Species delimitation and phylogeny of Doto (Nudibranchia: Dotidae) from the Northeast Atlantic, with a discussion on food specialization
Contributing authors: Klas Malmberg ([email protected]), Torkild Bakken ([email protected]), Tatiana Korshunova ([email protected]), Alexander Martynov ([email protected]; [email protected]) and Kennet Lundin ([email protected])
Abstract
The nudibranch genus Doto is taxonomically problematic in particular, and some species are described on the notion of strict monophagy. Here we perform species delimitation on NE Atlantic species, as well as placing them phylogenetically, using two markers: the mitochondrial COI and the nuclear H3. We also study the morphology of the species including radular ultrastructure and review food specificity. Specimens were first divided into potential species using ABDG on both markers, these groups were used as input species for species delimitation analyses using BPP, and analyses were performed with both markers combined and on H3 only. The analyses delimit 11 and eight species, respectively. With the exception of one species for which only COI was available, the differences are found in D. fragilis, which is split into three groups when COI is included and lumped into one with only H3. Doto hystrix is nested within these groups. We also found that specimens from Sweden seemingly close to D. maculata in external morphology have identical sequences as D. coronata. Analysis of food preferences of the species involved in the study contradicts the notion of strict monophagy within Doto.
1 INTRODUCTION
The genus Doto Oken, 1815 comprises over 90 species of small nudibranchs (Molluscabase, 2021), seldom over a centimeter in length, often only half a centimeter or less. They are generally difficult to identify, with taxonomic uncertainties and species complexes (Lemche, 1976; Morrow et al., 1992; Pola & Gosliner, 2010; Shipman & Gosliner, 2015; Thompson & Brown, 1984), coupled with variation in pattern and a scarcity of defining morphological features. The genus is therefore often viewed as the one of taxonomically most problematic groups among the nudibranchs. The family Dotidae, including four genera, is placed within the Dendronotoidea, combining presence of slender body, smooth oral veil, cone-shaped dorsolateral appenadges and absence of the cuticular lining of stomach (Korshunova, Bakken, et al., 2020; Korshunova & Martynov, 2020; Wägele & Willan, 2000).
The species of Doto typically have stout dorsolateral appendages, forming pairs along the body. These are not cerata sensu stricto because of radically different structure and function. The appendages have neither cnidosacs nor apical pore, but instead have small tubercles in several circlets, placed one above the other. Some species have small, simple pseudobranchs placed close to the base of the dorsolateral appendages, on the side facing the dorsal midline. They can be distinguished from the tubercles by being transparent and having a different shape. The front of the head has a smooth and often inconspicuous oral veil that usually has two rounded lateral flap-like extensions. The rhinophores are smooth, fingerlike and are placed in distinct, often flared sheaths. The radula is narrow and uniseriate, and the jaws are frail and reduced, or even absent (Lemche, 1976; Lundin et al., 2020; Odhner, 1936; Thompson & Brown, 1984).
Species of Doto feed on thecate and athecate hydrozoans and are often claimed as specialized on certain hydrozoan species or genera (Morrow et al., 1992; Picton & Brown, 1981). They may have a notable impact on the marine benthic ecosystem because of the predatory pressure on hydroids, but to our knowledge no studies have been made on this. They do not prey upon the hydroid polyps per se, but slice through the perisarc of the hydroid stalks below the polyps and feed on the caenosarc fluid (Thompson & Brown, 1984). In the North Sea area, fully grown specimens are predominantly observed during late winter to early summer, but some species can be observed later in the season. Because of the taxonomic uncertainties of the species complexes, it is difficult to estimate more precise seasonal variation.
The type species of the genus, Doto coronata (Gmelin, 1791), was originally described by Johann Gmelin as Doris coronata (Gmelin, 1791, p. 3105). Lorenz Oken (1815) introduced the genus name Doto Oken, 1815 (Oken, 1815). Nils Odhner made the first attempt at a revision of the species of Doto in the world and suggested a division of the genus into three main subgroups, based on the coloration of the tubercles on the dorsolateral appendages (Odhner, 1936). Doto coronata had a prominent position in one of these subgroups and Doto fragilis (Forbes, 1838) in another. In the 1970, Henning Lemche discovered that Doto coronata constitutes a species complex, based on observations from samples of Doto collected at numerous localities in the North Atlantic, mainly in the northeast (Lemche, 1976). Based on morphology and species-specific preferences for hydroid prey, Lemche (1976) described five new species from the coronata complex; D. dunnei Lemche, 1976, D. eireana Lemche, 1976, D. koenneckeri Lemche, 1976, D. millbayana Lemche, 1976, and D. tuberculata Lemche, 1976; as well as redescribing older species and establishing a neotype for D. maculata (Montagu, 1804), thus doubling the number of known species from the North East Atlantic area. Picton and Brown (1981) acknowledged that Doto fragilis constitutes a species complex and described the species Doto hystrix Picton & Brown, 1981. Thompson and Brown (1984) indicated that Doto fragilis feeds on hydroids of several different genera such as Halecium Oken, 1815, Nemertesia Lamouroux, 1812, and Tubularia Linnaeus, 1758, but did not separate any new species from the D. fragilis complex, although “varieties” associated with food were mentioned. They concluded that D. fragilis was quote “typically found in association with Nemertesia and Halecium” (Thompson & Brown, 1984: 32); thus, the taxonomic value for prey preference was partially dismissed. Picton and Morrow (1994, 2016) did not separate any species from within D. fragilis either, but mentioned that D. fragilis in the British Isles consists of three different forms, feeding on different hydroids. Morrow et al. (1992) introduced genetic methods for studies of Doto and used electrophoresis to separate two new species from the D. coronata complex; Doto sarsiae Morrow et al., 1992 and Doto hydrallmaniae Morrow et al., 1992, also indicating that other morphs of D. coronata may represent yet other species. It would take another 18 years until genetic studies using Sanger sequencing methods were presented, including North Atlantic and Indo-Pacific Doto species (Pola & Gosliner, 2010; Shipman & Gosliner, 2015). Most species separated by Lemche were confirmed to be distinct, but not all, e.g., D. millbayana did not reveal significant molecular divergence from D. dunnei (Shipman & Gosliner, 2015). The North Atlantic Doto still contains unresolved species complexes with minor differences in morphology, but found on different hydroids, such as for D. fragilis, and more work needs to be done before the phylogeny of the whole group is elucidated. A promising method to untangle the species complexes is to delimit species using multi-locus analysis based on the multi-species coalescent (MSC) method (Knowles & Carstens, 2007; Yang & Rannala, 2010). This method has for nudibranchs recently been applied to the genus Amphorina (see Korshunova, Malmberg, et al., 2020).
