host switch, and life cycles of the parasites. While a recent phylogeographic pattern of some parasites supports contraction of their ranges to southern refugia (Mediterranean peninsulas) during the last ice age (Callejón et al., 2012; Nieberding et al., 2004, 2005), the genetic structure of others also indicates their survival in northern refugia (Nieberding et al., 2005, 2008) or does not support the existence of any refugial populations (Sakka et al., 2015).
In the present study, we investigated the genetic diversity and population structure of Icosiella neglecta (Diesing, 1851) (Spirurida, Onchocercidae), a common nematode parasite of Palearctic frogs of the family Ranidae (Anderson, 2000). This species was found in several taxa of water frogs (Günther, 1990; Okulewicz et al., 2014) and less often was reported from brown frogs (genus Rana) (Chikhlyaev & Ruchin, 2014, 2021). First stage (L1) larvae live in the host's blood (Anderson, 2000; Olsen, 1986), from where they enter the haematophagous arthropod vector (Forcipomyia velox, Ceratopogonidae and Sycorax silacea, Psychodidae in the case of I. neglecta) (Desportes, 1941, 1942; Linley, 1985) with blood meal. First stage larvae develop in a vector into the infectious stage L3 which is transmitted to the frog during the sucking of blood. Adult stages live in muscles and subcutaneous tissues of their definitive hosts.
Population-genetic structure and phylogeography of two known vector species of I. neglecta, a biting midge F. velox and a drain fly S. silacea, were not studied. However, it might be hypothesized that they also survived the Pleistocene glacial periods in climatically favorable regions from which they expanded at the beginning of the Holocene, as was evidenced for other dipteran species occupying temperate zones of Palearctic and Nearctic (Aransay et al., 2003; Porretta et al., 2011; Trájer et al., 2018; Solecki et al., 2019). Contrarily to the definitive hosts (the water frogs), dipteran vectors reveal higher dispersal capacity because they can passively spread long distances and thus might significantly contribute to the spread of the parasites they transmit (Erisoz Kasap et al., 2015; Mignotte et al., 2021).
Based on knowledge about life-history traits, life cycles, and evolutionary history of the parasite, hosts, and vectors, we predict (1) the overall high genetic variability and low inter-population differentiation of I. neglecta as a vector-transmitted species, parasitizing a relatively wide spectrum of hosts, and (2) higher genetic variability in southern populations which might have served as glacial refugia for the parasite and its hosts and vectors and lower variability in northern regions as a result of the postglacial expansion. To test these hypotheses, we employed a phylogenetic and demographic approach using various molecular markers and investigated I. neglecta populations from localities covering most of its distribution range in the western Palearctic.
2 MATERIAL AND METHODS
2.1 Sampling, microscopic examination and prevalence
Ten species of the genus Pelophylax (n = 1007 individuals), specifically P. cf. bedriagae, P. cf. cerigensis, P. cypriensis, P. epeiroticus, P. esculentus, P. kurtmuelleri, P. lessonae, P. perezi, P. ridibundus, and P. shqipericus, were sampled in 19 European and western and central Asian countries (Table 1, Figure 1). A single water frog individual can host a high number of I. neglecta individuals, and thus, those individuals represent an infrapopulation of a parasite. A single water frog locality may encompass multiple parasite infrapopulations and is marked as a parasite component population (or metapopulation).
| Locality | Country | N | E | Host Species | n | P (%) (CI−95%) | 28S | COI |
|---|---|---|---|---|---|---|---|---|
