Second Sahelian amphibian endemism suggested by phylogeography of Groove crowned Bullfrog (Hoplobatrachus occipitalis) in western Sahel and hints of polyploid species formation
Abstract
Several studies have assessed the phylogeographic patterns of vertebrates in North Africa and Sahara–Sahel, but most of the phylogeographic knowledge on amphibians comes from the Mediterranean region while the southern Sahara and Sahel remain poorly studied. Here, we assess the phylogeography of the African Groove crowned frog Hoplobatrachus occipitalis, with a focus on western Sahel in order to better understand the biogeographic patterns of semi-aquatic species in this arid region. Using mitochondrial and nuclear markers, we have assessed the species’ genetic structure, distribution of genetic diversity, and the presence of cryptic diversity. We found evidence of a recent (re-)colonization of the mountains in its northernmost distribution, but also for the role of southern Mauritanian mountains and large rivers as refugia. Two major lineages were detected, one perhaps endemic to Mauritania and the other widespread in Africa. The first lineage possibly constitutes the second Sahelian amphibian endemic; the latter may have originated through an allopolyploidy event, with the Mauritanian lineage being one of the parental ones.
1 INTRODUCTION
Several studies have assessed the phylogeographic patterns of vertebrates in North Africa and Sahara–Sahel, but most of the phylogeographic knowledge on amphibians in North Africa comes from the Mediterranean region while the southern Sahara and Sahel remain mostly unassessed (Brito et al., 2014; Padial, Crochet, Geniez, & Brito, 2013). Amphibians in general are considered poor dispersers, usually leading to a high spatial genetic structure (Zeisset & Beebee, 2008), thus usually being good candidates for inferring biogeographic scenarios. Available studies have detected varying phylogeographic patterns in North Africa: While some species exhibit structured genetic diversity, such as Hyla meridionalis, Bufotes boulengeri, or Discoglossus pictus (Recuero, Iraola, Rubio, Machordom, & García-París, 2007; Stöck et al., 2006; Vences et al., 2014; Zangari, Cimmaruta, & Nascetti, 2006), others show minimal intraspecific variation, like Sclerophrys mauritanica (Harris & Perera, 2009) or Sclerophrys xeros (Froufe, Brito, & Harris, 2009). Relict populations have also been found, as for instance, Pelophylax saharicus or Bufotes boulengeri in southern Algeria mountains, likely isolated during the humid–arid cycles in the Pleistocene (Nicolas et al., 2017; Nicolas, Mataame, Crochet, Geniez, & Ohler, 2015).
In this study, we aim to assess the phylogeography of the African Groove crowned frog Hoplobatrachus occipitalis (Günther, 1858) and to better understand the biogeographic patterns of amphibian species in the Sahara–Sahel. Hoplobatrachus is a genus of widespread frogs that originated in Asia, whose only African representative, H. occipitalis, resulted from a dispersal event that likely took place in the Miocene (Kosuch, Vences, Dubois, Ohler, & Böhme, 2001). Although it is still considered a widespread species, previous studies have identified tetraploid and diploid populations in Liberia (Bogart & Tandy, 1976). Given a polyploidy event can result in instant speciation due to chromosomal incompatibility, this raised the possibility of the existence of cryptic diversity in Liberia and throughout the range of H. occipitalis, but to this day, no other tetraploid population was described and taxonomy remains unchanged (Bogart & Tandy, 1976, 1981). Hoplobatrachus occipitalis is found throughout North-eastern sub-Saharan Africa (except in the tropical rainforest) and a few isolated localities within Sahara (Figure 1). It occurs almost exclusively in permanent water bodies (Rödel, 2000), dispersing and reproducing only when there is a large amount of rainfall (Spieler & Linsenmair, 1997, 1998).
This species likely suffered huge range shifts during the humid–arid cycles affecting North Africa during the Plio-Pleistocene. Similar in duration to glacial cycles, these had major impacts on species distribution ranges and have been linked to population vicariance and cryptic diversity in mountain regions (reviewed in Brito et al., 2014). It is therefore possible that studying the genetic structure of populations in topographically heterogeneous regions in Sahara fringes might reveal undescribed diversity. Especially, considering the taxonomy of some amphibians currently described as broadly distributed, including Ptychadena spp. (Ptychadenidae), Hoplobatrachus occipitalis (Dicroglossidae), Tomopterna spp. (Pyxicephalidae), and Phrynobatrachus spp. (Phrynobatrachidae) may be unreliable (Padial et al., 2013). Increasing data on the distribution, genetic diversity, and phylogeographic patterns is considered priority for amphibian conservation in the Sahel (Padial et al., 2013) and contributes toward explaining the evolutionary processes at play.
We combined sequences from throughout the species distribution with samples mostly from the outer fringes of its range in Mauritania to answer the following questions: (a) How is genetic variability spatially structured?; (b) Where are the areas of higher genetic diversity?; and (c) Is there cryptic diversity? Considering the strong water requirements, we expect the climatic cycles to have had a major impact in the recovered genetic signature. As such, the phylogeographic structure of the species is expected to be related with the hydrographic network, with larger and more permanent rivers housing more lineages and higher diversity, due to being less prone to local extinctions. Populations in the northern fringes of the distribution and in the lowlands are expected to show less genetic diversity than those near Senegal River and those in permanent mountain rock pools, since the regions where the latter ones are climatically more stable and may work as refugia (Vale, Pimm, & Brito, 2015).
