Lack of congruence of genetic and niche divergence in Podarcis hispanicus complex
Abstract
Niche divergence among closely related lineages can be informative on the ecological and evolutionary processes involved in differentiation, particularly in the case of cryptic species complexes. Here we compared phylogenetic relationships and niche similarity between pairs of lineages included in the Podarcis hispanicus complex to examine patterns of niche divergence and its role in the organization of current diversity patterns, as allopatric, parapatric, and sympatric lineages occur in the Western Mediterranean Basin. First, we used ecological niche models to characterize the realized climatic niche of each Podarcis hispanicus complex lineage based on topographic and climatic variables, to identify important variables, and to test for niche conservatism or divergence between pairs of lineages. Variables related to precipitation generally exhibited the highest contribution to niche models, highlighting the importance of rainfall levels in shaping distributions of Podarcis wall lizards. We found that most forms have significant differences in realized climatic niches that do not follow the pattern of mitochondrial divergence. These results lend support to the hypothesis that genetic divergence across Podarcis hispanicus complex most likely occurred in allopatric conditions, mostly with significant niche divergence. Competition after secondary contact is also suggested by the common occurrence of niche overlap between lineages that exhibit strictly parapatric distribution. The almost continuous distribution of Podarcis lizards in the study area appears to be a result of a combination of complementary suitable niches and competition, which seem two important mechanisms limiting geographic distributions and restricting the existence of extensive contact zones.
1 INTRODUCTION
The geographic distribution of organisms is the result of three main limiting factors: determinants to dispersion (species’ intrinsic or historical constraints), abiotic factors, and biotic interactions: Species can live in climatically favorable regions where they are able to disperse and from where they are not excluded by biotic interactions (Barbosa, Sillero, Martínez-Freiría, & Real, 2012; Soberón, 2007). Range limits are thus determined by the areas where ecological conditions are favorable: The species’ range is the geographic expression of the species’ ecological niche. Grinnell (1917) defined the ecological niche as a portion of the habitat containing the environmental conditions that enable individuals of a species to survive and reproduce based on broad-scale variables (climate) that are not affected by species density. Instead, Elton (1927) emphasized the functional role of a species in a community, especially its position in food webs, based on fine-scale variables representing resources that may be consumed or modified by the species. The fundamental niche is the multidimensional environmental space where each dimension is a variable describing the conditions under which a species can maintain a viable population and persist over time (Hutchinson, 1957). Jackson and Overpeck (2000) defined the concept of potential niche as the part of the fundamental niche that is currently available for the species and Pearson (2007) introduced the concept of occupied niche, to which species distributions are limited by historical, geographic, and biotic factors. The realized niche is the part of the fundamental niche where the species is not excluded by biotic interactions and dispersal factors, and it may inform us about environmental variables driving range limits (Holt, 2003; Soberón & Nakamura, 2009). Because of this multidimensionality, it is not possible to investigate into detail each and every one of the dimensions that influence the niche, but climatic and physical factors merit particular attention, as they are recognized to affect profoundly the distributions of species at small scales (Soberón & Peterson, 2005).
We here follow Sillero (2011), who proposed the term realized niche model when predicting the species’ realized niche, for those correlative models using presence/true-absence, presence/pseudo-absence, or presence-only species records. When using presence-only data, like the analyses in this study, the model represents the realized niche and not the fundamental one, because historical/dispersal factors are in fact included in the presence data. Here we use mainly climatic variables associated with presence data, so we effectively model the realized climatic niche, while the projected distributions are the geographic extent of the realized climatic niche.
In view of the importance of the realized niche for understanding the environmental conditions that favor the occurrence of a species, comparing the realized climatic niche between pairs of species may also provide insights about other factors limiting distributions, as well as about the location and extent of contact zones (Rieseberg et al., 2003; Swenson, 2006; Swenson, Fair, & Heikoop, 2008). When two species have widely overlapping realized climatic niches, but exhibit parapatric or allopatric distributions in the geographic space, we can infer that their distributions are limited by other factors (e.g., biotic, historical, dispersal: Warren et al. 2014), under the condition that the studied variables are important for describing the species’ niche. Movement abilities and dispersal limitations play an important role in this topic (Holt, 2003). Considering the aforementioned parapatric example, biotic factors like a competition between the two species may also contribute to explaining distribution limits (Barbosa et al., 2012; Soberón & Peterson, 2005). Similarly, reduced dispersal capabilities and the existence of geographic barriers may favor or facilitate the allopatric occurrence of two species (Barbosa et al., 2012; Holt, 2003; Soberón & Peterson, 2005).