| Bënjë | Albania | 40.24 | 20.43 | P. kurtmuelleri | 34 | 50.00 (33.67–66.33) | 28S-1 | COI-1, 2 |
| Cekrezi | Albania | 41.00 | 20.10 | P. kurtmuelleri | 7 | — | 28S-1 | — |
| Divjake | Albania | 40.99 | 19.50 | P. kurtmuelleri, P. shqipericus | 42 | 0.00 (0.00–8.92) | — | — |
| Lin | Albania | 41.06 | 20.61 | P. kurtmuelleri | 20 | 20.00 (7.14–42.35) | 28S-1 | COI-1 |
| Nishaj | Albania | 41.69 | 19.59 | P. shqipericus | 20 | 0.00 (0.00–16.68) | — | — |
| Poçem | Albania | 40.51 | 19.70 | P. kurtmuelleri | 24 | 12.50 (3.50–31.00) | 28S-1 | — |
| Qazim Pali | Albania | 40.05 | 19.84 | P. kurtmuelleri | 28 | 67.86 (48.19–82.51) | 28S-1, 2 | COI-1, 2, 7, 8, 21 |
| Shelegur—Gërmenj | Albania | 40.18 | 20.67 | P. kurtmuelleri | 12 | 50.00 (24.27–75.73) | 28S-1 | COI-1 |
| Stjar | Albania | 39.93 | 20.05 | P. kurtmuelleri, P. epeiroticus | 27 | 0.00 (0.00–12.38) | — | — |
| Garni Canyon | Armenia | 40.11 | 44.73 | P. cf. bedriagae | 5 | — | — | COI-1, 6 |
| Lozenets | Bulgaria | 42.21 | 27.80 | P. ridibundus | 2 | — | — | COI-11 |
| Pravo Bardo | Bulgaria | 41.43 | 23.10 | P. kurtmuelleri | 1 | — | — | COI-2 |
| Episkopi | Cyprus | 34.80 | 32.53 | P. cypriensis | 12 | 16.67 (3.05–45.71) | — | COI-1 |
| Albrechtičky | Czechia | 49.71 | 18.10 | P. ridibundus | 1 | — | — | COI-27 |
| Košatka | Czechia | 49.74 | 18.15 | P. esculentus | 1 | — | 28S-1 | — |
| Havraníky | Czechia | 48.81 | 16.01 | P. ridibundus | 2 | — | 28S-1 | COI-1 |
| Boosen | Germany | 52.38 | 14.48 | P. esculentus | 2 | — | 28S-1 | — |
| Kolympia, Rhodes | Greece | 36.25 | 28.11 | P. cf. cerigensis | 10 | 0.00 (0.00–29.08) | — | — |
| Geoponika | Greece | 40.30 | 23.11 | P. kurtmuelleri | 37 | 5.41 (0.97–18.48) | — | COI-25 |
| Igoumenitsa | Greece | 39.54 | 20.20 | P. kurtmuelleri, P. epeiroticus | 39 | 0.00 (0.00–8.59) | — | — |
| Ioannina | Greece | 39.69 | 20.86 | P. kurtmuelleri, P. epeiroticus | 33 | 3.03 (0.16–16.11) | 28S-1 | — |
| Kalogria | Greece | 38.16 | 21.37 | P. kurtmuelleri, P. epeiroticus | 10 | 0.00 (0.00–29.08) | — | — |
| Ropa Valley, Corfu | Greece | 39.63 | 19.79 | P. kurtmuelleri, P. epeiroticus | 11 | 0.00 (0.00–26.45) | — | — |
| Földsziget | Hungary | 47.70 | 17.18 | P. ridibundus | 55 | 3.64 (0.65–12.45) | — | COI-2 |
| Miklósmajor | Hungary | 47.67 | 17.01 | P. ridibundus | 14 | 57.14 (31.72–79.39) | — | COI-24 |
| Osli | Hungary | 47.63 | 17.08 | P. ridibundus | 14 | 50.00 (23.82–76.18) | 28S-1 | COI-1, 18, 21, 22 |
| Dokan | Iraq | 35.95 | 44.98 | P. cf. bedriagae | 3 | — | 28S-1 | COI-15 |
| Okosnicë | Kosovo | 42.62 | 21.26 | P. kurtmuelleri | 10 | 0.00 (0.00–29.08) | — | — |
| Gjonaj | Kosovo | 42.26 | 20.65 | P. kurtmuelleri | 15 | 26.67 (9.67–53.42) | — | COI-1, 4, 5 |
| Spartak | Kyrgyzstan | 43.05 | 74.12 | P. cf. bedriagae | 11 | 27.27 (7.89–59.55) | — | COI-1 |
| Salima | Lebanon | 33.59 | 35.55 | P. cf. bedriagae | 10 | 80.00 (44.65–96.32) | 28S-1 | COI-12, 13, 14 |
| Daychouniyeh | Lebanon | 33.84 | 35.58 | P. cf. bedriagae | 1 | — | 28S-1 | — |
| Besa | Montenegro | 42.25 | 19.08 | P. kurtmuelleri, P. shqipericus | 27 | 25.93 (12.39–46.22) | 28S-1 | COI-1, 9, 10 |
| Virpazar | Montenegro | 42.23 | 19.09 | P. kurtmuelleri, P. shqipericus | 15 | 0.00 (0.00–22.22) | — | — |
| Crkvino | N. Macedonia | 41.66 | 21.82 | P. kurtmuelleri | 1 | — | 28S-1 | COI-3 |
| Mrdaja | N. Macedonia | 41.17 | 22.74 | P. kurtmuelleri | 14 | 7.14 (0.37–31.71) | — | COI-1 |
| Pretor | N. Macedonia | 40.96 | 21.07 | P. kurtmuelleri | 15 | 40.00 (19.09–66.76) | 28S-1 | COI-1 |
| Poiana Mărului | Romania | 45.43 | 22.47 | P. ridibundus | 16 | 68.75 (43.62–86.78) | 28S-1 | COI-1 |
| Lacul Pojarnia | Romania | 45.08 | 29.28 | P. ridibundus | 7 | — | — | — |
| Caraorman | Romania | 45.08 | 29.39 | P. ridibundus | 12 | 0.00 (0.00–24.26) | — | — |