2 MATERIALS AND METHODS
A total of 151 samples from 75 localities best representing the geographic range of H. occipitalis were selected for this study (Figure 1; Appendix). DNA was extracted from ethanol-preserved tissue using DNeasy Blood & Tissue Kit (Qiagen Iberia). Four genes were amplified: 16S rRNA (16S, mitochondrial), neurotrophin 3 (NTF3, nuclear), proopiomelanocortin (POMC, nuclear), and recombination activating 1 (RAG1, nuclear). Primers sequences were 16S forward—CGCCTGTTTAYCAAAAACAT and reverse—CCGGTYTGAACTCAGATCAYGT (Bossuyt & Milinkovitch, 2000); NTF3 forward—ATGTCCATCTTGTTTTATGTGATATTT and reverse—ACRAGTTTRTTGTTYTCTGAAGTC (Wiens et al., 2008); POMC forward—ATATGTCATGASCCAYTTYCGCTGGAA and reverse—GGCRTTYTTGAAWAGAGTCATTAGWGG (Vieites, Min, & Wake, 2007); and RAG1 forward—AAATWCTCRGAMTGGAAGTTYAARCT and reverse—TCACCWYCTTCTTCYTTBTCDGCRAA (Kotaki et al., 2010). Amplicon lengths were 555–556, 680, 485, and 889 nucleotides, respectively. Amplifications were performed in 10 μl of 2x MyTaq™ Mix and 0.5 μM each primer. PCR conditions were pre-denaturation at 95°C (15 min); 40 cycles with denaturing at 95°C (30 s), annealing range of 48–52°C (40 s), and extension at 72°C (45 s); and final extension at 60°C for 12 min. PCR products were sequenced using cycle sequencing on an automated sequencer (Applied Biosystems 3730xl). Sequences were deposited in GenBank (Appendix).
Twenty-five additional 16S sequences covering a significant portion of H. occipitalis distribution were retrieved from GenBank, together with six sequences from three Asian Hoplobatrachus species and two out-groups from the close genus Fejervarya (Appendix). DNA sequences were aligned with mafft v7 (Katoh & Standley, 2013), using default parameters and the Q-INS-i option. Protein coding markers were translated, and no stop codons were found. Final alignments of 16S, NTF3, POMC, and RAG1 were 465, 603, 481, and 888 nucleotides long, respectively, and were deposited in OSF (osf.io/chjq6). The most appropriate model of molecular evolution was selected using jModeltest 2 (Darriba, Taboada, Doallo, & Posada, 2012).
Phylogenies were inferred based on the 16S sequences. This was done firstly due to the low variation in nuclear genes; and second, in order to obviate the pattern found in each nuclear marker and contrast it with the mitochondrial phylogeny, comparisons that are crucial for understanding and discussing the observed phylogeographic pattern. We used Bayesian inference (BI) and maximumlikelihood (ML) methods implemented, respectively, in MrBayes v3.2.6 (Ronquist et al., 2012) and PhyML (Guindon et al., 2010). The best model according to jModeltest was TIM2+ I + G; but since TIM2 is not implemented, we used the closest over-parameterized model, GTR. We ran two independent chains in MrBayes. Burn-in was determined by assessing ESS values in Tracer v1.7 (Rambaut, Suchard, Xie, & Drummond, 2014), and a 50% majority rule tree generated using MrBayes sumt command. PhyML was set to run 1,000 bootstrapping replicates.
Haplotypes for the nuclear sequences were inferred using phase 2.1 (Stephens, Smith, & Donnelly, 2001), implemented in DnaSP (Rozas et al., 2017). phase ran for 104 iterations with a burn-in of 1,000, thinning interval of 5, and the default threshold of 0.9 (-p and –q, phases and genotypes). Haplotype networks were produced for all individual markers using tcs v1.21 (Clement, Posada, & Crandall, 2000) with gaps as missing data and 95% connection limit. Graphic representations were obtained using tcsBU (Santos, Cabezas, Tavares, Xavier, & Branco, 2015).
Uncorrected p-distances within and among species and lineages were calculated in mega6 (Tamura, Stecher, Peterson, Filipski, & Kumar, 2013) for the mitochondrial marker. Sequence and nucleotide diversity measures and demographic statistics were calculated in DnaSP v6 for all markers except RAG1 due to a lower sample size. Demographic changes were further analyzed through mismatch distribution in 16S using Arlequin 3.5 (Excoffier & Lischer, 2010).
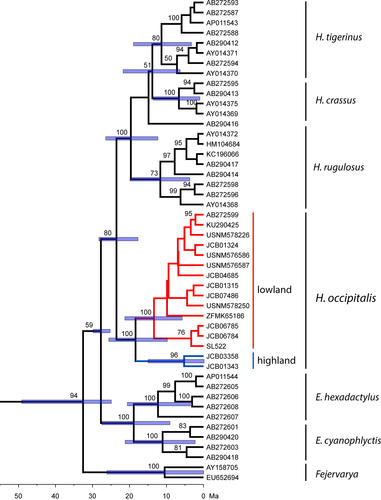
Time of divergence between the mitochondrial lineages was estimated with beast v1.10 (Drummond, Suchard, Xie, & Rambaut, 2012) in CIPRES gateway (Miller, Pfeiffer, & Schwartz, 2010). We used the same calibration points as Chen et al. (2017): 59.1–120.6 Ma for the Dicroglossidae-Ranidae split, 52.1–76.3 Ma separating the tribe Paini and Hoplobatrachus + Fejervarya, and 28.7–47.5 Ma between Nanorana + Feirana and Quasipaa + Yerana, plus the divergence of 30–25 Ma between Hoplobatrachus and Euphlyctis used by Alam et al. (2008). The calibration alignment was composed of each of the H. occipitalis 16S haplotypes detected above, the 16S sequences extracted from the mitogenomes in Chen et al. (2017), and others from Alam et al. (2008) (Figure 2, osf.io/chjq6). We ran beast for 5 x 107 generations, sampling every 5,000, with an uncorrelated lognormal relaxed clock (Drummond, Ho, Phillips, & Rambaut, 2006), and Yule speciation tree prior (Gernhard, 2008; Yule, 1925). Burn-in was determined using Tracer v1.6 (Rambaut et al., 2014). A maximum credibility tree was generated with TreeAnnotator (in the beast package).