The role of ecological divergence in driving the origin and maintenance of distinct lineages has been the subject of long-standing interest in evolutionary biology (Wright, 1921; Mayr, 1963; Coyne and Orr, 2004). Divergence in ecological niche has been hypothesized to constitute one of the main mechanisms of speciation (Schluter, 2001, 2009). Under this hypothesis, adaptation to different environments is a major agent of the evolution of reproductive isolation. Divergent selection in ecology may occur in sympatry, as well as in a situation of allopatric or parapatric speciation. If the realized climatic niches of allopatric or parapatric sister lineages do not overlap, then the ecological adaptation with barriers to dispersal may be inferred to have played an important role in lineage differentiation (Graham, Ron, Santos, Schneider, & Moritz, 2004; Wiens, 2004; Wiens & Graham, 2005). Alternatively, if realized climatic niches overlap significantly, speciation was not accompanied by divergence in environmental preferences and probably occurred in allopatry or parapatry (incidental divergence, e.g., genetic drift, Peterson, Soberón, & Sanchez-Cordero, 1999; Graham et al., 2004; Wiens, 2004; Wiens & Graham, 2005). In cases of non-sister lineages occupying similar niches, these may be inferred to have retained the ancestral niche through time or, alternatively, they may have independently derived the same niche through convergent evolution (Knouft, Losos, Glor, & Kolbe, 2006). Understanding the evolutionary patterns of niche diversification can thus disclose valuable insights about the distribution of lineages and its role in lineage divergence.
Wall lizards of the genus Podarcis evolved and diversified in the Mediterranean Basin (Arnold, Arribas, & Carranza, 2007), occupying a wide variety of habitats. They are distributed from Central Europe to the northern limits of the Sahara and from the Iberian Peninsula to the Black Sea (Sillero et al., 2014). Species of Podarcis from the Iberian Peninsula and Northern Africa, with the exception of Podarcis muralis (Laurenti, 1768), form a monophyletic clade (Harris & Arnold, 1999; Oliverio, Bologna, & Mariottini, 2000), which constitutes the Podarcis hispanicus (Steindachner, 1870) complex (Harris & Sá-Sousa, 2002). They are widely distributed in the Iberian Peninsula and often abundant in Northern Africa, where they exhibit high diversity of ecological situations, and several cases of sympatric, parapatric and allopatric distributions occur throughout these regions. The Podarcis hispanicus complex is thus an interesting group to study patterns of niche divergence and configuration of species’ distributions.
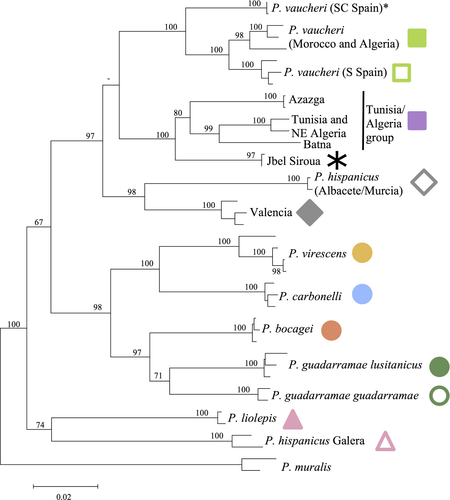
The current range of Iberian and North African Podarcis lizards is mostly parapatric, with several restricted contact zones between parapatric lineages currently being investigated (e.g., Podarcis bocagei (Seoane, 1884) with Podarcis carbonelli Pérez-Mellado, 1981, Pinho, Kaliontzopoulou, Carretero, Harris, & Ferrand, 2009; P. carbonelli with Podarcis vaucheri (Boulenger, 1905), Podarcis guadarramae guadarramae (Boscá, 1916) with Podarcis liolepis (Boulenger, 1905), pers. obs.). Two lineages are mostly sympatric with other lineages (P. bocagei with Podarcis guadarramae lusitanicus (Geniez, Sá-Sousa, Guillaume, Cluchier, & Crochet, 2014), and P. carbonelli with P. g. guadarramae, Podarcis virescens (Geniez et al., 2014), or P. vaucheri, Kaliontzopoulou, Pinho, Harris, & Carretero, 2011). Despite the increasing number of genetic studies on this complex of species, the ecological niche and distribution limits of some lineages are still poorly known. During the last decade, the Podarcis hispanicus species complex has been subject to several phylogenetic studies (e.g., Harris & Sá-Sousa, 2002; Kaliontzopoulou et al., 2011; Lima et al., 2009; Pinho, Ferrand, & Harris, 2006) which have uncovered cryptic variation and lead to the discovery of 16 mitochondrial lineages, including eleven lineages in the Iberian Peninsula and five in North Africa (Kaliontzopoulou et al., 2011; Figure 2). Eight lineages, identified on the basis of multilocus genetic data (see Pinho, Harris, & Ferrand, 2007), morphology and ecology, are currently recognized as valid species or subspecies (see Pérez-Mellado, 1981; Sá-sousa, 2001; Sá-Sousa & Harris, 2002; Busack, Lawson, & Arjo, 2005; Geniez, Cluchier, Sá-Sousa, Guillaume, & Crochet, 2007; Renoult, Geniez, Bacquet, Benoit, & Crochet, 2009; Geniez et al., 2014), while others still lack taxonomic revision. Lastly, one mitochondrial (mtDNA) lineage (called “Valencia” by Renoult et al., 2009 or “hispanicus sensu stricto” by others) has so far only been identified as an introgressed lineage in all southern populations of P. liolepis and some population of P. hispanicus (Renoult et al., 2009). In this work, when we mention P. liolepis or P. hispanicus Galera, we refer only to Liolepis and Hispanicus mtDNA lineages mentioned by Renoult et al. (2009), respectively.