| Baka | Slovakia | 47.90 | 17.47 | P. ridibundus, P. esculentus | 61 | 39.34 (27.75–52.47) | 28S-1 | COI-1, 26, 28, 30, 31 |
| Petržalka | Slovakia | 48.10 | 17.13 | P. ridibundus, P. esculentus, P. lessonae | 48 | 95.83 (85.75–99.25) | 28S-1 | COI-1, 19 |
| Číčov | Slovakia | 47.79 | 17.75 | P. esculentus, P. lessonae | 30 | 3.33 (0.18–17.72) | — | COI-17 |
| Devín | Slovakia | 48.17 | 16.98 | P. ridibundus, P. esculentus | 18 | 38.89 (18.53–62.53) | 28S-1 | — |
| Gbelce | Slovakia | 47.86 | 18.51 | P. ridibundus | 26 | 50.00 (30.38–69.62) | 28S-1 | COI-29 |
| Istragov | Slovakia | 47.85 | 17.56 | P. esculentus, P. lessonae | 23 | 21.74 (8.99–43.34) | 28S-1 | — |
| Kalinkovo | Slovakia | 48.06 | 17.25 | P. ridibundus | 4 | — | — | COI-23 |
| Rusovce | Slovakia | 48.06 | 17.15 | P. ridibundus, P. esculentus, P. lessonae | 72 | 75.00 (63.82–84.22) | 28S-1 | COI-1, 5, 20, 32 |
| Sap | Slovakia | 47.81 | 17.63 | P. esculentus, P. lessonae | 3 | — | — | COI-18 |
| Šulianske jazero | Slovakia | 47.94 | 17.43 | P. ridibundus | 15 | 40.00 (19.09–66.76) | — | COI-1 |
| Šúr | Slovakia | 48.23 | 17.20 | P. ridibundus | 20 | 90.00 (68.01–98.19) | — | COI-1 |
| Zlatná na Ostrove | Slovakia | 47.75 | 17.93 | P. esculentus, P. lessonae | 16 | 25.00 (9.03–50.00) | — | COI-1 |
| Tysaahtelek | Ukraine | 48.46 | 22.32 | P. ridibundus, P. esculentus | 3 | — | 28S-1 | COI-1 |
| Nevyts'ke | Ukraine | 48.68 | 22.40 | P. ridibundus | 5 | — | 28S-1 | COI-2 |
| Sintra | Portugal | 38.82 | −9.40 | P. perezi | 1 | — | — | COI-1 |
| Diyarbakır | Turkey | 38.01 | 40.25 | P. cf. bedriagae | 6 | — | 28S-1 | COI-1 |
| Başkavak | Turkey | 37.57 | 40.89 | P. cf. bedriagae | 18 | 55.56 (33.03–76.35) | 28S-1, 3 | COI-33 |
| Ankara | Turkey | 40.01 | 32.33 | P. cf. bedriagae | 6 | — | 28S-1 | — |
| Günyüzü | Turkey | 39.38 | 31.81 | P. cf. bedriagae | 4 | — | 28S-1 | COI-2 |
| Kayatepe | Turkey | 37.52 | 40.93 | P. cf. bedriagae | 9 | — | 28S-1 | COI-13 |

The taxonomy of Pelophylax water frogs follows Plötner (2005) and Frost (2021). Each individual was assigned to the species. The sex of the water frogs was determined according to the secondary sexual traits (i.e., vocal sacs and nuptial pads in males). Young specimens without recognizable secondary sexual traits were considered juveniles. Water frog species were identified according to morphological traits (Günther, 1990; Plötner, 2005), and species designation was also evaluated using genetic markers, sequences of the mitochondrial ND2 fragment, and in areas of sympatric occurrence and interspecific hybridization (i.e., Central Europe and a part of the Balkans), as well as by microsatellites. Details of ND2 and microsatellite laboratory analyses and the genetic identification of water frogs are given in studies of Plötner et al. (2008), Hoffmann et al. (2015), and Papežík et al. (2021).
A drop of blood taken from the frogs’ facial vein (Forzán et al., 2012) was used for a blood smear. The rest of the blood was stored in 96% ethanol. Blood smears were fixed for 3–5 min in methanol and subsequently stained for 20 min in Giemsa solution. Stained smears were examined using a Leica DM2500 microscope at 1000× magnification.
The prevalence of I. neglecta and its 95% confidence intervals were calculated only for localities with ≥10 microscopically examined frogs, using QP 3.0 (Reiczigel et al., 2019). In seven localities, we found a syntopic occurrence of two water frog species, which allowed us to compare the rate of the prevalence between the species pairs P. ridibundus and P. esculentus (localities Baka, Bratislava - Chorvátske rameno, Devín), P. esculentus and P. lessonae (localities Rusovce, Číčov), and P. kurtmuelleri and P. epeiroticus (localities Stjar, Ioannina). The comparison was carried out using a 2 × 2 contingency table and Fisher's exact test.