3 RESULTS
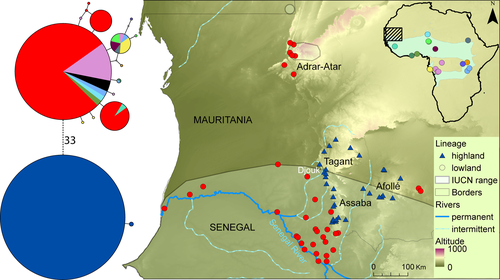
The mitochondrial phylogenetic tree of Hoplobatrachus was clearly divided into two divergent lineages (Figure 1; Figures S1 and S2) with a genetic distance of 7.1 ± 1.1%, and an average divergence time of 17.8 Ma (Figure 2). Their distribution is parapatric, with one lineage restricted to southern Mauritanian table mountains of Tagant, Assaba, and Afollé (“highland” lineage, occurring from 70 to 400 m), and the other one distributed throughout Africa (Figure 1; Figure S2), including the isolated population in Adrar-Atar and the Sahelian lowlands (“lowland” lineage). In southern Mauritania, the “lowland” lineage occurs only below 100 m, but in the rest of the Sahelian range, it occurs up to at least 400 m. Despite the overlap, we chose these labels since they are informative in the region where both lineages occur. Both lineages had little genetic variability (Table S2). The “highland” one had only two alleles, one of which restricted to a single locality in southern Djouk valley (Figure S2). In the lowland lineage, fourteen closely related alleles were detected throughout Africa, five of which in Mauritania. The most common one stretches from the northernmost, isolated populations in Saharan Mountains (Adrar-Atar in Mauritania and Aïr in Niger) to Tanzania and the Republic of Congo. The remaining haplotypes in Mauritania occurred only in or near Senegal River (Figure S2).
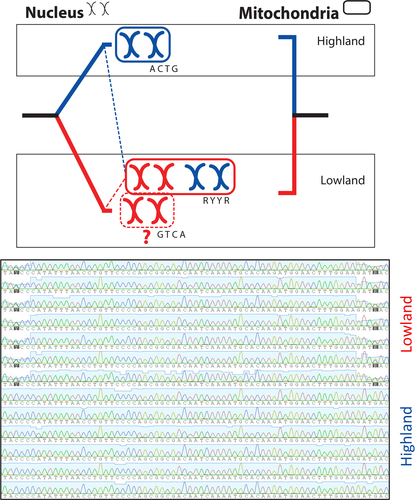
Nuclear genetic diversity exhibited a very peculiar pattern that could be indicative of a genetic duplication: In POMC and NTF3, all “highland” samples were homozygous and shared the same allele, while all “lowland” samples had two alleles and were heterozygous in all variable positions (Figure 3).The heterozygous state always coded the highland allele plus a second one (except in Adrar-Atar for POMC). In NTF3, only two alleles were recovered in the 49 sequenced samples (Figure S3). In POMC, six alleles were found in 134 samples (Figure 4). The Adrar-Atar populations shared no nuclear allele with the “highland” populations, but they were all also bi-allelic: All specimens shared a common allele, plus a second one. In RAG1, we found the same pattern as in POMC and NTF3, but in this case, the highland population had two alleles. The “lowland” specimens had those “highland” alleles plus another, 17 substitutions away, being heterozygous in all those 17 positions (Figure S3).

4 DISCUSSION
Here, we present one of the first phylogeographic studies on Sahelian amphibians (but see Froufe et al., 2009; Vasconcelos, Froufe, Brito, Carranza, & Harris, 2010), adding another contribution to this poorly studied region. We have identified two lineages of H. occipitalis, whose divergence (7% in 16S) is above sister-species distances reported for the close genus Fejervarya (Kotaki et al., 2010), and above the average distance among other Hoplobatrachus species (3.4%–6.1%; Table S1). The split between both lineages was prior to the Plio-Pleistocene climatic fluctuations (average 17.8 Ma, Figure 2). Not a single specimen from a mitochondrial lineage was found with the other lineage's nuclear pattern, in spite of both existing in the same rivers, indicating lack of gene flow. If confirmed, “highland” may be the second Sahelian amphibian endemic (the first one being Kassina wazae, known only from the type locality in northern Cameroon).
The very low intra-lineage genetic diversity, both for mitochondrial and nuclear markers (Table S2), is a signal of a very strong recent demographic expansion in the region, as also reflected by the mismatch distribution plots (Figure S4). This is similar to the patterns found in other western Sahelian taxa, for instance, in the toad Sclerophrys xeros (Froufe et al., 2009) and in mammals (e.g., Mouline et al., 2008), and that were explained with retreats to southern regions (Froufe et al., 2009) or humid refugia (Mouline et al., 2008) during unfavorable periods, subsequently followed by range expansions during wetter phases. Given the strong water dependency in amphibians, the same pattern is likely for H. occipitalis as well (Brito et al., 2014; Vale, Tarroso, & Brito, 2014). In the north-western side of the Sahara, some amphibians species also present low genetic diversity, probably due to similar climatic constraints (Nicolas et al., 2015). Although an apparently common pattern in arid species, this shallow phylogeographic structure contrasts with the general pattern of amphibians retaining a strong phylogeographic signal and having low dispersal capacity (Zeisset & Beebee, 2008). This could mean that the presence of amphibian species in some climatically dynamic arid regions is more related with high dispersal and population recovery ability than just resilience to aridity.