The application of ecological niche modeling (ENM) together with Geographical Information Systems (GIS) allows the development of robust models that relate biological diversity with environmental factors in a geographically explicit framework (Guisan & Zimmermann, 2000; Sillero, 2011; Warren, 2012, 2013). The integration of phylogenetic information allows assessing if closely related lineages evolved in similar or distinct ecological conditions (e.g., Kidd & Ritchie, 2006; Pearman et al., 2014; Rissler & Apodaca, 2007) either in sympatry, parapatry or allopatry. In this study, we employ ENM based on topographic and climatic variables, to (i) characterize the realized climatic niche (sensu Sillero, 2011) of each member of the Podarcis hispanicus species complex using genetically confirmed occurrence records and (ii) to identify which are the main topographic and climatic variables influencing the niche and how they are related to the presence of each lineage. Then, based on the inferred ecological niche models, we (iii) test for niche conservatism or divergence between lineages to assess the similarity (or lack thereof) in realized climatic niche and to incorporate these results with previously determined phylogenetic relationships within this group (see Kaliontzopoulou et al., 2011). Finally, we want to (iv) infer how niche divergence has contributed to current patterns of spatial organization among lineages of the Podarcis hispanicus complex and how it has shaped the potential for geographic co-occurrence between pairs of lineages.
2 MATERIAL AND METHODS
2.1 Study area and presence records
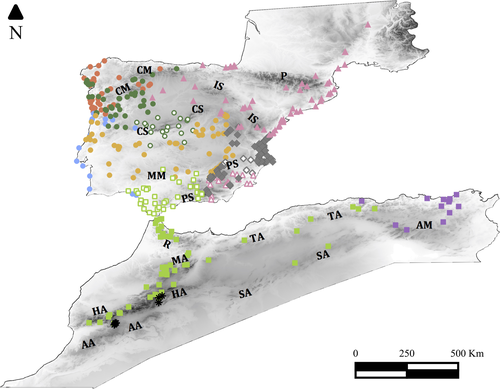
Our study area includes Southern France, the Iberian Peninsula, and the Mediterranean region of Northern Africa (Figure 1), encompassing the known distribution range of the Podarcis hispanicus species complex. Due to the low number of available records for some lineages, we modeled together the monophyletic group formed by the three Algerian and Tunisian lineages (see Figure 2), which we will refer to as Tunisia/Algeria group hereafter. The most recently discovered lineage of P. vaucheri from southern Spain (P. vaucheri SC Spain, Kaliontzopoulou et al., 2011) was excluded due to the very low number of geographic records. Because field morphological identification is still unsafe for several lineages, in this study we use mitochondrial lineages as a proxy for evolutionary lineages as this is the only cost-effective way to gather numerous genetically confirmed presence data. We thus assembled a total of 518 presence records for 13 Podarcis lineages identified on the basis of mtDNA. We have included 93 new samples specifically analyzed for this study (GenBank accession numbers KY461834 to KY461930) as well as 425 samples published in other studies, with mtDNA sequences and detailed geographic information available (Busack et al., 2005; Arntzen & Sá-Sousa, 2007; Carranza, Arnold, & Amat, 2004; Castilla et al., 1998; Dias, Luis, Pinho, & Kaliontzopoulou, 2016; Harris & Arnold, 1999; Harris, Batista, Carretero, Pinho, & Sá-Sousa, 2002; Harris, Carranza, Arnold, Pinho, & Ferrand, 2002; Harris & Sá-Sousa, 2001, 2002; Kaliontzopoulou et al., 2011; Lima et al., 2009; Oliverio et al., 2000; Pinho, Harrid, & Ferrand, 2007; Pinho, Harris, & Ferrand, 2008; Pinho et al., 2006; Renoult, 2006; Sanz-Azkue, García-Etxebarria, Gosá, Rubio, & Jugo, 2006). For all individuals, we identified the mtDNA lineage through the analysis of partial fragments of mitochondrially encoded 12S ribosomal RNA region (12S), 16S ribosomal RNA region (16S), NADH dehydrogese subunit 4 (ND4), control region (D-loop), or cytochrome c oxidase subunit I (COI). Identification was performed by comparison of the target sequences with our extensive database of reference sequences available for all gene fragments employed using tree-building approaches like in Kaliontzopoulou et al. (2011). Individual identifications were all unambiguous given the good separation of all lineages in each of these gene fragments. For fragments specificaly sequenced for this study we used the primers and conditions described in Pinho, Ferrand & Harris (2006). The details on fragments sequenced for each sample are in Table S1. For several individuals, the identification was based in more than one fragment (individuals with more than one fragment amplified are indicated with a “+” in Table S1). Presence records were either geolocalized in the field with a GPS or the coordinates were retrieved from Google Earth for samples that were collected without GPS, with an accuracy according to the spatial resolution of environmental variables used for modeling purposes (see below). Detailed information about all used samples, as their respective assignments to mtDNA lineages and geographic coordinates, is listed in Table S1.