2.2 DNA extraction, PCR amplification and sequencing
Parasite DNA was extracted from blood samples of host frogs. Before extraction, 20 to 50 μl of a mixture of ethanol and blood was taken and centrifuged for 2 min at a speed of 5000 × g. After centrifugation, the ethanol was removed and the blood sample was dried in a thermoblock at 55°C. DNA was extracted using a commercial E.Z.N.A Tissue DNA kit (Omega Bio-tek) following standard protocol provided by the manufacturer.
Initially, we amplified seven molecular markers, specifically 12S ribosomal RNA (12S), cytochrome c oxidase subunit I (COI), the myosin heavy chain (myoHC), 70 kilodalton heat shock proteins (hsp70), RNA polymerase II large subunit (Rbp1), 18S ribosomal RNA (18S), and 28S ribosomal RNA (28S), according to Floyd et al. (2005) and Lefoulon et al. (2015). Out of these markers, only 18S, 28S, and COI were successfully amplified, yielding a clear band on an agarose gel without numerous unspecified fragments, and therefore were used in this study. Other markers were not successfully amplified, probably because the parasite DNA was extracted from host blood samples and was therefore in the minority. Primers and polymerase chain reaction (PCR) conditions were as follows: 18S (NEM_18S_F 5′-CGCGAATRGCTCATTACAACAGC-3′, NEM_18S_R 5′-GGGCGGTATCTGATCGCC-3′; Floyd et al., 2005; annealing temperature 54°C, 35 cycles), 28S (F28SF1 5′-CCTCAACTCAGTCGTGATTACC-3′, F28SR2 5′-CTCTGGCTTCATCCTGCTCA-3′; Lefoulon et al., 2015; annealing temperature 60°C, 33 cycles), COI (COIintF 5′-TGATTGGTGGTTTTGGTAA-3′, COIintR 5′-ATAAGTACGAGTATCAATATC-3′; Lefoulon et al., 2015; annealing temperature 55°C, 30 cycles). Polymerase chain reactions were processed in a final volume of 15 μl using a GoTaq Long PCR Master Mix with 1.5 mM MgCl2 (Promega, Madison, Wisconsin, USA) and 0.3 μM of each primer. PCR products were checked on 1% agarose gel and subsequently purified using a combination of enzymes alkaline phosphatase (CIP; 0.2 μl for 10 μl of PCR product) and exonuclease I (ExoI; 0.1 μl; New England Biolabs, Ipswich, Massachusetts, USA) which were incubated with PCR product for 30 min at 37°C and then denatured for 15 min at 80°C. An approximate length of the amplicons was ~750 bp (18S), ~1150 bp (28S), and ~650 bp (COI).
Extracted parasite DNA from the whole host blood sample might contain a high number of L1 larvae carrying different haplotypes. However, we did not observe any heterozygous nucleotide positions in sequences which indicates that only one haplotype likely predominated in a blood isolate and this was preferentially amplified during PCR. Sanger sequencing was carried out by the Macrogene Europe Service Centre (Amsterdam, the Netherlands) and in the Comenius University Science Park (Bratislava, Slovakia) and is deposited in the NCBI GenBank under the accession numbers OL351798–OL351829.
2.3 Phylogenetic and population-genetic analyses
For phylogenetic and population-genetic analyses, 28S and COI genes were used. The sequences of the 28S fragment were used for taxonomical evaluation of I. neglecta and included 54 new sequences and two sequences of Icosiella from GenBank (KP760379 and KP760380) (Table S1). To assess proximity and relative position to other phylogenetically related taxa, sequences of three Ochoterenella spp. (KP760393–KP760395) and two Oswaldofilaria spp. (KP760402, KP760403) were retrieved from GenBank and included in the analyses. Moreover, the representative homolog sequences of Brugia malayi (KP760362), Dipetalonema robini (KP760374), and Onchocerca cervipedis (KX853345), selected from the phylogeny of Lefoulon et al. (2015), served as the outgroup for rooting the phylogram. DNA sequences were aligned using the fast Fourier transform algorithm in MAFFT (Katoh et al., 2002) and subsequently manually trimmed to equalize the length of individual sequences. Phylogenetic trees were inferred by means of Bayesian inference (BI) and maximum likelihood (ML), carried out in MrBayes 3.2 (Ronquist et al., 2012) and RaxML 8.1.10 (Stamatakis, 2014), respectively. BI analysis used the Metropolis-coupled Markov chain Monte Carlo algorithm with two parallel runs of one cold and three hot chains and ran for 107 generations, sampling trees every 100 generations. Initial 30% of all saved trees were discarded as “burn-in” period after checking that the standard deviation split frequency fell below 0.01. Whether the runs converged was checked using Tracer 1.7.1 (Rambaut et al., 2018). Posterior probabilities for each tree node were calculated as the frequency of samples recovering given clade. The clade bootstrap supports for ML trees were assessed by simulating 103 pseudo-replicates. For both analyses, all parameters specifying the evolutionary model were a priori set as variable, simulating a GTR model.