The parapatric distribution of both lineages, in light of a recent expansion (Figure S4), suggests that during at least the last climatic cycles, populations of the “highland” lineage were isolated and persisted in refugia in the southern mountains, while the “lowland” lineage has retracted its range to even more southern regions, more humid and climatically stable (Brito et al., 2014; Vale et al., 2014), and subsequently recolonized the areas around the southern mountains and the Adrar-Atar during favorable periods. The species distribution and previously published ecological models (Vale et al., 2014) indicate the southern part of the study area as more stable and more widely suitable to the occurrence of the species, therefore supporting the possibility of other southern refugia.
The Djouk valley between Assaba and Tagant seems to be of particular importance for species persistence during arid phases, since it is where “highland” diversity is highest, but also considering similar evidence found in lizards (Gonçalves et al., 2018) and crocodiles (Velo-Antón, Godinho, Campos, & Brito, 2014). As for the “lowland” lineage, the fact that only one mitochondrial allele is found in most of Mauritania (including Afollé and Adrar-Atar) is consistent with a recent expansion, but the fact that all nuclear POMC alleles in Adrar-Atar are private also suggests a refugium there. Previous studies with fishes and odonates have proposed that an extended drought in the Holocene led to local population extirpation in Central Saharan Mountains, including Adrar des Ifoghas in Mali-Algeria, Aïr in Niger, Tibesti and Ennedi in Chad, and Adrar-Atar. The latter three seem to have been subject to recolonization events during a humid spell later in the Holocene (Dumont, 1982; van Bocxlaer, Verschuren, Schettler, & Kröpelin, 2011), which may have included H. occipitalis. Isolated populations of H. occipitalis can be found in Aïr (2014 & S. A. S. G., 22014), a mountain system that presently is devoid of fish fauna (UNEP, 2007), indicating that they can indeed persist through severe arid periods (like the present). In conclusion, both persistence and recolonization are likely to have occurred. Rapid range expansions in semi-arid landscapes are probably explained by high individual dispersal (Marko Spieler & Linsenmair, 1998), high fertility (Alam et al., 2012), aggressive intraspecific interactions that encourage the occupation of marginal habitats (Spieler & Linsenmair, 1998), and rapid development allowed by tadpole carnivory (Grosjean, Vences, & Dubois, 2004). This strategy contrasts with the one found, for instance, in Sclerophrys or Tomopterna, which survive by burrowing (Loveridge, 1976), but phylogeographic patterns in those and other Sahelian amphibians are still poorly known and do not allow further discussion.
The most interesting finding was that all “lowland” specimens were heterozygous for all variable positions in all nuclear markers. This suggests an event of duplication, either in three genes, or genome duplication, but biogeographical and mating experiment evidence seems to favor the latter. Tetraploid populations of Hoplobatrachus occipitalis have already been described in Liberia, while the ones studied in Cameroon and East Africa revealed diploidy (Bogart & Tandy, 1976, 1981). The fact that all heterozygotes belonged to one mitochondrial lineage while all homozygotes belong to the other indicates reproductive isolation between both lineages, which could be caused by ploidy incompatibility. Since both lineages occur in proximity (a few km away) across several river systems around Assaba Mountains, the importance of the lack of admixture signal is further stressed. This also decreases the likelihood that the observed pattern was caused by hybridization. While hybridization between two divergent lineages could cause all those heterozygous positions, not a single specimen of “highland” or “lowland” was found with the nuclear pattern of the other lineage. Diploid hybridization could only explain this if there was a geographic separation between lineages immediately after a very brief contact and that separation was maintained; which does not seem to be the case. While studying post-mating isolation among Dicroglossidae species, Alam et al. (2012) have forayed into the possibility of tetraploid formation from hybrid triploids, which they found to have higher fitness than diploid hybrids, for example, H. chinensis X H. tigerinus. However, they avoided drawing further conclusions regarding the origin of species in the Hoplobatrachus, Euphlyctis, and Fejervarya genera.
Polyploidy events can happen in nature through several genetic mechanisms, occurring repeatedly, and have multiple maternal origins (Schmid, Evans, & Bogart, 2015; Stöck et al., 2006), and it is an event that is made more likely during periods of climatic instability (Mable, Alexandrou, & Taylor, 2011). Considering polyploidy is common in Dicroglossidae, the fact that most “lowland” specimens possess the “highland” nuclear haplotypes suggests that the “highland” could have been one of two parental diploid species in an allopolyploid hybrid origin (Figure 3).The mitochondrial lineage split could hypothetically be an indicator of ploidy; however, we only had nuclear data from the north-western distribution of H. occipitalis, so it is possible some “lowland” specimens are actually diploids from the other ancestral species (e.g., the populations described by Bogart and Tandy (1981)). Clarification of this issue will likely require karyotyping specimens from throughout the species distribution.
ACKNOWLEDGEMENTS
BIODESERTS group members assisted the fieldwork. D. Gower and A. Ohler provided access to samples under the scope of a Synthesys 3 grant (FR-TAF-4220/GB-TAF-3882). P.A. Crochet provided valuable comments. This work was also funded by National Geographic Society (CRE 7629-04/8412-08), MBZ Species Conservation Fund (11052499), FCT (PTDC/BIA-BEC/099934/2008, PTDC/BIA-BIC/2903/2012), and by ERDF through COMPETE (FCOMP-01-0124-FEDER-008917/028276). Individual support was given by FCT (SFRH/BD/78402/2011, IF/00459/2013) within QREN-POPH-T4.1 funded by ESF and Portuguese MEC. Logistic support for fieldwork was given by Pedro Santos Lda (Trimble GPS), Off Road Power Shop, P.N. Banc d'Arguin (Mauritania), and Ministère de l'Environnement et du Développement Durable of Mauritania.