2.2 Environmental variables
A set of 21 variables was initially considered for the study area. Nineteen climatic variables were obtained from the WorldClim database (Hijmans, Cameron, Parra, Jones, & Jarvis, 2005; http://www.worldclim.org/). Two topographic variables were also obtained: Altitude was retrieved from the Shuttle Radar Topography Mission (USGS 2006; http://glcf.umd.edu/data/srtm/), and slope was calculated in QGIS version 2.4.0 (QGIS Development Team 2013). All layers had a spatial resolution of 1 km2. To avoid the possible statistical effects of collinearity among predictor variables on niche modeling, we calculated pairwise Pearson correlations, and for each correlation higher than 0.75, we excluded the variable that correlates with the higher number of other variables, but always trying to keep the variables with a higher biological importance for the species. This resulted in a final set of seven variables considered for modeling purposes, including altitude (Alt), temperature seasonality (TSea), maximum temperature of the warmest month (MaxT), minimum temperature of the coldest month (MinT), annual precipitation (APre), precipitation of the driest month (PreDM), and precipitation seasonality (PreSea). All spatial data operations and analyses were performed using QGIS version 2.4.0 (QGIS Development Team, 2013).
2.3 Niche modeling and characterization
To model the realized climatic niche (sensu Sillero, 2011) of each lineage, we used the maximum entropy approach implemented in Maxent version 3.4.1 (available from https://biodiversityinformatics.amnh.org/open_source/maxent/). Maxent was designed for presence-only data (Phillips, Anderson, & Schapired, 2006) and is particularly efficient for datasets including a small number of records (Pearson, Raxworthy, Nakamura, & Peterson, 2007). We enabled linear, quadratic, and hinge features. Because of the small number of records available for some groups, we did not divide the datasets into training and testing subsets. As Maxent is a probabilistic method, each iteration of the modeling process results in slightly different models. To account for model uncertainty, we calculated 50 replicated, independent Maxent models for each lineage, which were then averaged to obtain a consensus model (Araújo & New, 2007). For other parameters, we kept the default values. An important issue was whether our dataset was geographically biased or not. So we calculated models with and without correction for sampling bias. To take sampling bias into account when calculating Maxent models, we used a 100 km buffer area around each occurrence point where we randomly sampled 10,000 background samples as advocated by Phillips (2008) and Fourcade, Engler, Rödder, and Secondi (2014). We tested model performance by considering the area under the curve (AUC) of the receiver operating characteristics (ROC) plots (Liu, Berry, Dawson, & Pearson, 2005). Variable importance was determined by jack-knife resampling of the model gain and AUC. Gain is a measure of how much better the predicted distribution fits the sample points as compared to a uniform distribution (Phillips & Dudík, 2008; Phillips, Dudík, & Schapire, 2004; Phillips et al., 2006). For this purpose, Maxent excludes each variable in turn and creates a model with the remaining variables; then, it also creates a series of models using each variable separately. Maxent determines the contribution of each environmental variable to the final model by randomly permuting the values of each variable among the presence points and measuring the relative decrease in AUC, normalized to percentage (Permutation importance).
In addition to Maxent models, we performed an ecological niche factor analysis (ENFA; Hirzel, Hausser, Chessel, & Perrin, 2002) to obtain a multivariate representation of the realized climatic niche of the different lineages by comparing the statistical distribution of climatic variables for each presence dataset (the lineage niche) and for the whole study area. This analysis was performed using Biomapper 4.0 (Hirzel, Hausser, & Perrin, 2006) following Hirzel et al. (2002). ENFA summarizes climatic variation into uncorrelated factors in a similar manner to Principal Component Analysis. The first factor, marginality, expresses the average difference between the species niche and the total available conditions (Hirzel et al., 2002). Marginality varies most often from 0, for species living in average habitat conditions, to 1, for species inhabiting very particular conditions relatively to the studied area. Larger values indicate that the mean variable values where species are present are fairly different from the average variable values across the studied area. The other factors are called specialization and represent the variance of the species niche compared with available conditions, ranging from 1 in generalist species, to infinite in specialist species (Hirzel et al., 2002). Comparisons between species are usually performed using the inverse of specialization, tolerance, which varies from 0 for specialist species to 1 for generalist species. The robustness of the models was evaluated with the continuous Boyce index using a Spearman rank correlation coefficient that measures how much model predictions differ from a random distribution of the observed presences across the prediction gradients (Boyce, Vernier, Nielsen, & Schmiegelow, 2002).
2.4 Niche differentiation
We analyzed climatic niche differentiation among lineages using the “ENMTools” 0.1 R package (Warren, 2016) in R 3.2.3 (R Development Core Team, 2015). We measured the predicted climatic niche overlap between pairs of lineages by calculating Schoener's D statistics from probability surfaces of the climatic niche models obtained from Maxent as described by Warren, Glor, and Turelli (2008). D ranges from 0 if niche models have no overlap to 1 if niche models fully overlap. To obtain a clustering representation of the difference in lineage climatic niche, we used the UPGMA method from “stats” 3.3.0 R package (R Development Core Team, 2015) in R.