A dataset of the partial COI sequences, including 89 newly generated and two GenBank (KP760188, KP760189) sequences, was used to assess the genetic structure of I. neglecta. All sequences were trimmed manually to unify their length and translated into amino acids to avoid any signal misreads. The level of DNA polymorphism (i.e., haplotype diversity (Hd), nucleotide diversity (π), number of unique haplotypes, number of variable sites, and normalization tests) was assessed using DnaSP 5 (Librado & Rozas, 2009).
Population-genetic structure based on COI haplotypes was analyzed using median-joining haplotype network performed in PopART (Leigh & Bryant, 2015), and the principal component analysis (PCA) carried out in the adegenet package (Jombart, 2008) from R statistical environment (R Core Team, 2021).
2.4 Demographic analyses
The past population dynamics of the mtDNA lineages were inferred using the COI sequence variability and the Bayesian coalescent-based approach of the Bayesian skyline plots (BSPs) (Drummond et al., 2005) as implemented in BEAST 2.6.3 (Bouckaert et al., 2019) and run on XSEDE using the CIPRES Science Gateway 3.3 (Miller et al., 2010). This analysis estimates the effective population size through time and does not require a specific a priori assumed demographic model (Drummond et al., 2005). Both strict and uncorrelated log-normal relaxed molecular clocks were used in several independent BSP analyses. Since the runs with both molecular clocks provided concordant results, the final analyses were run, enforcing a strict molecular clock model. A uniform prior to the substitution rate with the initial value 0.0433 substitution/site/Ma was applied according to Jorge et al. (2018). The TN93 substitution model (Tamura & Nei, 1993), one of the four substitution models offered by BEAST, was chosen as the best-fit model according to the Bayesian information criterion (BIC) by jModelTest 2.1 (Darriba et al., 2012). The base frequencies were set as empirical. The analyses were run repeatedly to check for consistency between the runs, each for 60 million generations sampled every 6000 generations. Convergence, effective sample size (ESS), stationarity, and the appropriate number of generations to be discarded as burn-in (10%) were assessed using Tracer 1.7.1. The resulting BSPs were also summarized in Tracer 1.7.1 with the maximum times as the median of the root height parameter.
In addition to BSPs, the occurrence of historical demographic changes was also assessed by the neutrality-test statistics of Fu's FS (Fu, 1997) and Tajima's D (Tajima, 1989) calculated in DnaSP 5 with estimation of the statistical significance using 10,000 coalescent simulations. Significant negative values of D and FS indicate an excess of low-frequency mutations relative to expectations under the standard neutral model (i.e., strict selective neutrality of variants, constant population size, and lack of subdivision and gene flow) and thus indicate population expansion.
3 RESULTS
3.1 Distribution and prevalence of Icosiella neglecta
Icosiella neglecta parasitized eight species of water frogs, specifically P. cf. bedriagae, P. cypriensis, P. esculentus, P. kurtmuelleri, P. lessonae, P. perezi, P. ridibundus, and P. shqipericus. Its presence was confirmed in the Iberian Peninsula (Portugal), Central and Eastern Europe (Czech Republic, Germany, Hungary, Slovakia, Ukraine), in the Balkans (Albania, Bulgaria, Greece, Kosovo, Montenegro, Northern Macedonia, Romania), the Transcaucasia (Armenia), the Middle East (Cyprus, Iraq, Lebanon, Turkey), and in Central Asia (Kyrgyzstan) (Figure 1).
The prevalence of I. neglecta in water frog populations varied from 3.03% (Ioannina, Greece) to 95.83% (Bratislava - Chorvátske rameno, Slovakia) (Table 1). The parasite was found almost exclusively in adult frogs, as only 2.31% of juveniles (n = 173) was infected. Differences in the prevalence between the water frog species in seven syntopic localities (Stjar, Ioannina, Baka, Bratislava - Chorvátske rameno, Devín, Rusovce, Číčov) (Table 1) were not significant (Fisher's exact tests, p = 0.266–1.000).
3.2 Phylogeny, population structure and demographic analyses
All 17 sequences of 18S from Slovakia, Hungary, Albania, and Northern Macedonia did not show any variability, and this marker was thus considered to be non-informative and unsuitable for population-genetic study.