APPENDIX A: List of samples used in this study. Coordinates are in decimal degrees
| SAMPLE | 16S | NTF3 | POMC | RAG1 | LAT | LONG | ORIGIN |
|---|---|---|---|---|---|---|---|
|
H. cf. occipitalis (“highland”) |
|||||||
| JCB01343 | MK864190 | MK879075 | MK879124 | - | 15.883 | −12.036 | Assaba, Mauritania |
| JCB01385 | MK864191 | MK879076 | - | MK879258 | 16.640 | −11.056 | Assaba, Mauritania |
| JCB01399 | MK864192 | MK879077 | MK879125 | MK879259 | 17.267 | −12.199 | Tagant, Mauritania |
| JCB01400 | MK864193 | - | MK879126 | - | 17.267 | −12.199 | Tagant, Mauritania |
| JCB01401 | MK864194 | MK879078 | MK879127 | - | 17.267 | −12.199 | Tagant, Mauritania |
| JCB02022 | MK864195 | - | MK879128 | - | 17.835 | −11.558 | Tagant, Mauritania |
| JCB02060 | MK864229 | - | MK879129 | - | 17.635 | −11.324 | Assaba, Mauritania |
| JCB02069 | MK864196 | MK879079 | MK879130 | - | 17.250 | −10.668 | Hodh El Gharbi, Mauritania |
| JCB02083 | MK864197 | MK879080 | MK879131 | - | 17.635 | −11.324 | Assaba, Mauritania |
| JCB02084 | MK864198 | MK879081 | MK879132 | - | 17.635 | −11.324 | Assaba, Mauritania |
| JCB02091 | MK864199 | - | MK879133 | - | 17.261 | −10.690 | Hodh El Gharbi, Mauritania |
| JCB02111 | MK864200 | MK879082 | MK879134 | - | 17.032 | −10.245 | Hodh El Gharbi, Mauritania |
| JCB02126 | MK864201 | MK879083 | MK879135 | - | 17.032 | −10.245 | Hodh El Gharbi, Mauritania |
| JCB02231 | MK864202 | MK879084 | MK879136 | - | 16.516 | −10.453 | Hodh El Gharbi, Mauritania |
| JCB02232 | MK864203 | MK879085 | MK879137 | - | 16.516 | −10.453 | Hodh El Gharbi, Mauritania |
| JCB02233 | MK864204 | MK879086 | MK879138 | - | 16.516 | −10.453 | Hodh El Gharbi, Mauritania |
| JCB02235 | MK864243 | - | MK879139 | - | 16.516 | −10.453 | Hodh El Gharbi, Mauritania |
| JCB02242 | MK864252 | - | MK879140 | - | 16.516 | −10.453 | Hodh El Gharbi, Mauritania |
| JCB02343 | MK864205 | MK879087 | MK879141 | - | 16.538 | −10.742 | Assaba, Mauritania |
| JCB02344 | MK864206 | MK879088 | MK879142 | - | 16.538 | −10.742 | Assaba, Mauritania |
| JCB02345 | MK864207 | MK879089 | MK879143 | - | 16.538 | −10.742 | Assaba, Mauritania |
| JCB02376 | MK864208 | MK879090 | MK879144 | - | 16.579 | −10.705 | Assaba, Mauritania |
| JCB02378 | MK864209 | - | - | - | 16.579 | −10.705 | Assaba, Mauritania |
| JCB02379 | MK864210 | - | MK879145 | - | 16.579 | −10.705 | Assaba, Mauritania |
| JCB02384 | MK864230 | - | - | - | 16.579 | −10.705 | Assaba, Mauritania |
| JCB02456 | MK864211 | MK879091 | MK879146 | MK879262 | 15.933 | −12.011 | Guidimaka, Mauritania |
| JCB02457 | MK864212 | - | MK879147 | - | 15.933 | −12.011 | Guidimaka, Mauritania |
| JCB02458 | MK864239 | - | - | - | 15.933 | −12.011 | Guidimaka, Mauritania |
| JCB02553 | MK864213 | - | MK879148 | - | 16.547 | −12.010 | Assaba, Mauritania |
| JCB02554 | MK864214 | MK879092 | MK879149 | - | 16.547 | −12.010 | Assaba, Mauritania |
| JCB02555 | MK864215 | MK879093 | MK879150 | - | 16.547 | −12.010 | Assaba, Mauritania |
| JCB02586 | MK864216 | MK879094 | MK879151 | - | 16.889 | −12.185 | Assaba, Mauritania |
| JCB02609 | MK864217 | - | - | - | 17.401 | −12.364 | Tagant, Mauritania |
| JCB02610 | MK864218 | - | - | - | 17.401 | −12.364 | Tagant, Mauritania |
| JCB02611 | MK864219 | - | - | - | 17.401 | −12.364 | Tagant, Mauritania |
| JCB02613 | MK864244 | - | MK879152 | - | 17.401 | −12.364 | Tagant, Mauritania |
| JCB02669 | MK864220 | MK879095 | MK879153 | MK879263 | 17.738 | −12.245 | Tagant, Mauritania |
| JCB02670 | MK864221 | - | MK879154 | - | 17.738 | −12.245 | Tagant, Mauritania |
| JCB03129 | MK864222 | MK879096 | MK879155 | - | 17.887 | −12.111 | Tagant, Mauritania |
| JCB03331 | MK864223 | MK879097 | MK879156 | - | 16.756 | −11.997 | Assaba, Mauritania |
| JCB03358 | MK864260 | MK879098 | MK879157 | - | 17.070 | −12.208 | Assaba, Mauritania |