2 MATERIALS AND METHODS
One of the aims of the present study is to test the species limits of the Doto species occurring in the North East Atlantic area, by using samples from Sweden, Norway and Northern Ireland (see Table 1, Table S1 and Figure 1 for sampling localities). We particularly wanted to investigate potential species hidden within our current understanding of D. fragilis and D. maculata, in relation to D. coronata. For this analysis, one mitochondrial and one nuclear gene were used. In addition, we are placing the species phylogenetically by analyzing our data together with already published data. Further, we test the results from the molecular analysis against morphology and radular ultrastructure for congruence, using the unified species concept (de Queiroz, 2007). Distance-based single-locus species delimitation was used to generate primary species hypotheses, which then were tested using a MSC-based multi-locus species delimitation method. In this model, genes evolve inside a species phylogeny, the branches are species, and their properties restrict the gene trees. One of these restrictions is that the divergence times between species have to be more recent than the coalescent times for any genes shared between them, assuming no genetic transfer after speciation (Rannala & Yang, 2003). This model can be used for statistical testing of species assignments (Fujita et al., 2012; Rannala, 2015) and has been shown to outperform distance methods (Yu et al., 2017). Thus, we could achieve a more robust model of taxonomy and phylogeny of the genus Doto in the Northeast Atlantic than previous studies, as a step on the way to a more conclusive model on a larger scale.
| Species | Museum voucher no. | COI cluster | H3 cluster | PSH | Country | GenBank accession no. | |
|---|---|---|---|---|---|---|---|
| COI | H3 | ||||||
| Doto coronata | Gastr.9344 | — | 2 | 4 | SE | — | MZ926919 |
| Doto coronata | Gastr.9345 | — | 2 | 4 | SE | — | MZ926920 |
| Doto coronata | Gastr.9067 | 4 | 2 | 4 | NI | MZ902283 | MZ926915 |
| Doto coronata | Gastr.9068 | 4 | 2 | 4 | NI | MZ902284 | MZ926916 |
| Doto coronata | NTNU-VM-63030 | 4 | — | 4 | NO | MZ902294 | — |
| Doto coronata | NTNU-VM-62587 | 4 | — | 4 | NO | MZ902295 | — |
| Doto coronata | NTNU-VM-65472 | 4 | — | 4 | NO | MZ902285 | — |
| Doto coronata | NTNU-VM-65471 | 4 | — | 4 | NO | MZ902286 | — |
| Doto coronata | NTNU-VM-67969 | 4 | — | 4 | NO | MZ902292 | — |
| Doto coronata | NTNU-VM-67970 | 4 | — | 4 | NO | MZ902293 | — |
| Doto coronata | NTNU-VMc76152 | 4 | — | 4 | NO | MZ902315 | — |
| Doto coronata | NTNU-VM-76153 | 4 | — | 4 | NO | MZ902317 | — |
| Doto coronata | NTNU-VM-66933 | 4 | — | 4 | NO | MZ902300 | — |
| Doto coronata | NTNU-VM-66932 | 4 | — | 4 | NO | MZ902301 | — |
| Doto coronata | NTNU-VM-66924 | 4 | — | 4 | NO | MZ902306 | — |
| Doto coronata | NTNU-VM-66931 | 4 | — | 4 | NO | MZ902307 | — |
| Doto coronata | NTNU-VM-66926 | 4 | — | 4 | NO | MZ902305 | — |
| Doto coronata | NTNU-VM-76154 | 4 | — | 4 | NO | MZ902319 | — |
| Doto coronata | NTNU-VM-76180 | 4 | — | 4 | NO | MZ902321 | — |
| Doto coronata | NTNU-VM-76024 | 4 | — | 4 | NO | MZ902313 | — |
| Doto fragilis | NTNU-VM−76183 | 1 | — | 1 | NO | MZ902316 | — |
| Doto fragilis | NTNU-VM-76182 | 1 | — | 1 | NO | MZ902318 | — |
| Doto cuspidata | Gastr.9057 | — | 7 | 10 | NI | — | MZ926900 |
| Doto cf. cuspidata (Doto sp.) | NTNU-VM-66937 | 6 | — | 6 | NO | MZ902297 | — |
| Doto cf. cuspidata (Doto sp.) | NTNU-VM-66936 | 6 | — | 6 | NO | MZ902299 | — |
| Doto cf. Cuspidata (Doto sp.) | NTNU-VM-66935 | 6 | — | 6 | NO | MZ902298 | — |
| Doto dunnei | Gastr.9058 | 5 | 2 | 5 | NI | MZ902276 | MZ926901 |
| Doto dunnei | Gastr.9490 | 5 | 2 | 5 | SE | MZ902269 | — |
| Doto dunnei | Gastr.9491 | 5 | 2 | 5 | SE | MZ902270 | MZ926890 |
| Doto dunnei | Gastr.9492 | 5 | 2 | 5 | SE | MZ902271 | MZ926891 |
| Doto dunnei | Gastr.9493 | 5 | 2 | 5 | SE | MZ902272 | MZ926892 |
| Doto dunnei | Gastr.9494 | 5 | 2 | 5 | SE | MZ902273 | MZ926893 |
| Doto dunnei | Gastr.9495 | 5 | 2 | 5 | SE | MZ902274 | MZ926894 |
| Doto dunnei | Gastr.9496 | — | 2 | 5 | SE | — | MZ926895 |
| Doto fragilis neotype | Gastr.9061 | 1 | 1 | 1 | NI | MZ902275 | MZ926897/MZ926898 |
| Doto fragilis | Gastr.9060 | — | 1 | 1 | NI | — | MZ926896 |
| Doto fragilis white morph | Gastr.9473 | 1 | 1 | 1 | SE | MZ902244 | MZ926922 |
| Doto fragilis white morph | Gastr.9474 | 1 | 1 | 1 | SE | MZ902245 | MZ926923 |
| Doto fragilis white morph | Gastr.9475 | 1 | 1 | 1 | SE | MZ902246 | MZ926924 |
| Doto fragilis white morph | Gastr.9476 | — | 1 | 1 | SE | — | MZ926925 |
| Doto fragilis white morph | Gastr.9477 | — | 1 | 1 | SE | MZ902247 | MZ926926 |
| Doto fragilis white morph | Gastr.9478 | 1 | 1 | 1 | SE | MZ902248 | MZ926927 |
| Doto fragilis white morph | Gastr.9479 | 1 | 1 | 1 | SE | MZ902249 | MZ926928 |
| Doto fragilis | NTNU-VM-76138 | 1 | — | 1 | NO | MZ902314 | — |
| Doto fragilis | NTNU-VM-65470 | 1 | — | 1 | NO | MZ902287 | — |
| Doto fragilis | NTNU-VM-65538 | 1 | — | 1 | NO | MZ902289 | — |
| Doto fragilis | NTNU-VM-65537 | 1 | — | 1 | NO | MZ902290 | — |
| Doto fragilis | NTNU-VM-65539 | 1 | — | 1 | NO | MZ902291 | — |
| Doto fragilis | NTNU-VM-66940 | 1 | — | 1 | NO | MZ902302 | — |
| Doto fragilis | NTNU-VM-67129 | 1 | — | 1 | NO | MZ902308 | — |
| Doto fragilis | NTNU-VM-67128 | 1 | — | 1 | NO | MZ902309 | — |
| Doto fragilis | NTNU-VM-76223 | 1 | — | 1 | NO | MZ902320 | — |
| Doto fragilis | NTNU-VM-76222 | 1 | — | 1 | NO | MZ902322 | — |
| Doto cf. fragilis | Gastr.9499 | 3 | 1 | 3 | SE | MZ902280 | MZ926906 |
| Doto cf. fragilis | Gastr.9500 | 3 | 1 | 3 | SE | MZ902281 | MZ926907 |
| Doto cf. fragilis | Gastr.8671 | 3 | 1 | 3 | SE | MZ902277 | MZ926902 |
| Doto cf. fragilis | Gastr.9497 | 3 | 1 | 3 | SE | MZ902278 | MZ926903/MZ926904 |
| Doto cf. fragilis | Gastr.9498 | 3 | 1 | 3 | SE | MZ902279 | MZ926905 |
| Doto cf. fragilis | Gastr.9343 | 2 | 1 | 2 | SE | MZ902257 | MZ926918 |
| Doto cf. fragilis | NTNU-VM-62667 | 2 | — | 2 | NO | MZ902296 | — |
| Doto cf. fragilis | NTNU-VM-66941 | 2 | — | 2 | NO | MZ902304 | — |