The final dataset of the 28S encompassing 56 sequences of I. neglecta (Icosiellinae), three Ochoterenella spp. (Waltonellinae), two Oswaldofilaria spp. (Oswaldofilariinae), and outgroup spanned 800 unambiguously aligned nucleotide positions (Alignment S1). A single genetic variant vastly prevailed in almost all investigated I. neglecta sequences. Only sequences from the infrapopulations MIK3208 from Albania and S4 from Turkey differed in three (99.6% of similarity) and one (99.9% of similarity) nucleotide substitutions, respectively. BI and ML analyses generated trees with congruent topologies, and therefore, only BI tree with both posterior probabilities (PP) and bootstrap support (BS) is presented in Figure 2. All investigated I. neglecta sequences formed a well-supported clade and were placed in the sister position to Ochoterenella spp. (PP =1, BS =100).
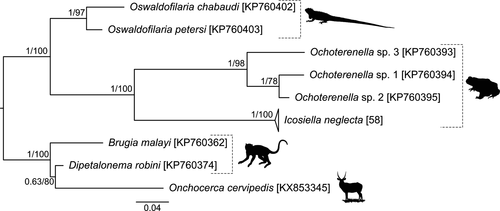
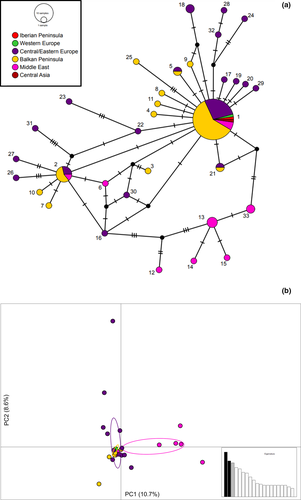
A total of 33 unique COI haplotypes were recognized among 91 I. neglecta sequences. The final length alignment spanned 521 nucleotide positions with 39 polymorphic sites (Alignment S2). The overall haplotype diversity (Hd) was 0.729, and nucleotide diversity (π) reached 0.4%. To compare parameters of genetic diversity among regions, we pooled samples from Central and Eastern Europe, from Transcaucasia and the Middle East, and from the Balkan countries. Since only one COI sequence was analyzed from Western Europe (France; GenBank sequence) and the Iberian Peninsula (Portugal), these regions were not included in the comparison. Genetic diversity reached the highest values in the Transcaucasia and the Middle East (Hd = 0.910, π = 0.7%), followed by populations in Central and Eastern Europe (Hd = 0.848, π = 0.5%), and in the Balkans (Hd = 0.630, π = 0.2%).
Forty-seven infrapopulations from various geographic regions (extending from the Iberian Peninsula to Central Asia) shared the same haplotype 1 (Table 1 and Table S1), which was considered ancestral (Figure 3). Contrastingly, a high number of unique haplotypes were recognized only in a single infrapopulation, suggesting they evolved relatively recently and locally. A group of similar haplotypes (12–15 and 33), genetically diverged from other samples, was observed in eastern Turkey, Lebanon, and Iraq (Figure 3). No obvious geographic structure was found in the rest of the range. PCA produced similar results to those obtained with a haplotype network (Figure 3). A plot of two principal components (accounting for 10.7% and 8.6% of the variance, respectively) showed a separation of only five samples (haplotypes 12–15 and 33) from eastern Turkey, Lebanon, and Iraq.
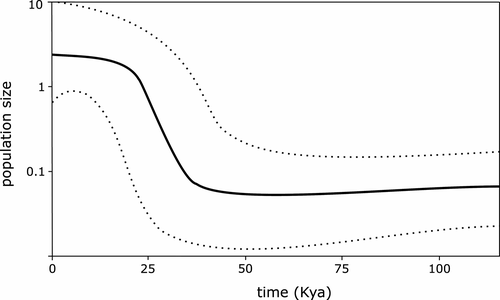
Bayesian Skyline Plots (BSPs) suggested a significant past population expansion during the last ca 30 Kya (Figure 4). A recent population expansion was also indicated by negative and statistically significant values of Fu's FS (−3.944, p < 0.02) and Tajima's D (−2.371, p < 0.01).
4 DISCUSSION
Parasites of amphibians and reptiles have generally been neglected compared to parasites of fish and endothermic vertebrates (Poulin & Morand, 2000; Cole & Viney, 2018; Carlson et al., 2020). In part, this is because that they have no obvious socio-economic or epidemiological importance for humans or livestock. However, their study contributes to the knowledge of biological diversity and the principles that shape it. This study focused on the distribution, prevalence, and genetic diversity of a nematode species I. neglecta, a common parasite of Ranidae frogs. Using a non-lethal sampling and a DNA-barcoding approach, we found that I. neglecta is a widely distributed species, exhibiting high genetic variability in the mitochondrial COI fragment, but has the weak spatial population-genetic structure.