| JCB03359 | MK864261 | MK879099 | MK879158 | - | 17.070 | −12.208 | Assaba, Mauritania |
| JCB03360 | MK864234 | - | - | - | 17.070 | −12.208 | Assaba, Mauritania |
| JCB03393 | MK864241 | - | MK879159 | - | 17.152 | −12.199 | Assaba, Mauritania |
| JCB03394 | MK864224 | - | MK879160 | - | 17.152 | −12.199 | Assaba, Mauritania |
| JCB03395 | MK864225 | MK879100 | MK879161 | - | 17.152 | −12.199 | Assaba, Mauritania |
| JCB03409 | MK864226 | MK879101 | MK879162 | - | 17.188 | −12.248 | Assaba, Mauritania |
| JCB03417 | MK864227 | MK879102 | MK879163 | - | 17.188 | −12.248 | Assaba, Mauritania |
| JCB04779 | MK864228 | MK879103 | MK879164 | MK879266 | 15.957 | −12.010 | Guidimaka, Mauritania |
| JCB04807 | MK864251 | - | MK879165 | - | 15.901 | −11.939 | Guidimaka, Mauritania |
| JCB06017 | MK864245 | - | MK879166 | - | 17.887 | −12.111 | Tagant, Mauritania |
| JCB06018 | MK864253 | - | MK879167 | - | 17.887 | −12.111 | Tagant, Mauritania |
| JCB06036 | MK864242 | - | MK879168 | MK879268 | 18.053 | −11.943 | Tagant, Mauritania |
| JCB06086 | MK864256 | - | MK879169 | - | 16.540 | −10.801 | Assaba, Mauritania |
| JCB06087 | MK864236 | - | MK879170 | - | 16.540 | −10.801 | Assaba, Mauritania |
| JCB06088 | MK864254 | - | MK879171 | - | 16.540 | −10.801 | Assaba, Mauritania |
| JCB06113 | MK864250 | - | MK879172 | - | 17.152 | −12.199 | Assaba, Mauritania |
| JCB07694 | MK864237 | - | MK879173 | MK879271 | 16.339 | −11.978 | Assaba, Mauritania |
| JCB07706 | MK864259 | - | MK879174 | - | 16.339 | −11.978 | Assaba, Mauritania |
| JCB07707 | MK864235 | - | MK879175 | - | 16.339 | −11.978 | Assaba, Mauritania |
| JCB07719 | MK864238 | - | MK879177 | - | 16.297 | −12.005 | Assaba, Mauritania |
| JCB07720 | MK864255 | - | MK879178 | - | 16.297 | −12.005 | Assaba, Mauritania |
| JCB07724 | MK864231 | - | MK879179 | MK879272 | 15.935 | −12.000 | Guidimaka, Mauritania |
| JCB07745 | MK864246 | - | MK879180 | - | 16.003 | −11.872 | Assaba, Mauritania |
| JCB07746 | MK864232 | - | MK879181 | - | 16.003 | −11.872 | Assaba, Mauritania |
| JCB07747 | MK864258 | - | MK879182 | - | 16.003 | −11.872 | Assaba, Mauritania |
| JCB07772 | MK864257 | - | MK879183 | MK879273 | 15.945 | −11.929 | Guidimaka, Mauritania |
| JCB07773 | MK864247 | - | MK879184 | - | 15.945 | −11.929 | Guidimaka, Mauritania |
| JCB07774 | MK864233 | - | MK879185 | - | 15.945 | −11.929 | Guidimaka, Mauritania |
| JCB07782 | MK864248 | - | MK879186 | - | 15.949 | −11.682 | Guidimaka, Mauritania |
| JCB07783 | MK864249 | - | MK879187 | - | 15.949 | −11.682 | Guidimaka, Mauritania |
| JCB07867 | MK864240 | - | MK879188 | - | 16.763 | −11.223 | Assaba, Mauritania |
|
H. occipitalis (“lowland”) |
|||||||
| JCB01315 | MK864111 | MK879104 | MK879189 | - | 16.142 | −13.477 | Gorgol, Mauritania |
| JCB01324 | MK864124 | MK879105 | MK879190 | - | 15.996 | −12.723 | Gorgol, Mauritania |
| JCB01325 | MK864125 | MK879106 | MK879191 | - | 15.996 | −12.723 | Gorgol, Mauritania |
| JCB01644 | MK864126 | - | MK879192 | - | 20.323 | −13.142 | Adrar, Mauritania |
| JCB01645 | MK864127 | MK879107 | MK879193 | - | 20.323 | −13.142 | Adrar, Mauritania |
| JCB01646 | MK864128 | - | MK879194 | - | 20.323 | −13.142 | Adrar, Mauritania |
| JCB01668 | MK864129 | - | MK879195 | - | 20.534 | −13.044 | Adrar, Mauritania |
| JCB01669 | MK864130 | MK879108 | MK879196 | - | 20.534 | −13.044 | Adrar, Mauritania |
| JCB01670 | MK864131 | MK879109 | MK879197 | - | 20.534 | −13.044 | Adrar, Mauritania |
| JCB01714 | MK864132 | MK879110 | MK879198 | - | 20.581 | −13.136 | Adrar, Mauritania |
| JCB01715 | MK864133 | - | MK879199 | - | 20.581 | −13.136 | Adrar, Mauritania |
| JCB01737 | MK864152 | - | MK879200 | MK879260 | 20.253 | −13.088 | Adrar, Mauritania |
| JCB01738 | MK864134 | MK879111 | MK879201 | - | 20.253 | −13.088 | Adrar, Mauritania |
| JCB01739 | MK864135 | MK879112 | MK879202 | - | 20.253 | −13.088 | Adrar, Mauritania |