| Doto cf. fragilis | NTNU-VM-65469 | 2 | — | 2 | NO | MZ902288 | — |
| Doto cf. fragilis | NTNU-VM-66939 | 2 | — | 2 | NO | MZ902303 | — |
| Doto hystrix | NTNU-VM-67896 | 1 | — | 1 | NO | MZ902310 | — |
| Doto hystrix | NTNU-VM-67895 | 1 | — | 1 | NO | MZ902311 | — |
| Doto hystrix | NTNU-VM-76126 | 1 | — | 1 | NO | MZ902312 | — |
| Doto hystrix | Gastr.9480 | 1 | 1 | 1 | SE | MZ902250 | MZ926929 |
| Doto hystrix | Gastr.9062 | 1 | 1 | 1 | NI | MZ902266 | MZ926910 |
| Doto hystrix | Gastr.9481 | 1 | 1 | 1 | SE | MZ902251 | MZ926930 |
| Doto koenneckeri | Gastr.9063 | 7 | 4 | 7 | NI | MZ902265 | MZ926911 |
| Doto maculata | Gastr.9488 | — | 3 | 11 | NI | — | MZ926934/MZ926935 |
| Doto cf. maculata (D. coronata) | Gastr.8990 | 4 | 2 | 4 | SE | MZ902260 | MZ926908 |
| Doto cf. maculata (D. coronata) | Gastr.9086 | 4 | 2 | 4 | SE | MZ902261 | MZ926917 |
| Doto cf. maculata (D. coronata) | Gastr.9448 | 4 | 2 | 4 | SE | MZ902262 | MZ926921 |
| Doto cf. maculata (D. coronata) | Gastr.9486 | 4 | 2 | 4 | SE | MZ902259 | MZ926931 |
| Doto cf. maculata (Doto sp.) | Gastr.9489 | 5 | 2 | 5 | NI | MZ902263 | MZ926936 |
| Doto cf. maculata (D. coronata) | Gastr.9487 | 4 | 2 | 4 | SE | MZ902258 | MZ926932/MZ926933 |
| Doto millbayana | Gastr.8951 | — | 2 | 5 | SE | — | MZ926899 |
| Doto pinnatifida | Gastr.9064 | 9 | 6 | 9 | NI | MZ902267 | MZ926912 |
| Doto pinnatifida | Gastr.9065 | 9 | 6 | 9 | NI | MZ902268 | MZ926913 |
| Doto pinnatifida | Gastr.9066 | 9 | 6 | 9 | NI | MZ902282 | MZ926914 |
| Doto tuberculata | Gastr.9056 | 8 | 5 | 8 | NI | MZ902264 | MZ926909 |
| Aeolidia filomenae | Gastr.9482 | SE | MZ902252 | MZ926937 | |||
| Aeolidia filomenae | Gastr.9483 | SE | MZ902253 | MZ926938 | |||
| Aeolidia filomenae | Gastr.9484 | SE | MZ902254 | MZ926939 | |||
| Dendronotus lacteus | Gastr.9446:1 | SE | MZ902255 | MZ926940 | |||
| Dendronotus lacteus | Gastr.9446:2 | SE | MZ902256 | MZ926941 | |||
Note
- Names given in brackets are identifications and corrections as a result of the analysis. For more information see Table S1.
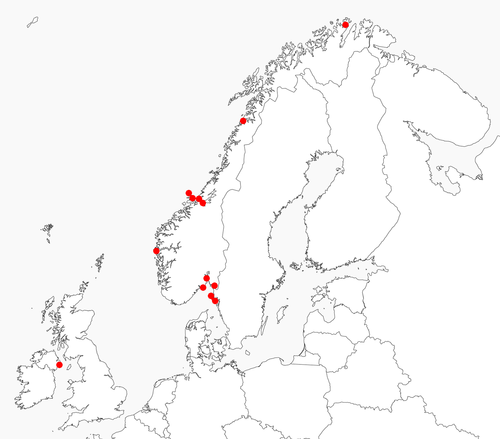
In the molecular study, a total of 82 specimens were included, representing 11 morphospecies of Doto, from the Skagerrak, the Irish Sea, the North Sea, the Norwegian Sea and the Barents Sea. Also, five out-group specimens from two species were used; Aeolidia filomenae Kienberger et al., 2016 and Dendronotus lacteus (Thompson, 1840) (see Table 1 and Table S1 for details). The phylogeny was estimated using our data combined with previously published data.
The Swedish specimens were collected at four different locations on the Swedish west coast, from south to north at the mouth of the Gullmar fjord close to Lysekil, the archipelago outside Smögen, the Väderö archipelago and finally at the Ide fjord at the border to Norway. The specimens from Norway were collected from south to north at the Oslo and Larvik area, at the mouth of the Sognefjord north of Bergen, at the Trondheimsfjord area, at Saltstraumen in Nordland and finally at the Finnmark area in the Arctic. The specimens from Northern Ireland were collected at Strangford Lough close to the Queens University Marine Laboratory at Portaferry. Specimens were deposited at the Gothenburg Natural History Museum (GNM), Gothenburg, Sweden, and at the NTNU University Museum (NTNU-VM) (Bakken et al., 2021), Trondheim, Norway.
2.1 Morphological analysis
The external and internal morphology of 16 specimens of 14 species was studied under a MBS-10 stereomicroscope, using a Nikon D-810 digital camera.
The fine structure of the radula of 14 species from both Sweden and Northern Ireland was studied to cover regional variations at different parts of the Northeast Atlantic. The coated radulae were examined and photographed using a scanning electron microscope (CamScan Series II and JSM 6380).
2.2 Molecular analysis
2.2.1 DNA extraction, amplification and sequencing
DNA was extracted from a small tissue sample taken from the lateral side of the foot or the tail end of the foot on small specimens, using Qiagen's DNeasy Blood & Tissue Kit. Two molecular markers, the mitochondrial gene Cytochrome c oxidase subunit I (COI) and nuclear gene Histone H3 (H3), were amplified using the primers and PCR programs listed in Table 2. Sequencing was carried out by Eurofins MWG Operon (Ebersberg, Germany), Sequences were assembled into consensus sequences using Geneious v.8.1.9 (Biomatters Ltd). The Norwegian specimens were handled by the Canadian Centre for DNA bar coding (CCDB) (Guelph), following their workflow for DNA bar coding, and only COI was sequenced. All new sequences are deposited in GenBank (see Table 1 and Table S1 for accession numbers).
| Gene | Amplicon length | Primer | Sequence 5′-3′ | Reference | Amplification program |
|---|---|---|---|---|---|
| COI | 709 (658) | LCO1490 | GGTCAACAAATCATAAAGATATTGG | Folmer et al. (1994) | 95°C for 5 min, 35 cycles each of 95°C for 40 s, 45°C for 45 s and 72°C for 60 s, finally, 72°C for 8 min |
| HCO2198 | TAAACTTCAGGGTGACCAAAAAATCA | Folmer et al. (1994)) | |||
| H3 | 374 (328) | H3F | ATGGCTCGTACCAAGCAGACVGC | Brown et al. (1999) | 95°C for 5 min, 35 cycles each of 95°C for 30 s, 50°C for 30 s and 72°C for 90 s, finally, 72°C for 8 min. |
| H3R | ATATCCTTRGGCATKATRGTGAC | Brown et al. (1999) |
Note
- The amplicon length is followed, in parentheses, by the length of the fragment with primers removed.
The sequences of each marker were aligned using MAFFT v7.017 (Katoh et al., 2002) as implemented in Geneious. In the H3 dataset, several individuals showed clear signs of heterozygosity, i.e., distinct double peaks at certain positions in the chromatograms. Due to this, we separated the H3 alleles using the PHASE algorithm (Stephens & Donnelly, 2003; Stephens et al., 2001) as implemented in DNAsp v.5.10 (Librado & Rozas, 2009), the phasing was run for 100 iterations after 100 initial burn-in iterations, with a thinning interval of 1 using default settings. For homozygous specimens, only one of the two identical alleles was kept. The phased dataset was used in all subsequent analyses.