4.1 Distribution of Icosiella neglecta
Our results confirm that I. neglecta has a wide range, from the Iberian Peninsula to Central Asia (Kyrgyzstan). According to the literature survey, the species was also detected in countries that were not involved in this study, specifically France (Desportes, 1941; Lefoulon et al., 2015), including Corsica (Barta et al., 1989), Italy (Comas et al., 2014), Spain (Jiménez et al., 2001; Navarro & Lluch, 2006), United Kingdom [Icosiella neglecta (Diesing, 1851) in GBIF Secretariat 2019], Poland (Okulewicz et al., 2014; Starzynska, 1958), the European part of Russia (Chikhlyaev & Ruchin, 2014, 2021), and Moldova (Gherasim, 2020) (Figure 1). Anderson (2000) reported I. neglecta from North Africa as well but did not specify any localities. According to our knowledge, only two studies dealt with helminth communities of water frogs in North Africa. Specifically, Navarro and Lluch (2006) examined populations of P. saharicus in northern Morocco, and Layla et al. (2018) studied the same species in Libya, but neither of the studies reported I. neglecta. The species was neither reported from Northern Europe despite the occurrence of hosts (Ranidae frogs) and both known vectors, S. silacea and F. velox, in this region (Kvifte et al., 2011; Navai et al., 2017). Whether the absence of I. neglecta in North Africa and Northern Europe reflects unsuitable ecological conditions or a gap in our knowledge remains unclear.
4.2 Population-genetic structure of Icosiella neglecta
Out of three molecular markers, two showed no (18S) or low (28S) intraspecific variability and therefore were not suitable for a population-genetic study. Contrarily, a mitochondrial COI fragment was highly variable and reflected a high mutation rate in this marker (Armenteros et al., 2014; Derycke et al., 2010; Ferri et al., 2009; Jorge et al., 2018).
We found only weak spatial population-genetic structure of I. neclecta across the study area, a pattern which was also observed in some other generalist nematode parasites with low host specificity (Archie & Ezenwa, 2011; Belaich et al., 2015; Blouin et al., 1995; Dubey & Shine, 2008; Sakka et al., 2015). Indeed, there was no obvious association of COI haplotypes with geography, except haplotypes 12–15 and 33 from eastern Turkey, Lebanon, and Iraq, which formed a homogeneous group in a haplotype network and a PCA plot, albeit only weakly differentiated from other haplotypes. It could be assumed that I. neglecta diverged in this region (as was observed in other nematode parasitic species) (Callejón et al., 2012; Nguyen et al., 2019; Nieberding et al., 2008), but is not totally isolated from other regions as is evidenced of the occurrence of a widely distributed haplotype 1 in eastern Turkey.
In general, a COI haplotype network formed a star-like pattern with the most frequent, widely distributed, and centrally located haplotype 1, which is ancestral. Other haplotypes differed from haplotype 1 by a small number of substitutions, suggesting that they evolved only relatively recently and locally. The structure of a haplotype network reflects high haplotype but low nucleotide diversity (i.e., the occurrence of a high number of closely related haplotypes with minor nucleotide differences). This pattern agrees with our first prediction, which postulates high genetic variability and low inter-population differentiation of I. neglecta as a representative of a vector-transmitted species, parasitizing a relatively wide spectrum of hosts (cf. Barrett et al., 2008). By contrast, we did not corroborate the biogeographic pattern of the “southern richness” and the “northern purity” (our second prediction), which we could expect if I. neglecta expanded primarily with its hosts from southern European refugia. High genetic variability of I. neglecta was, however, found in the Middle East and the Transcaucasia which, together with differentiation of the COI haplotypes from these regions, indicates a long-term allopatric divergence. The lack of a significant geographic structure was found, for example, in Heligmosomum mixtum, a nematode parasite whose most common host is the bank vole (Myodes glareolus). Contrarily, in other nematode parasites (e.g., genus Heligmosomoides), the geographic distribution of genetic diversity indicates the existence of southern and northern glacial refugia similar to their hosts (Apodemus mice species) (Nieberding et al., 2004, 2005, 2008).
Since the component populations of I. neglecta in Europe are not geographically structured, we can assume that the current phylogeographic pattern of this nematode might be explained rather by the historical dispersion of the dipteran vectors than their water frog hosts. We can hypothesize that the vectors might have survived harsh conditions in climatically favorable southern regions where transmitted I. neglecta among water frog populations and after the last ice age contributed to the spread of the parasite to the north. Unfortunately, the molecular data to validate this hypothesis are still lacking, but if this hypothesis were valid, we could expect a similar phylogeographic pattern, as we observe in I. neglecta, also in its vectors. Even though high genetic variability and the absence of geographic population structure were observed in other Diptera (e.g., in Phlebotomus and Aedes species) (Esseghir et al., 1997; Porretta et al., 2011, 2012), F. velox and S. silacea were not yet genetically studied.