We tested our models for niche identity based on D statistics, that is, whether the niches of each pair of lineages, as inferred by ENM, are more different than expected if they were drawn from the same underlying distribution (Warren et al., 2008). To test niche identity for each pair of lineages X and Y, pseudoreplicate datasets were created by randomly partitioning the pooled set of X and Y occurrences, maintaining the same number of samples as those originally available for each lineage. A niche model was created from each pseudoreplicate, and its corresponding D statistic (De) was obtained. We used 100 replications to create a null distribution of similarity values. The observed values of D were compared to the percentiles of these null distributions of De in a one-tailed test to evaluate the null hypothesis that the niche models are not statistically significantly different. If D falls in the 95% of the null distribution, the null hypotheses that niche models are identical should not be rejected. Of course, tests of niche identity between allopatric species are not fully meaningful, as climatic niches of species with allopatric distributions are expected to differ, especially in areas like the Iberian Peninsula where different regions experience very different climatic conditions.
Finally, we tested for background similarities, that is, to determine whether ENMs are more similar than expected by chance, even if they occur in different regions (Warren et al., 2008). This type of analysis partly overcomes the problem that allopatric species often inhabit regions with distinct distributions of environmental variables, hence generating distinct realized climatic niche even without a difference in habitat selection patterns. This test compares the actual similarity of the models (assessed by Schoener's D) between pairs of lineages X and Y with a null distributions of 100 expected similarities (Ds) between a randomized dataset for Y based on random occurrence points drawn from within the range occupied by Y, and the symmetric randomized dataset for X. We defined the range available for each lineage using a 50km buffer zone around the occurrence points. Since most of the study area is covered with presence records, these buffer zones should be a good approximation of actual distributions without creating extensive false continuous occurrence areas between isolated populations. If D falls in the 95% of confidence limits of Ds, the null hypotheses of background similarity should not be rejected. Rejection of the null hypothesis indicates that the niche models of two Podarcis lineages are more similar or different than would be expected by chance given the existing differences between the environment in their distribution, hence that the observed niche differentiation between them is a function of habitat selection and/or suitability and not an artifact of the underlying environmental differences between the habitats available to each lineage.
To estimate the degree of geographic overlap of pairs of Podarcis lineages to understand whether niche divergence is accompanied, or not, by distinct geographic distributions, we first described the area of occurrence of each lineage and then calculated an index of pairwise geographic overlap. Because for some lineages we still lack accurate distribution limits, we described the current distribution range of each lineage by defining a 50 km buffer zone around the corresponding occurrence points, which should be a good approximation of actual distributions, as described before. We measured the distribution similarity of pairs of lineages using the binary Jaccard's similarity index (JSI) as implemented in the package “vegan” (Oksanen et al., 2017) for R. JSI ranges from 0, when two forms have no distribution grid squares in common, to 1 when all grid squares are shared. As for D, we used the UPGMA algorithm to obtain a clustering representation of global similarities in lineage distributions.
To test for correlations between both D, or JSI of geographic distribution, and the genetic distance, we performed two Mantel tests. To calculate the genetic distances between each pair of lineages, we used the most complete genetic dataset available in terms of mtDNA fragment length: the dataset used in Kaliontzopoulou et al. (2011). We calculated Tamura and Nei (1993) genetic distance between each individual using ape 4.1 R package (Paradis, Claude, & Strimmer, 2004), and then, we used the average genetic distance between each pair of lineages to compute the Mantel test with ade4 1.7-6 R package (Dray & Dufour, 2007) to test the null hypothesis that niche overlap, or geographic distribution, and genetic distance are not related, with 9,999 permutations for α = 0.05.
3 RESULTS
3.1 Accuracy of the ecological niche models and variable importance
The Maxent model results presented in this study are the results obtained with sampling bias correction as both results are similar (see the subsection “Some methodological remarks” in “Discussion” for further details). We obtained consensus Maxent models with very high average AUC values (0.93–0.99, Table 1). ENFA models also performed well, as indicated by a very high continuous Boyce index for most of models (0.79–0.99, Table 1). Only the ENFA models for P. hispanicus Albacete/Murcia and Tunisia/Algeria group had lower (0.49 and 0.25, respectively) but still positive values, meaning that ENFA models are good.