2.2.2 Single-locus clustering and distance analyses
Both the COI and the H3 datasets were analyzed with Automatic Barcode Gap Discovery (ABGD) (Puillandre et al., 2012) to divide the specimens into putative species, using the web version of ABGD, using simple distances and default settings. For these analyses, the out-groups were removed. Uncorrected pairwise distances were calculated for both datasets in MEGA 6.06 (Tamura et al., 2013), and missing data and gaps was excluded using pairwise deletions.
2.2.3 Multi-locus species delimitation
Multi-locus species delimitation analyses were performed using BPP v.3.3 (Yang, 2015). The analyses were performed with COI and H3, as well as with only H3, with out-groups excluded, the datasets were divided into 11 primary species hypotheses to be tested based on the result of the ABGD analyses. Joint Bayesian species delimitations and species tree estimations were conducted, a method using the MSC model to compare different arrangements of species delimitation and species phylogeny in a Bayesian framework, accounting for incomplete lineage sorting due to ancestral polymorphism and gene tree–species tree conflicts (Rannala & Yang, 2013; Yang & Rannala, 2010, 2014). Two analyses (A and B) with different population size (θs) and divergence time (τ0) priors were performed, using the same settings and priors as in Martinsson and Erséus (2018) (A: θ 2400, τ0 2200; B: θ 21000, τ0 2200). All analyses were performed three times to confirm consistency between runs. We considered species delimited with a PP>0.90 in all analyses to be well supported. For clusters with a PP<0.90, we accepted the best supported more inclusive species.
2.2.4 Haplotype network
To visualize the relation between different clades within the Doto fragilis complex (see results), a COI haplotype network was created, using the sequences belonging to this species complex, in PopART v1 (Leigh & Bryant, 2015) using the TCS algorithm (Clement et al., 2002; Templeton et al., 1992). Sites with missing data or gaps were masked and not included in the networks, and due to large amount of missing data, specimens Gastr. 9343 and Gastr. 9474 were excluded altogether.
2.2.5 Phylogenetic inference
Phylogenies were estimated using Bayesian Inference on both single gene and the concatenated dataset in MrBayes v.3.2.6 (Ronquist et al., 2012). In total, four analyses were performed: one for each marker, one for a concatenated dataset with our data and one for a concatenated dataset with added sequences of several species of Doto Oken, 1815 and Kabeiro Shipman & Gosliner, 2015 from Pola and Gosliner (2010), Shipman and Gosliner (2015) and Moles et al. (2016). For all analyses, the genes were partitioned according to codon position, and partitions were unlinked. Rate variation across sites was set to gamma distribution with a proportion of invariable sites; model jumping was implemented to integrate over substitution model space. All analyses ran for 10 million generations sampling every 10 000 generations, the first 25% were discarded as burn-in, and a majority rule consensus tree was constructed. Matrices and trees are available on TreeBASE (submission 26369). For the concatenated matrix with our data, a phylogeny was also estimated with maximum likelihood in PhyML 3.0 (Guindon et al., 2010), as implemented at the Montpellier Bioinformatics platform (http://www.atgc-montpellier.fr/), Smart Model Selection (Lefort et al., 2017) with Bayesian information criterion was used for automatic model selection; and nearest neighbor interchange were used for tree improvement. Branch support was calculated with the SH-like (Shimodaira–Hasegawa test-like) approximate likelihood ratio test (aLRT) (Anisimova & Gascuel, 2006) and 1000 bootstrap replicates.
2.3 Review of Doto prey specificity
Thompson and Brown (1984: 28–29) provided at least two different hydroid species as food for four out of 12 Doto species from the British Isles. Distantly related species such as D. fragilis and D. pinnatifida (Montagu, 1804) (Figure 2) were evidently indicated as feeding on the same hydroid species, i.e., Nemertesia antennina (Linnaeus, 1758) (Picton & Brown, 1981; Picton & Morrow, 2016; Thompson & Brown, 1984).
To test the for strict prey specificity within the genus Doto (e.g., Picton & Brown, 1981; Shipman & Gosliner, 2015), we compiled data on the hydroid associations of the Doto species from the available literature sources and own observations (Table 3). For pre-1976 sources (i.e., before Lemche in 1976 showed that “D. coronata” is a complex of different species), we generally did not include records of D. coronata from the same hydroids, as other species split by Lemche and other authors later on (Morrow et al., 1992; Picton & Brown, 1981) were claimed to be almost monophagous. In cases when a hydroid species was not indicated for some of these narrowly defined species, but for D. coronata or other pre-1976 species, we included such hydroids in Table 3. Clytia hemisphaerica (Linnaeus, 1767) was added to the list of food sources of D. coronata, and Nemertesia antennina was added to D. hystrix from our own observations (Table 3).
| Hydroid species | Doto species | Reference |
|---|---|---|
| Abietinaria abietina (Linnaeus, 1758) |
D. coronata (Gmelin, 1791) D. tuberculata Lemche, 1976 |
Lemche, 1976 Picton & Brown, 1981 Thompson & Brown, 1984 |
| Aglaophenia pluma (Linnaeus, 1758) |
D. coronata (Gmelin, 1791) D. koenneckeri Lemche, 1976 D. lemchei Ortea & Urgorri, 1978 |
Swennen, 1961 Picton & Brown, 1981 Thompson & Brown, 1984 Morrow et al., 1992 Present study |
| Aglaophenia sp. |
D. koenneckeri Lemche, 1976 D. lemchei Ortea & Urgorri, 1978 D. millbayana Lemche, 1976 D. paulinae Trinchese, 1881 D. pinnatifida (Montagu, 1804) |
Ortea & Urgorri, 1978 Urgorri & Besteiro, 1983 Just & Edmunds, 1985 Rudman, 2006 Shipman & Gosliner, 2015 Present study |
| Aglaophenia tubulifera (Hincks, 1861) | D. lemchei Ortea & Urgorri, 1978 |
Morrow et al., 1992 Picton & Brown, 1981 Thompson & Brown, 1984 |
| Amphisbetia operculata (Linnaeus, 1758) |
D. coronata (Gmelin, 1791) Doto eireana Lemche, 1976 D. pinnatifida (Montagu, 1804) |
Hecht, 1896 Cornet & Marche-Marchad, 1951 Ortea, 1978 Thompson & Brown, 1984 |
| Bougainvillia muscus (Allman, 1863) |
D. coronata (Gmelin, 1791) |
Miller, 1961 Thompson, 1964 |
| Clava multicornis (Forsskål, 1775) |
D. coronata (Gmelin, 1791) |
Larsen, 1925 Jaeckel, 1952 Miller, 1961 |
| Clytia hemisphaerica (Linnaeus, 1767) |
D. coronata (Gmelin, 1791) |
Present study |
| Coryne eximia Allman, 1859 |
D. coronata (Gmelin, 1791) |
Swennen, 1961 |
| Coryne muscoides (Linnaeus, 1761) |