| JCB01846 | MK864136 | - | MK879203 | - | 19.757 | −13.044 | Adrar, Mauritania |
| JCB01847 | MK864137 | - | MK879204 | - | 19.757 | −13.044 | Adrar, Mauritania |
| JCB01848 | MK864138 | - | MK879205 | - | 19.757 | −13.044 | Adrar, Mauritania |
| JCB01888 | MK864139 | - | MK879206 | - | 20.237 | −13.005 | Adrar, Mauritania |
| JCB01889 | MK864140 | MK879113 | MK879207 | - | 20.237 | −13.005 | Adrar, Mauritania |
| JCB01890 | MK864162 | - | MK879208 | - | 20.237 | −13.005 | Adrar, Mauritania |
| JCB02153 | MK864141 | MK879114 | MK879209 | MK879261 | 16.764 | −9.770 | Hodh El Gharbi, Mauritania |
| JCB02154 | MK864142 | MK879115 | MK879210 | - | 16.764 | −9.770 | Hodh El Gharbi, Mauritania |
| JCB02155 | MK864143 | MK879116 | MK879211 | - | 16.691 | −9.717 | Hodh El Gharbi, Mauritania |
| JCB02156 | MK864144 | MK879117 | MK879212 | - | 16.691 | −9.717 | Hodh El Gharbi, Mauritania |
| JCB02157 | MK864145 | - | MK879213 | - | 16.691 | −9.717 | Hodh El Gharbi, Mauritania |
| JCB02158 | MK864146 | MK879118 | MK879214 | - | 16.691 | −9.717 | Hodh El Gharbi, Mauritania |
| JCB02918 | MK864147 | - | - | MK879264 | 19.999 | −13.289 | Adrar, Mauritania |
| JCB02919 | MK864164 | - | - | - | 19.999 | −13.289 | Adrar, Mauritania |
| JCB04523 | MK864153 | - | MK879215 | - | 16.600 | −15.765 | Trarza, Mauritania |
| JCB04614 | MK864121 | MK879120 | MK879217 | MK879265 | 16.175 | −12.975 | Gorgol, Mauritania |
| JCB04536 | MK864148 | MK879119 | MK879216 | - | 16.810 | −15.416 | Trarza, Mauritania |
| JCB04656 | MK864149 | MK879121 | MK879218 | - | 15.507 | −12.970 | Gorgol, Mauritania |
| JCB04685 | MK864169 | - | MK879219 | - | 15.289 | −12.536 | Guidimaka, Mauritania |
| JCB04702 | MK864170 | - | MK879220 | - | 15.144 | −12.010 | Guidimaka, Mauritania |
| JCB04726 | MK864173 | MK879122 | MK879221 | - | 15.591 | −11.880 | Guidimaka, Mauritania |
| JCB04733 | MK864171 | - | MK879222 | - | 15.576 | −11.944 | Guidimaka, Mauritania |
| JCB04757 | MK864172 | - | MK879223 | - | 15.682 | −12.163 | Guidimaka, Mauritania |
| JCB04939 | MK864150 | MK879123 | MK879224 | MK879267 | 17.070 | −12.689 | Assaba, Mauritania |
| JCB04943 | MK864161 | - | MK879225 | - | 17.070 | −12.689 | Assaba, Mauritania |
| JCB04974 | MK864151 | - | MK879226 | - | 17.391 | −13.456 | Brakna, Mauritania |
| JCB06121 | MK864155 | - | - | - | 16.470 | −12.485 | Assaba, Mauritania |
| JCB06148 | MK864159 | - | MK879227 | - | 15.692 | −12.471 | Gorgol, Mauritania |
| JCB06158 | MK864158 | - | MK879228 | - | 15.635 | −12.433 | Guidimaka, Mauritania |
| JCB06169 | MK864183 | - | MK879229 | - | 15.484 | −12.271 | Guidimaka, Mauritania |
| JCB06631 | MK864184 | - | MK879230 | MK879269 | 13.700 | 9.530 | Zinder, Niger |
| JCB06784 | MK864189 | - | MK879231 | MK879270 | 13.966 | 9.281 | Zinder, Niger |
| JCB06785 | MK864185 | - | MK879232 | - | 13.966 | 9.281 | Zinder, Niger |
| JCB07393 | MK864157 | - | MK879233 | - | 16.234 | −16.430 | Trarza, Mauritania |
| JCB07457 | MK864112 | - | MK879234 | - | 15.476 | −12.934 | Gorgol, Mauritania |
| JCB07458 | MK864119 | - | MK879235 | - | 15.476 | −12.934 | Gorgol, Mauritania |
| JCB07459 | MK864114 | - | MK879236 | - | 15.476 | −12.934 | Gorgol, Mauritania |
| JCB07486 | MK864123 | - | MK879237 | - | 15.322 | −12.842 | Gorgol, Mauritania |
| JCB07487 | MK864122 | - | MK879238 | - | 15.322 | −12.842 | Gorgol, Mauritania |
| JCB07497 | MK864179 | - | MK879239 | - | 15.159 | −12.761 | Gorgol, Mauritania |
| JCB07498 | MK864117 | - | MK879240 | - | 15.159 | −12.761 | Gorgol, Mauritania |
| JCB07500 | MK864181 | - | MK879241 | - | 15.159 | −12.761 | Gorgol, Mauritania |
| JCB07546 | MK864174 | - | MK879242 | - | 15.047 | −12.450 | Guidimaka, Mauritania |
| JCB07547 | MK864180 | - | MK879243 | - | 15.047 | −12.450 | Guidimaka, Mauritania |
| JCB07586 | MK864115 | - | MK879244 | - | 14.853 | −12.396 | Guidimaka, Mauritania |