| Model | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pb | Pc | Pgl | Pgg | Pv | Pl | PhG | PhAM | V | PvSS | PvMA | TAg | JS | |
| N | 40 | 23 | 44 | 19 | 72 | 52 | 16 | 7 | 68 | 29 | 52 | 14 | 9 |
| AUC | 0.99 | 0.99 | 0.97 | 0.97 | 0.93 | 0.95 | 0.96 | 0.94 | 0.98 | 0.95 | 0.97 | 0.93 | 0.99 |
| B | 0.97 | 0.79 | 0.97 | 0.84 | 0.99 | 0.98 | 0.89 | 0.49 | 0.90 | 0.88 | 0.97 | 0.25 | 0.84 |
For all lineages, at least one of the three variables related to precipitation (annual precipitation, precipitation in the driest month, and precipitation seasonality) had a considerable importance for the final Maxent models (21%–71%; Figure 3). For four lineages, only one variable had a very high importance (more than 50%): precipitation in the driest month for P. hispanicus Albacete/Murcia and P. liolepis; annual precipitation for Tunisia/Algeria group; and altitude for Jbel Siroua. Generally, temperature-related variables had lower contributions to the models (Figure 3). The gains of most Maxent models considering each variable alone were higher for precipitation in the driest month, but it is evident the gain for Jbel Siroua when considering altitude and a minimum temperature of coldest month (Figure S1). However, when excluding only one variable at a time, the gain was quite similar across variables for all models. Only when excluding annual precipitation for Tunisia/Algeria group was the gain lower (Figure S2). Therefore, this variable provided exclusive information to the niche model of this lineage.
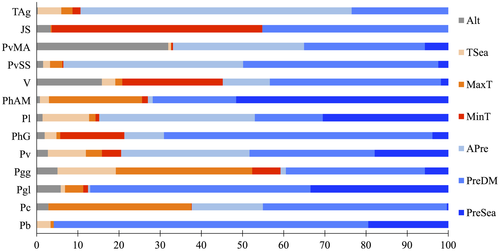
Variables’ response curves for each lineage are represented in detail in Figure S3. The probability of occurrence of almost all lineages is higher with medium levels of precipitation in the driest month and with moderate levels of precipitation seasonality, but the presence of P. bocagei, P. carbonelli, P. g. lusitanicus, and Tunisia/Algeria group is also more probable with high levels of annual precipitation. With some exceptions, the probabilities of occurrence of most lineages are lower with high maximum temperatures of the warmest month and low minimum temperatures of the coldest month. For Jbel Siroua, the probability of occurrence is very high with low values of minimum temperature of the coldest month and high altitudes.
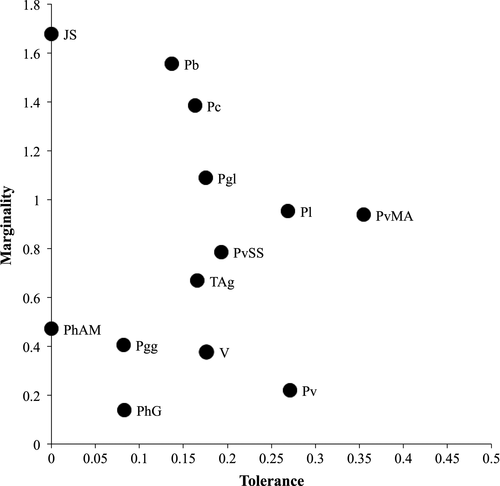
Examination of marginality versus tolerance values calculated by ENFA models (Figure 4) showed a general trend toward low tolerances, even for those forms inhabiting wider areas, such as P. liolepis, P. virescens, and P. vaucheri Morocco/Algeria, and a continuum trend from low to high marginality. The lineage with the highest tolerance was P. vaucheri Morocco/Algeria, although its value can be considered relatively low (0.35). Jbel Siroua and P. hispanicus Albacete/Murcia were the lineages with the lowest tolerance values, very close to zero. Jbel Siroua was also the lineage with the highest marginality (1.68), while P. hispanicus Galera had the lowest marginality (0.14). Still, we can identify a group of lineages occupying average habitat conditions with marginality lower than 0.5: P. hispanicus Galera, P. virescens, Valencia, P. g. guadarramae, and P. hispanicus Albacete/Murcia; a group of forms occupying marginal habitats in the study area with marginality higher than 1.0: Jbel Siroua (the highest value), P. bocagei, P. carbonelli, and P. g. lusitanicus; and an intermediate group with marginality between 0.5 and 1.0 comprising Tunisia/Algeria group, P. vaucheri Southern Spain, P. vaucheri Morocco/Algeria, and P. liolepis.
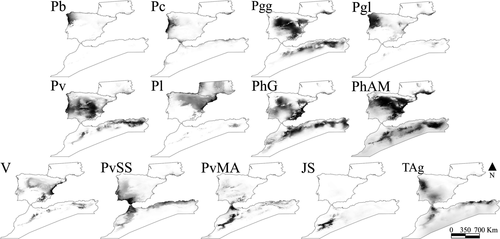
3.2 Predicted areas of suitability
According to Maxent results (Figure 5), some models occupy Central and Southern Iberian Peninsula (P. g. guadarramae, P. virescens, P. hispanicus Galera, P. vaucheri Spain and P. hispanicus Albacete/Murcia, Tunisia/Algeria group) as well as North of Africa. Lineages occurring in the north and west of the Iberian Peninsula (P. bocagei, P. carbonelli, P. g. lusitanicus, P. liolepis) and the two North African lineages mostly distributed in mountain regions (Jbel Siroua and P. vaucheri Morocco/Algeria) had more restricted predicted distributions (Figure 5), mostly identical with the current known distributions. Detailed maps with each model with the presence records depicted are represented in Figure S4.