D. coronata (Gmelin, 1791) |
Miller, 1961 |
| Diphasia fallax (Johnston, 1847) |
D. coronata (Gmelin, 1791) |
Present study |
| Diphasia rosacea (Linnaeus, 1758) | D. coronata (Gmelin, 1791) |
Picton & Brown, 1981 Thompson & Brown, 1984 |
|
Dynamena pumila (Linnaeus, 1758) |
D. coronata (Gmelin, 1791) |
Alder & Hancock, 1846 Lemche, 1976 Picton & Brown, 1981 Thompson & Brown, 1984 |
| Ectopleura larynx (Ellis & Solander, 1786) |
D. coronata (Gmelin, 1791) D. fragilis (Forbes, 1838) |
Walton, 1908 Miller, 1961 Hamond, 1972 Swennen, 1961 |
| Eudendrium ramosum (Linnaeus, 1758) | D. coronata (Gmelin, 1791) | Hamond, 1972 |
| Eudendrium spp. | D. coronata (Gmelin, 1791) | Picton & Morrow, 1994 |
| Garveia nutans Wright, 1859 | D. coronata (Gmelin, 1791) | Picton, 1978 |
| Halecium beanii (Johnston, 1838) | D. coronata (Gmelin, 1791) | Miller, 1961 |
| Halecium halecinum (Linnaeus, 1758) |
D. coronata (Gmelin, 1791) D. fragilis (Forbes, 1838) |
Farran, 1909 Miller, 1961 Thompson, 1964 Hunnam & Brown, 1975 Picton & Brown, 1981 Present study |
| Halecium muricatum (Ellis et Solander, 1786) | D. fragilis (Forbes, 1838) |
Picton, 1978 Picton & Brown, 1981 Thompson & Brown, 1984 |
| Halecium spp. |
D. coronata (Gmelin, 1791) D. fragilis (Forbes, 1838) D. pinnatifida (Montagu, 1804) |
Walton, 1908 Hamond, 1972 Brown & Picton 1979 Urgorri & Besteiro, 1983 Present study |
| Halopteris catharina (Johnston, 1833) | D. maculata (Montagu, 1804) |
Lemche, 1976 Picton & Brown, 1981 Thompson & Brown, 1984 |
| Kirchenpaueria pinnata (Linnaeus, 1758) | D. dunnei Lemche, 1876 |
Lemche, 1976 Picton & Brown, 1981 Thompson & Brown, 1984 |
| Laomedea flexuosa Alder, 1857 | D. coronata (Gmelin, 1791) |
Miller, 1961 Swennen & Dekker, 1987 |
| Lafoea dumosa (Fleming, 1820) | D. coronata (Gmelin, 1791) | Miller, 1961 |
| Nemertesia antennina (Linnaeus, 1758) |
D. coronata (Gmelin, 1791) D. fragilis (Forbes, 1838) D. pinnatifida (Montagu, 1804) D. hystrix Picton & Brown, 1981 |
Jaeckel, 1952 Miller, 1961 Kress, 1968 Thompson, 1964 Lemche, 1976 Picton & Brown, 1981 Thompson & Brown, 1984 Present study |
| Nemertesia norvegica (Sars, 1873) |
Doto fragilis, Doto cf. fragilis |
Present study |
| Nemertesia ramosa (Lamarck, 1816) |
D. cuspidata Alder & Hancock, 1862 D. fragilis (Forbes, 1838) D. millbayana Lemche, 1976 |
Miller, 1961 Thompson, 1964 Lemche, 1976 Just & Edmunds, 1985 Thompson & Brown, 1984 |
|
Obelia geniculata (Linnaeus, 1758) |
D. coronata (Gmelin, 1791) D. paulinae Trinchese, 1881 |
Miller, 1961 Lemche, 1976 Schmekel & Kress, 1977 Picton & Brown, 1981 Thompson & Brown, 1984 Lambert, 1991 Martynov et al., 2006 Rudman, 2006 Shipman & Gosliner, 2015 Present study |
| Obelia dichotoma (Linnaeus, 1758) | D. coronata (Gmelin, 1791) |
Swennen, 1961 Picton & Brown, 1981 Thompson & Brown, 1984 Lambert, 1991 Shipman & Gosliner, 2015 |
| Obelia longissima (Pallas, 1766) | D. coronata (Gmelin, 1791) |
Dekker, 1986 Urgorri & Besteiro, 1983 Martynov et al., 2006 Martynov & Korshunova, 2011 |
| Rhizocaulus verticillatus (Linnaeus, 1758) |
D. coronata (Gmelin, 1791) |
Miller, 1961 |
| Plumularia setacea (Linnaeus, 1758) | D. millbayana Lemche, 1976 |
Lemche, 1976 Picton & Brown, 1981 Just & Edmunds, 1985 Thompson & Brown, 1984 |
| Schizotricha frutescens (Ellis & Solander, 1786) | D. hystrix Picton & Brown, 1981 |
Picton & Brown, 1981 Thompson & Brown, 1984 Present study |
| Sertularia argentea (Linnaeus, 1758) |
D. coronata (Gmelin, 1791) D. dunnei Lemche, 1876 or D. millbayana Lemche, 1876 (recorded as Doto sp.) |
Miller, 1961 Picton & Brown, 1981 Thompson & Brown, 1984 Shipman & Gosliner, 2015 |
| Sertularia cupressina Linnaeus, 1758 | D. coronata (Gmelin, 1791) |
Swennen, 1961 Thompson, 1964 Thompson & Brown, 1984 Shipman & Gosliner, 2015 |
| Sertularella gayi (Lamouroux, 1821) | D. tuberculata Lemche, 1976 |
Lemche, 1976 Picton & Brown, 1981 Thompson & Brown, 1984 |
| Sertularia sp. | D. pinnatifida (Montagu, 1804) |
McMillan, 1944 Moore, 1950 Kress, 1968 |
| Symplectoscyphus tricuspidatus (Alder, 1856) | D. coronata (Gmelin, 1791) | Martynov et al., 2006 |
| Synthecium sp. | D. lemchei Ortea & Urgorri, 1978 |
Thompson et al., 1990 |
| Tamarisca tamarisca (Linnaeus, 1758) | Doto cf. fragilis | Present study |
| Thuiaria thuja (Linnaeus, 1758) | D. coronata (Gmelin, 1791) | Reid, 1846 |
| Tubularia indivisa Linnaeus, 1758 | D. coronata (Gmelin, 1791) |
Jeffreys 1869 Jaeckel, 1952 |
3 RESULTS
From the total 87 specimens, including the out-groups, COI sequence data were successfully recovered from 79 specimens, and H3 from 48 specimens after phasing in the H3 dataset consisted of 53 sequences. The COI alignment was 658 base pairs (bp) long and the H3 alignment 328 bp long.
3.1 Single-locus clustering analyses
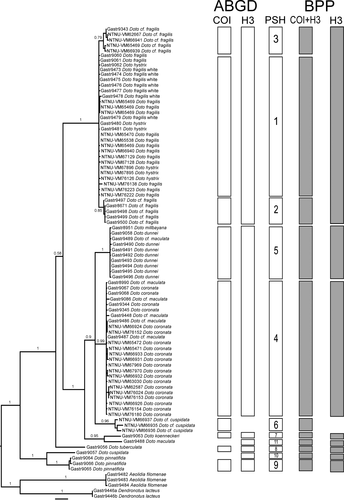
The COI dataset was divided into nine clusters using ABGD, with a prior maximal distance p = 0.0046, and with higher P, the number of clusters is lower, with both the initial and recursive partitioning (Figure S3a), whereas the H3 dataset was divided into seven clusters, with a prior maximal distance p = 0.0129, and a single cluster using a higher P (Figure S3b). The highest number of clusters in each marker was used, as downstream analysis only can merge clusters, not divide them further. In COI, Doto fragilis (Forbes, 1838) formed three clusters, D. hystrix Picton & Brown, 1981 grouped with all white morph D. fragilis from Sweden, as well as some D. fragilis from Norway and Northern Ireland. The other two clusters consist only of D. cf. fragilis. Specimens previously identified as “D. cf. maculata (Montagu, 1804)” from Sweden, grouped with D. coronata (Gmelin, 1791) and one “D. cf. maculata” from Northern Ireland clustered with D. dunnei Lemche, 1976, whereas D. koenneckeri Lemche, 1976, D. pinnatifida (Montagu, 1804), D. tuberculata Lemche, 1976, and the Norwegian D. cf. cuspidata forms separate clusters. In H3 all D. fragilis and D. hystrix clustered together, the D. coronata/Swedish “cf. maculata” group and D. dunnei group formed a single cluster also for the latter including D. millbayana Lemche, 1976. Further, D. cuspidata, D. koenneckeri and D. pinnatifida form separate clusters. When combining the results from both markers, we find a maximum set of 11 clusters or primary species hypotheses (Table 1, Figure 2). The maximum intracluster genetic distance varies between 0.0% and 2.0% in COI, and between 0.0% and 1.8% in H3. The minimal intercluster distance within Doto varies between 2.3% and 14.4% in COI and between 0.0% 12.5% in H3, and the distances between Doto spp. and the out-groups are generally higher than within Doto (Table 4).