| JCB07587 | MK864116 | - | MK879245 | - | 14.853 | −12.396 | Guidimaka, Mauritania |
| JCB07588 | MK864118 | - | MK879246 | - | 14.853 | −12.396 | Guidimaka, Mauritania |
| JCB07613 | MK864113 | - | MK879247 | - | 14.846 | −12.182 | Guidimaka, Mauritania |
| JCB07614 | MK864176 | - | MK879248 | - | 14.846 | −12.182 | Guidimaka, Mauritania |
| JCB07616 | MK864120 | - | MK879249 | - | 14.846 | −12.182 | Guidimaka, Mauritania |
| JCB07618 | MK864177 | - | MK879250 | - | 14.996 | −12.184 | Guidimaka, Mauritania |
| JCB07619 | MK864163 | - | MK879251 | - | 14.996 | −12.184 | Guidimaka, Mauritania |
| JCB07620 | MK864160 | - | MK879252 | - | 14.996 | −12.184 | Guidimaka, Mauritania |
| JCB07628 | MK864156 | - | MK879253 | - | 15.356 | −12.210 | Guidimaka, Mauritania |
| JCB07629 | MK864154 | - | MK879254 | - | 15.356 | −12.210 | Guidimaka, Mauritania |
| JCB07630 | MK864175 | - | MK879255 | - | 15.356 | −12.210 | Guidimaka, Mauritania |
| JCB07674 | MK864182 | - | MK879256 | - | 16.143 | −12.066 | Assaba, Mauritania |
| JCB07675 | MK864178 | - | MK879257 | - | 16.143 | −12.066 | Assaba, Mauritania |
| BMNH.1976.2490 | MK864186 | - | - | - | 14.5 | 33 | Wad Madani, Sudan |
| MNHN.1979.702 | MK864187 | - | - | - | 9.34 | 13.41 | Garoua, Cameroon |
| MNHN.1995.5751 | MK864165 | - | - | - | −4.22 | 15.26 | Brazzaville, RCongo |
| MNHN.1995.5753 | MK864166 | - | - | - | −4.22 | 15.26 | Brazzaville, RCongo |
| MNHN.1995.2261 | MK864167 | - | - | - | 18.11 | 8.78 | Aïr, Niger (Aïr) |
| MNHN.2003.761 | MK864168 | - | - | - | 6.37 | 2.43 | Cotonou, Benin |
| MNHN.1979.1143 | MK864188 | - | - | - | 9.34 | 13.41 | Garoua, Cameroon |
| USNM.580615 | KY080138 | - | - | - | −3.027 | 10.375 | Basse-Banio, Gabon |
| USNM.576587 | KY080137 | - | - | - | 1.088 | 17.307 | Likouala, RCongo |
| USNM.578250 | KY080136 | - | - | - | −2.743 | 9.994 | Ogooué-Maritime, Gabon |
| USNM.576586 | KY080135 | - | - | - | 1.088 | 17.307 | Likouala, RCongo |
| USNM.578226 | KY080134 | - | - | - | −2.730 | 9.974 | Ogooué-Maritime, Gabon |
| USNM.576593 | KY080133 | - | - | - | −4.45 | 14.77 | Pool, RCongo |
| USNM.576592 | KY080132 | - | - | - | −4.45 | 14.77 | Pool, RCongo |
| USNM.578222 | KY080131 | - | - | - | −2.748 | 10.001 | Ogooué-Maritime, Gabon |
| USNM.584158 | KY080130 | - | - | - | −2.667 | 13.596 | Lekoumou, RCongo |
| USNM.576591 | KY080129 | - | - | - | −4.45 | 14.77 | Pool, RCongo |
| USNM.576588 | KY080128 | - | - | - | 1.088 | 17.307 | Likouala, RCongo |
| USNM.576585 | KY080127 | - | - | - | 1.088 | 17.307 | Likouala, RCongo |
| USNM.580614 | KY080126 | - | - | - | −3.083 | 10.437 | Basse-Banio, Gabon |
| USNM.578221 | KY080125 | - | - | - | −2.784 | 10.106 | Ogooué-Maritime, Gabon |
| occ-afri-B | AB272600 | - | - | - | −3.83 | 32.59 | Tanzania |
| occ-afri-A | AB272599 | - | - | - | −3.83 | 32.59 | Tanzania |
| ZMB.79256 | KF991268 | - | - | - | 10.9 | −10.9 | Guinea |
| ZFMK.65186 | AY014374 | - | - | - | 1.3 | 32.5 | Uganda |
| Maur023 | AY014373 | - | - | - | - | - | Mauritania |
| Iso0537 | DQ347291 | - | - | - | - | - | Unknown (shop) |
| MVZ.234146 | EU979846 | - | - | - | −2.243 | 33.852 | Mwanza, Tanzania |
| MVZ.235754 | EU979845 | - | - | - | 20.253 | −13.088 | Terjit, Mauritania |
| SL522 | GQ183571 | - | - | - | 0.39 | 29.87 | Rwenzori, DRCongo |
| CMR.1058 | AF261263 | - | - | - | - | - | unknown |
| KU.290425 | DQ283059 | - | - | - | 5.354 | −0.703 | Winneba, Ghana |
| Out-groups | |||||||
| H. rugulosus | AY014368 | - | - | - | - | - | - |
| H. rugulosus | AY014372 | - | - | - | - | - | - |
| H. tigerinus | AY014370 | - | - | - | - | - | - |
| H. tigerinus | AY014371 | - | - | - | - | - | - |
| H. crassus | AY014369 | - | - | - | - | - | - |
| H. crassus | AY014375 | - | - | - | - | - | - |
| F. greenei | AY014378 | - | - | - | - | - | - |
| F. kirtisinghei | AY014380 | - | - | - | - | - | - |