3.3 Niche divergence in environmental and geographic spaces
The dendrogram constructed based on realized climatic niche divergence (D, Figure 6a) indicated the existence of four clusters, but even within groups, we found high divergence levels. Jbel Siroua, P. vaucheri Morocco/Algeria, P. vaucheri South Spain, and Tunisia/Algeria group were the first cluster to be segregated. Then, we can distinguish a cluster with P. carbonelli, P. bocagei, and P. g. lusitanicus. Finally, a group established by Valencia lineage, P. hispanicus Galera, P. g. guadarramae, and P. virescens was distinct from another cluster that includes P. liolepis, P. hispanicus Albacete/Murcia. Such results contrast with mtDNA phylogenetic relationships (Figure 2) and estimates of relationships based on geographic distribution areas (JSI, Figure 6b; see Table S2 for detailed results), which shows that most lineages have unique distributions, except for the two lineages from Northwest Iberia (P. bocagei and P. g. lusitanicus) and the three from southeast (Valencia lineage, P. hispanicus Galera, and P. hispanicus Albacete/Murcia).
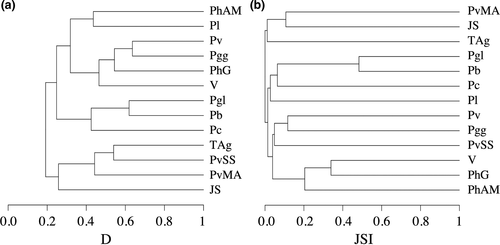
Niche overlap D varied between 0.015 and 0.636, but most pairs exhibited a restricted niche overlap (Table 2) as expected from their mostly allopatric distribution (Figure 6b). Of 78 pairs, only six had D > 0.5. When tested for niche identity, 66 of 78 pairs were significantly different (bold values in Table 2), and thus, the null hypothesis of identical niche could be rejected in these cases. Niche background similarity tests revealed that 45 pairs were less similar than expected based on random sampling of regional differences in environmental variables (gray shaded values in Table 2). For 33 comparisons, D did not significantly deviate from the expected distribution. No pairs of Podarcis lineages were more similar than what expected by chance.
| Pb | Pc | Pgl | Pgg | Pv | Pl | PhG | PhAM | V | PvSS | PvMA | JS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pc | 0.344 | |||||||||||
| Pgl | 0.619 | 0.506 | ||||||||||
| Pgg | 0.141 | 0.258 | 0.332 | |||||||||
| Pv | 0.178 | 0.370 | 0.338 | 0.636 | ||||||||
| Pl | 0.250 | 0.163 | 0.306 | 0.324 | 0.301 | |||||||
| PhG | 0.121 | 0.272 | 0.274 | 0.576 | 0.512 | 0.222 | ||||||
| PhAM | 0.250 | 0.163 | 0.324 | 0.306 | 0.301 | 0.436 | 0.222 | |||||
| V | 0.167 | 0.228 | 0.328 | 0.483 | 0.493 | 0.436 | 0.419 | 0.435 | ||||
| PvSS | 0.111 | 0.458 | 0.192 | 0.231 | 0.402 | 0.109 | 0.304 | 0.109 | 0.191 | |||
| PvMA | 0.081 | 0.301 | 0.184 | 0.308 | 0.351 | 0.115 | 0.316 | 0.037 | 0.299 | 0.469 | ||
| JS | 0.015 | 0.083 | 0.044 | 0.140 | 0.111 | 0.037 | 0.187 | 0.067 | 0.102 | 0.157 | 0.399 | |
| TAg | 0.163 | 0.418 | 0.279 | 0.222 | 0.359 | 0.067 | 0.268 | 0.115 | 0.136 | 0.540 | 0.416 | 0.217 |
The results from the Mantel tests show no significant correlation between niche overlap and genetic distances (r = −.206, p-value = .914) or geographic distribution and genetic distances (r = .055, p-value = .280).
4 DISCUSSION
We used ecological niche modeling in the Podarcis hispanicus species complex to examine which climatic factors influence species’ distributions and to assess how climatic niche divergence may contribute to lineage divergence and shape the geographic distribution of these lineages. We documented a high variation in niche similarity between pairs of lineages, ranging from almost no similarity (D = 0.015) to important niche similarity (D = 0.636), but without congruence between niche divergence, or geographic overlap, and mtDNA divergence. These results may be attributed to multiple events of allopatric divergence and shed more light on the ecological processes potentially involved in shaping lineage divergence and geographic space occupancy patterns in this group. Indeed, this lack of congruence may be the result of the divergence of closely related lineages in markedly distinct environments, while other more closely related lineages diverged in allopatric regions but with more similar environmental conditions.