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. D. fragilis/hystrix | 2.0 | 0.6 | 0.0 | 0.0 | 5.5 | 5.8 | — | 6.4 | 6.1 | 9.8 | 10.4 | 7.3 | 14.0 | 11.0 | ||||||||||||
| 2. D. fragilis | 2.8 | 0.5 | 0.3 | 0.0 | 5.8 | 6.1 | — | 6.7 | 6.1 | 10.4 | 11.0 | 7.6 | 14.3 | 11.3 | ||||||||||||
| 3. D. fragilis | 2.8 | 2.3 | 1.7 | — | 5.8 | 6.1 | — | 6.7 | 6.1 | 10.4 | 11.0 | 7.6 | 14.3 | 11.3 | ||||||||||||
| 4. D. coronata/maculata | 10.0 | 11.5 | 11.0 | 1.9 | 0.8 | — | 3.4 | 6.7 | 11.0 | 11.6 | 4.9 | 11.3 | 11.9 | |||||||||||||
| 5. D. dunnei | 9.9 | 11.3 | 10.4 | 4.7 | 0.8 | 0.0 | — | 3.5 | 6.6 | 10.8 | 11.2 | 4.6 | 11.2 | 12.2 | ||||||||||||
| 6. D. cf. cuspidata | 10.7 | 12.4 | 11.6 | 6.3 | 6.5 | 2.0 | — | — | — | — | — | — | — | — | ||||||||||||
| 7. D. koenneckeri | 11.9 | 12.7 | 12.1 | 8.7 | 8.8 | 9.4 | — | — | 7.0 | 11.3 | 11.6 | 3.4 | 10.1 | 12.2 | ||||||||||||
| 8. D. tuberculata | 10.1 | 12.0 | 11.9 | 10.5 | .4 | 10.9 | 12.0 | — | — | 10.4 | 11.6 | 8.5 | 15.9 | 12.8 | ||||||||||||
| 9. D. pinnatifida | 12.4 | 14.4 | 13.9 | 11.8 | 10.5 | 13.1 | 14.1 | 12.0 | 0.5 | 0.0 | 7.3 | 11.3 | 15.5 | 13.7 | ||||||||||||
| 10. D. cuspidata | — | — | — | — | — | — | — | — | — | — | — | 12.5 | 15.9 | 14.3 | ||||||||||||
| 11. D. maculata | — | — | — | — | — | — | — | — | — | — | — | 1.8 | 11.3 | 11.3 | ||||||||||||
| 12. Aeolidia filomenae | 19.8 | 21.1 | 21.4 | 19.0 | 17.0 | 19.4 | 19.9 | 19.0 | 16.3 | — | — | 0.5 | 0.3 | 10.7 | ||||||||||||
| 13. Dendronotus lacteus | 17.9 | 19.5 | 18.1 | 19.9 | 18.5 | 19.0 | 17.1 | 18.5 | 17.6 | — | — | 21.7 | 0.0 | 0.0 | ||||||||||||
3.2 Phylogenetic inference
In both the COI and H3 gene trees (Figure S1a,b), Doto is monophyletic with maximal support, and the groups found in the respective ABGD analyses are recovered. In the H3 tree, there are no signs of hybridization, and all phased alleles of the same individual are closely related.
In the combined Bayesian tree (Figure 2, Figure S1), Doto is monophyletic with maximum support and is divided into four clades: (1) D. cuspidata + D. pinnatifida, which is the sister to the remaining Doto, (2) D. tuberculata, (3) D. hystrix + D. fragilis and (4) D. coronata, Doto sp. (= “D. cf. cuspidata”), D. dunnei, D. maculata, D. millbayana and D. koenneckeri. In the last clade, D. koenneckeri and one “D. cf. maculata” from Northern Ireland are found as sisters, but well separated, and the remaining forms a trichotomy consisting of one clade including D. dunnei, D. millbayana and one D. cf. maculata, another consisting of all D. coronata and all of the Swedish D. cf. maculata specimens, and the last clade with all Norwegian D. cf. cuspidata.
The concatenated Bayesian tree including the extended dataset of Doto and Kabeiro sequences (Figure 3) is similar to Shipman and Gosliner (2015, fig. 4) and Moles et al. (2016, fig. 7). Doto cuspidata is sister to D. pinnatifida, and D. formosa Verrill, 1875, is placed together with D. hystrix and D. fragilis. Our D. coronata + “D. cf. maculata” clade is placed together with additional D. coronata sequences (Figure 3, Figure S1), and our D. dunnei, D. millbayana and the Northern Ireland D. maculata are placed together with additional specimens of D. dunnei, D. millbayana, D. sp. A and the D. coronata specimens from Moles et al. (2016), our D. maculata specimen are placed close to one D. maculata and one D. africoronata Shipman & Gosliner, 2015 specimen, our Doto sp. (= “D. cf. cuspidata”) are placed together with D. paulinae Trinchese, 1881. The trees with only our data and the tree based on the extended data are mainly congruent; the only difference in the species included in both trees is the position of D. tuberculata, which is sister to the D. fragilis complex in the extended tree, and found in a tricotomy with the D. fragilis complex and D. coronata complex.

The ML tree (Figure S2) is congruent with the Bayesian trees.
3.3 MSC delimitation
All of the 11 input species are well supported and accepted as well-delimited species when both COI and H3 are included (Table 5). However, when only H3 was used the species in the D. fragilis/hystrix complex were combined (Table 5).
| Species | COI+H3 | H3 only | ||
|---|---|---|---|---|
| A (PP) | B (PP) | A (PP) | B (PP) | |
| 1. D. fragilis/hystrix | 1.000 | 1.000 | 0.023 | 0.016 |
| 2. D. fragilis | 0.999 | 1.000 | 0.020 | 0.013 |
| 3. D. fragilis | 0.999 | 1.000 | 0.010 | 0.006 |
| 4. D. coronata/maculata | 0.999 | 0.955 | 1.000 | 1.000 |
| 5. D. dunnei/millbayana | 0.999 | 0.955 | 1.000 | 1.000 |
| 6. D. cf. cuspidata | 0.998 | 0.955 | — | — |
| 7. D. koenneckeri | 0.990 | 0.955 | 0.972 | 0.996 |
| 8. D. tuberculata | 1.000 | 0.994 | 1.000 | 1.000 |
| 9. D. pinnatifida | 1.000 | 1.000 | 0.999 | 1.000 |
| 10. D. cuspidata | 0.999 | 1.000 | 0.999 | 1.000 |
| 11. D. maculata | 0.990 | 0.955 | 0.972 | 0.996 |
| 1+2+3 | 0.000 | 0.000 | 0.955 | 0.968 |
| 1+2 | 0.000 | 0.000 | 0.007 | 0.004 |
| 1+3 | 0.000 | 0.000 | 0.016 | 0.012 |
| 2+3 | 0.001 | 0.000 | 0.019 | 0.015 |
| 4+5+6+7+11 | 0.000 | 0.039 | 0.000 | 0.000 |
Note
- Posterior probabilities are means of three runs. Posterior probabilities in bold are considered significant and species in bold are accepted.