The hypothesis of postglacial vector-transmitted dispersal of I. neglecta is further supported by demographic analyses. Both Fu's FS and Tajima's D reached significant negative values, indicating an excess of low-frequency mutations relative to expectations under the standard neutral model and thus population expansion. Bayesian skyline plots (BSPs) revealed the imprint of a sudden demographic expansion in the recent evolutionary history of I. neglecta. The estimated time for the beginning of this expansion is roughly 30 Kya. This is just before the last glacial maximum (LGM) in Europe (23–18 Kya) (Hewitt, 2004), and from then on, the species appears to have continued to expand until very recently. This demographic trend closely matches that observed in many other western Palearctic species, for which the demographic expansion has been associated with a postglacial range expansion (Hewitt, 2004, 2011). However, it is necessary to note that the estimated time of the population growth strongly depends on the mutation rate in the analyzed gene, which is unknown in the case of COI in I. neglecta and was based on the mutation rate for other nematode species (Jorge et al., 2018) in this study. Thus, the estimated expansion time of I. neglecta should be taken with caution.
Relatively recent dispersal of I. neglecta also indicates the occurrence of the most common and widely distributed COI haplotype 1 in Cyprus. Water frogs on this island, described recently as a separate Cypriot endemic species P. cypriensis (Plötner et al., 2012), are isolated from the mainland (Anatolia and the Levant region) at least since the early Pleistocene (ca 2.4 Mya) (Poulakakis et al., 2013) or even the late Miocene (ca 5.3 Mya) (Akın et al., 2010; Plötner et al., 2012), according to the different points used for molecular clock calibration. The haplotype 1, therefore, could not have survived on the island unchanged for such a long time and could have been introduced there either by vectors (cf. Depaquit et al., 2008; Erisoz Kasap et al., 2015) or human-mediated introduction of frogs, which was evidenced by Plötner et al. (2015) in the case of Anatolian water frogs recently introduced to Cyprus.
The last factor which might shuffle the genetic diversity and population-genetic structure of I. neglecta, especially in some western and southern European countries, is the human-mediated introduction of its hosts. Many water frog taxa, including those analyzed in our study (P. cf. bedriagae, P. kurtmuelleri, P. ridibundus, and P. shqipericus), have been introduced outward their natural ranges (e.g., to Belgium, France, Italy, and Switzerland), where they became invasive (Domeneghetti et al., 2013; Dubey et al., 2014; Dufresnes et al., 2017; Holsbeek et al., 2008, 2010). Recently, Çiçek et al. (2021) have indicated that Turkey is an important country for the water frog trade. For more than 40 years, live animals have been exported from Turkey to European countries, mainly to France, Italy, Switzerland, and Albania, less to Greece, Spain, and Lebanon (Çiçek et al., 2021). During the export, the live animals can carry their parasites and, after successful introduction, can spread them to non-native areas. Although this threat is real, we do not assume that the human-mediated introduction of hosts has affected the results of our population-genetic study because in our water frog dataset we did not detect mtDNA haplotypes of allochthonous species (P. Papežík, unpublished data). However, this factor should be considered when analyzing parasites of water frogs, especially in some western and southern European countries.
5 CONCLUSIONS
Despite parasitic nematode infections are being ubiquitous in natural populations, principles shaping the distribution of genetic diversity in nematodes parasitizing amphibians are still insufficiently known (Cole & Viney, 2018). In this study, we investigated the distribution and phylogeographic pattern of I. neglecta, a common nematode parasite of Ranidae frogs. Using molecular markers, we found that the distribution of I. neglecta ranges from the Iberian Peninsula to Central Asia and reaches a locally high prevalence. Moreover, observed mitochondrial variability is high, but populations are genetically homogeneous. This population-genetic structure does not reflect the population-genetic structure of its hosts, the water frogs, but might be significantly influenced by dispersal of its dipteran vectors, which, as we hypothesize, might have contributed to relatively recent parasite spread. The future phylogeographic and population-genetic studies focused on I. neglecta vectors could clarify this hypothesis. High genetic variability and low inter-population differentiation of I. neglecta likely reflect its life-history traits as a vector-transmitted species, parasitizing a relatively wide spectrum of hosts. Further research on this parasite should focus on areas where genetic diversity has either not been or has been insufficiently evaluated, such as North Africa, the Iberian and Apennine Peninsula, and Central Asia. Special attention should be projected to the Middle East region, including southeastern Anatolia, Iraq, and Lebanon, where we found unique, albeit weakly differentiated haplotypes, which indicates that this region played an important role in the evolutionary history of I. neglecta.
ACKNOWLEDGMENTS
We are grateful to J. Christophoryová, R. Dankovics, D. Gruľa, D. Hegner, T. Husák, A. Javorčík, D. Koleška, S. I. Mohammed, S. Papežíková, R. A. Sadek, R. Smolinský, I. Stolárik, J. Vorel, and Ž. Živčicová for assistance in the field. We also thank L. Choleva and M. Doležálková-Kaštánková for providing water frog samples, and J. Poláková for technical assistance. The study was supported by the Scientific Grant Agency of the Slovak Republic VEGA 1/0286/19, and the Slovak Research and Development Agency under contract APVV-19-0076.