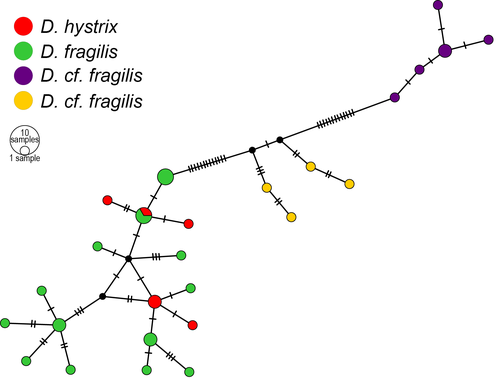
3.4 Haplotype network
The network (Figure 4) shows that the three groups within D. fragilis are separated and that one of the groups also contains D. hystrix. In this group, the two mainly have separate haplotypes, only sharing one, but there is no clear structure within the group.
3.5 Review of prey specificity
Out of 42 species of hydroids, 11 species were reported to be associated with at least two different Doto species (Table 3). This contradicts the notion of monophagy as a major trend of the evolution within the genus Doto. Half of the number of hydroid species (21) was recorded solely for Doto coronata, D. dunnei and D. millbayana were recorded from three different hydroids (Kirchenpaueria pinnata for D. dunnei and Plumularia setosa growing upon Nemertesia antennina and N. ramosa for D. millbayana, see Table 3), although these do not show significant genetic divergence (Shipman & Gosliner, 2015; present study).
4 DISCUSSION
In this study, we use a combination of data sources and methods to test the species limits of northeast Atlantic Doto species. For a majority of the species the results are clear-cut. The exceptions are mainly in D. fragilis, where we find three groups, one that do not show any genetic separation from D. hystrix despite large morphological differences. The three groups are separated in COI, but not H3. It is possible that these groups represent separate species; however, more data are needed to confirm this.
This study is one of few studies on nudibranchs that delimits species using multi-locus analysis based on the MSC method. MSC analyses have been used successfully on a wide variety of taxa (e.g., Delić et al., 2017; Fossen et al., 2016; Leache & Fujita, 2010; Martinsson & Erséus, 2018), and it has been shown to outperform distance methods (Yu et al., 2017), which so far has been the standard for molecular species delimitation in nudibranchs. However, in this study we use the same data for the initial division of specimen into species hypothesis, and for the testing of them using MSC analysis, this is not optimal as it introduces issues of circularity (see e.g., Martinsson & Erséus, 2018; Yang, 2015), and the results should be interpreted with caution.
Specimens of Doto fragilis are found in three closely related clades, one that also includes D. hystrix. In the D. fragilis/D. hystrix clade, the D. fragilis from Northern Ireland groups with the Swedish D. fragilis “white morph” and the D. hystrix from Northern Ireland, Norway and Sweden. The spiky tubercles on the dorsolateral appendages of D. hystrix are distinctly different from the rounded tubercles of D. fragilis. Doto hystrix has until now only been reported from the hydroid Schizotricha frutescens, but are reported here also observed on Nemertesia antennina (Table 3). Species of Doto fragilis complex has been found on hydroids from the genera Halecium, Nemertesia and Tubularia, but so far not on Schizotricha frutescens (see Table 3). The second D. fragilis clade consists of most often red-colored specimens from Sweden. The D. fragilis from Shipman and Gosliner (2015) are also found in this clade (Mn33151). That specimen was from Wales and was feeding on the hydroid Nemertesia ramosa. The third D. fragilis clade consists of five specimens in the analysis: one brownish light white specimen from Sweden and four specimens from Norway.
Our review of the prey species recorded for Doto species shows that most species of the genus Doto feed on several different hydroids, and hence, the previously mentioned notion of general strict food specialization within Doto (e. g. Picton & Brown, 1981; Morrow et al., 1992, 1994) should be abandoned. Not even the species D. hystrix show strict monophagy, as previously thought (Picton & Brown, 1981), but has been documented feeding on both Schizotricha frutescens and Nemertesia antennina (present study, Table 3). It is possible that different populations within a species of Doto have specialized in feeding on separate hydroids. However, the composition of hydroid species is similar along most of the European Atlantic coast, and the fauna is dominated by widely distributed species (Medel & López-González, 1998), which points against differentiation between populations.
There are other examples of nudibranchs with similarly small genetic distances as the ones found between the clusters in D. fragilis, such as between some Indo-Pacific Chromodoris species, with interspecies COI distances as low as 2.0% (Layton et al., 2018) or between species of Felimare along the American Pacific coast with a minimal interspecific COI distance of 2.5% (Hoover et al., 2016), all of these represent cases of recent speciation. It is therefore possible that the clusters we recovered are separate species, but as we have no support from nuclear markers, and the BPP analysis with only H3 did not support a separation, we do not describe them in this paper. More data and analysis are needed to further test the separation of them. “Doto fragilis” is a species complex here found in three closely related clades, one that also includes D. hystrix, and all, except D. hystrix, are externally similar. These potential new species will be described following ongoing revisions of more Doto groups. Here, to clearly indicate the clade with true D. fragilis, we designate a neotype for it (GNM Gastr. 9061. Strangford lough, Portaferry, United Kingdom, date 20 May 2014, coordinates lat 54°22,00′ long 05°32,00′, depth 20–30 m, collector: Klas Malmberg, Figure 5). The specimen is deposited at the Gothenburg Natural History Museum in Sweden. The original types of D. fragilis are lost (Mollusca Type in Great Britain, 2020; Natural History Museum, 2020). Therefore, to define true D. fragilis we here designate a neotype for this species. According to ICZN 1999 article 75.3.6. “…evidence that the neotype came as nearly as practicable from the original type locality...” The neotype of D. fragilis originates from the Northern Ireland near Portaferry, just 60 km from the original type locality of D. fragilis on the western coast of Isle of Man (Forbes, 1838), thus fulfilling the ICZN requirements. According to the original description (Forbes, 1838), D. fragilis does not contain red-colored morphs.

The genetic markers used in our study show no separation between D. hystrix and D. fragilis, despite them being distinct morphologically and ecologically. Doto hystrix has long, pointed tubercles on the rim of the rhinophore sheaths (Figure 6) and, mostly feeds on the hydroid Schizotricha frutescens (Picton & Brown, 1981, Table 3), whereas the food preferences are wider for D. fragilis. There are also differences in the spawning between the two species. This can be seen as a parallel to what is reported from, e.g., some blue butterflies (Lepidoptera: Lycaenidae) where distinct species are mixed in COI (Wiemers & Fiedler, 2007) and also from recently described species from the eubranchid nudibranchs of the genus Amphorina (Korshunova, Malmberg, et al., 2020). However, the differences in ecology and morphology are large and points toward D. fragilis and D. hystrix being separate entities, in conflict with the genetic data. Therefore, we do not synonymize them in this study. Further studies are needed to better test whether the lack of genetic separation in combination with ecological and morphological differences are due to recent speciation, with a rapid evolution of morphological characters, or interspecific differences, possibly due to adaptation to different prey species.

The northwest Atlantic species Doto formosa groups with D. fragilis and D. hystrix, but unfortunately only H3 was available for D. formosa on GenBank. Closer studies of D. formosa are needed not only in sequence data, but in morphology and ecology. The original description of D. formosa by Verrill in 1875 is very meager and could as well cover D. fragilis as well. The results from our study show that these species form a clade, but more studies are needed to resolve the taxonomical status of the taxa involved.
Prey species is an indirect method for indicating morphologically similar species of nudibranch molluscs, as the species are partially specific in their choice of prey, through coevolution with the prey (see Goodheart et al., 2017). In some cases, this is justified, as, for example, the prey of D. maculata is the hydroid Halopteris catharina, but the concept of one Doto species/one hydroid species (with the exception of D. coronata) needs to be reconsidered (see below and Table 3). We also illuminate some problems in the D. coronata complex (see Shipman & Gosliner, 2015 for a discussion about this complex).
The analysis of the hydroid associations suggests that the evolution of morphological traits and that of food preference within the genus Doto were not associated processes. For example, the morphologically and genetically well-supported species D. pinnatifida, D. fragilis and D. coronata were found associated with the same hydroid species Nemertesia antennina, whereas D. koenneckeri and D. lemchei were found on the same hydroid species Aglaophenia pluma (Table 3). The present analysis also implies that possibly other species from D. coronata complex than solely D. coronata s.str. already were recorded from several various hydroids, but these records can still be hidden under the “D. coronata” name, as a precise species identification within this complex is a considerable challenge, even for experts.
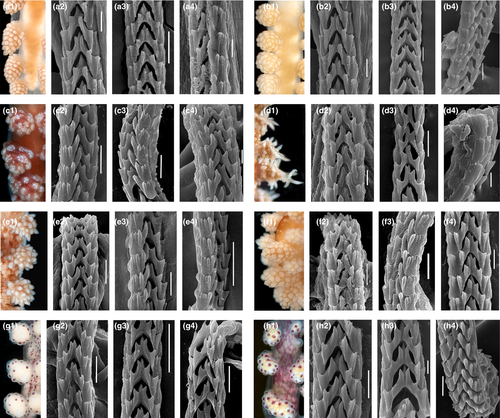
The morphology of the radula is linked to the prey specialization in any molluscs, including nudibranchs. All the studied species of Doto have the radula formula 0.1.0 (Figures 7 and 8). Though the radula in the genus Doto does not show significant differences, our data on various species reveal several promising patterns. For every species, there is some minor differences in the details of the general shape of the teeth as well as the number and arrangement of the lateral denticles. The radula of Doto millbayana (Figure 8d2–d4) has a very distinct, sharp outline of the denticles and readily distinguishes from the majority of studied here species (Figures 7 and 8), supporting its status as a separate species. In addition, there is a slight similarity between D. coronata and the D. cf. maculata from Sweden in the small central cusp.
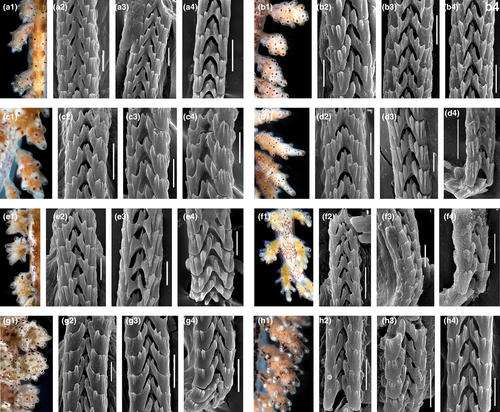
The specimens collected in Sweden and identified as Doto cf. maculata based on coloration, were never found on Halopteris catharina even though this hydroid species occurs in both the Skagerrak and the Kattegat areas, but instead on hydroids that could be identified during the scuba dives as belonging to the family Plumulariidae. There were also morphological characters that did not fully match D. maculata. This led us to suspect that the Swedish specimens could be a separate species, or at least not D. maculata, and this was confirmed by the genetic analysis. Contrary to the D. fragilis/D. hystrix case, between putative “D. cf. maculata” and real D. coronata there are no reliable morphological and ecological differences. The dorsolateral appendages of “D. cf. maculata” from Sweden is slightly different from that of D. coronata in the absence of a pigment dot on the apical tubercle. Further, the “D. cf. maculata” specimens lack the red markings at the inner side (toward the midline of the dorsal side of the animal) of the base of the dorsolateral appendages (Figure 6) that is commonly mentioned to be “typical” for D. coronata. However, D. coronata is a highly variable species (Martynov & Korshunova, 2011; Martynov et al., 2006) (Figure 6) and for D. maculata the apparent “key features” were ambiguously indicated in the redescription by Lemche (1976: 697 “Round reddish or dark brownish spots are placed on the tips of the tubercles on the cerata, except in many cases the end one”). Hence, coloration patterns should be used with care and always be checked against other morphological characters. Doto coronata evidently feeds on a variety of hydroids (Lemche, 1976; Picton & Brown, 1981; Table 3). This was supported by the molecular analysis (Shipman and Gosliner (2015), which showed that the true D. coronata has a wide prey specificity, as it was collected from the hydroids Sertularia cupressina, Obelia geniculata and O. dichotoma. The two species D. hydrallmaniae Morrow et al., 1992 and D. sarsiae Morrow et al., 1992 were not included in the study. According to an earlier work of Morrow et al. (1992), electrophoretic methods show that they represent distinct species, but are closely related to the D. coronata species complex; however, this is not yet confirmed by other molecular phylogenetic studies.
The study by Shipman and Gosliner (2015) could not resolve any genetic difference between Doto dunnei and Doto millbayana from sequences of COI, H3 and 16S, which led them to suggest that the two could be the same species. In our analysis, we got similar result from H3, but that does not provide any further support to synonymize the two species. However, we found minor radular characters supporting the separation. In the review of hydroid prey they have only been reported from their supposed prey species, so they appear monophagous. There is a distinct difference in the shape of the pseudobranchs (rounded in D. dunnei, pointed in D. millbayana). The dark red pigment spots on the dorsum are often numerous and partly fusing in D. dunnei compared to D. millbayana in which the pigment spots are fever and generally more separated. We therefore suggest that the two should remain separate species until further study.
One D. maculata specimen from Northern Ireland groups with D. africoronata Shipman & Gosliner, 2015 and the D. maculata from Shipman and Gosliner (2015) (Figure 6), unfortunately both specimens of D. maculata lack one of the genes, which could explain why they are not forming a monophyletic group, rather than being nested with D. africoronata.
As a conclusion, the multi-locus species delimitation is shown as a valuable tool in nudibranch systematics, and that D. fragilis is a species complex, including D. hystrix and D. formosa. Doto fragilis and D. hystrix have mixed COI haplotypes, possibly due to recent speciation. The present results are concordant with the concept of multi-level organismal diversity (Korshunova, Bakken, et al., 2020; Korshunova et al., 2019; Martynov et al., 2020).
ACKNOWLEDGEMENTS
KL and KM wish to thank the staff at the dive centers at Smögen, Hamburgsund and Lysekil on the Swedish west coast, and the staff at the marine station at Portaferry, Northern Ireland, for support during the collection work; TB in Norway to Jussi Evertsen, Torjus Haukvik, Erling Svensen, Bernard Picton and Christian Skauge for support during field work. Bernard Picton is thanked for valuable input and discussion on earlier versions of the manuscript. The Royal Society of Arts and Sciences in Gothenburg supported the molecular work by grant to SM. DNA bar code data of Norwegian specimens in this publication was generated in collaboration with the Norwegian Barcode of Life Network (NorBOL) funded by the Research Council of Norway (226134/F50) and the Norwegian Biodiversity Information Centre (14-14, 70184209). The Electron Microscopy Laboratory MSU in Moscow is thanked for support with electron microscopy. The work of AM was conducted under the research project of MSU Zoological Museum (18-1-21 №121032300105-0). The work of TK was conducted under the IDB RAS Government basic research program in 2021 № 0088-2021-0008. For KL and KM, the field work was supported by the foundation Birgit och Birger Wåhlströms Minnesfond för den bohuslänska havs- och insjömiljön. For TB, TK and AM the study was supported by the Norwegian Taxonomy Initiative project #sneglebuss Barents Sea (19-18, 70184240).
Open Research
DATA AVAILABILITY STATEMENT
Specimens were deposited at the Gothenburg Natural History Museum (GNM), Gothenburg, Sweden, and at the NTNU University Museum (NTNU-VM) (Bakken et al., 2021), Trondheim, Norway. All new sequences are deposited in GenBank (see Table 1 for accession numbers). Matrices and trees are available on TreeBASE (submission 26369), matrices are also available as Additional Data S1–S5.