4.1 Biogeographic and ecological affinities
The overall low niche tolerance observed across lineages suggests that Podarcis lineages have some degree of climatic specialization, and their marginality values follow a gradient of habitat from marginal to average conditions (Figure 4). Low marginalities correspond mostly to forms occupying Mediterranean conditions (e.g., P. hispanicus Galera, P. g. guadarramae, or P. virescens), while high marginalities correspond to Atlantic or mountainous conditions (e.g., P. bocagei, P. g. lusitanicus, or Jbel Siroua lineage). Such results point to distinct realized climatic niches that are, however, not completely divergent between several pairs of lineages.
Variables related to precipitation generally have the highest contribution to the models but their influence on distribution varies among lineages. For almost all lineages, we found that environmental suitability responds to precipitation in the driest month in a very similar way, with a peak of suitability around similar values of the lower end of the distribution of values for this variable, suggesting that all lineages select areas with similar and moderate values of rain in the driest month. This is not surprising as in general Podarcis lizards occupy areas in the study region with two well demarcated seasons, a humid season with higher levels of precipitation and a dry season with lower levels of precipitation more prominent across Mediterranean climatic regions. Still, species like P. bocagei, P. g. lusitanicus and P. liolepis exhibit larger standard deviations in the response curve for this variable (Figure S2), as they can also occupy regions with higher precipitation levels. However, high values of annual precipitation seem to be important for P. bocagei, P. carbonelli, P. g. lusitanicus, P. liolepis, and Tunisia/Algeria group. Preference for habitats with relatively high humidity has been previously reported for several Podarcis species (e.g., P. cretensis, Herkt, 2007), including some of the lineages included in this study (e.g., P. carbonelli, Sá-sousa, 2001; Román, Ruiz, Delibes, & Revilla, 2006; Sillero & Carretero, 2013; north African Podarcis, Kaliontzopoulou, Brito, Carretero, Larbes, & Harris, 2008). High maximum temperatures during the warmest month hinder the occurrence of most lineages, for example, P. liolepis, P. hispanicus Galera, P. bocagei, P. carbonelli, P. virescens, Valencia, P. vaucheri Spain. Indeed, Podarcis lizards are known to avoid extremely arid, hot, and dry regions associated with desert environments in North Africa, which is most likely linked to the origin of the genus in more temperate Mediterranean environments (see Harris, Batista, et al., 2002; Harris, Carranza, et al., 2002; Lima et al., 2009). Podarcis bocagei, P. carbonelli, and P. g. lusitanicus seem more related to less marked temperature seasonality. This is congruent with a previous study where P. bocagei and P. carbonelli were associated with Atlantic bioclimatic regions, characterized by mild summers (Sillero, Brito, Skidmore & Toexopeus, 2009).
The six lineages with the lowest marginality, that is, inhabiting environments closest to the average conditions available (Mediterranean climate), have high proportions of predicted suitable niches both in the Iberian Peninsula and in North Africa (P. hispanicus Galera, P. g. guadarramae, P. virescens, P. hispanicus Albacete/Murcia, P. vaucheri Spain, and Tunisia/Algeria group). Because these lineages are not monophyletic and their most recent common ancestor is the ancestor of the entire group, it is likely that a niche related to Mediterranean climate is the ancestral state, while specialization to Atlantic or Atlantic-like marginal climates evolved later, independently.
4.2 Climatic niche divergence in a phylogenetic context
Our data did not reveal any relationships between climatic niche divergence and mtDNA divergence as shown by Mantel test and by the comparison of maximum likelihood estimates of mtDNA relationships and niche overlap among Podarcis hispanicus complex (Figures 2 and 6a). Despite methodological differences, several examples of closely related taxa that diverge greatly in niche have been revealed in previous studies (e.g., on dendrobatid frogs, Graham et al., 2004; Anolis sagrei group, Knouft et al., 2006). Several Podarcis sister mtDNA lineages have distinct distributions as shown by low JSI and no significant niche similarities (e.g., Jbel Siroua with Tunisia/Algeria group, JSI = 0, D = 0.217; and P. carbonelli with P. virescens, JSI = 0.028, D = 0.370; divergence from 4 to 5.5 Mya, Kaliontzopoulou et al., 2011). This may be attributed to allopatric divergence in distinct environments. It is thus possible that significant realized climatic niche divergence occurred associated with several differentiation events under the influence of local adaptation (Warren et al., 2008). On the other hand, some highly divergent non-sister lineages with distinct distributions do not exhibit significant realized climatic niche differences (e.g., P. hispanicus Galera with P. g. guadarramae, JSI = 0; D = 0.576–10.4 Mya to the most recent common ancestor, Kaliontzopoulou et al., 2011). Convergence or long-term niche conservatism are both possibilities to explain such patterns (Knouft et al., 2006). Furthermore, some allopatric sister lineages without significant realized climatic niche differences (P. vaucheri Spain with P. vaucheri Morocco/Algeria, JSI = 0, D = 0.469) reflect cases with substantial environmental space overlap. Since similar conditions exist on both sides of the Strait of Gibraltar, these lineages have diverged in allopatry but under more or less similar environmental conditions.
By conducting analyses of background similarity, we found that 58% of pairs of lineages’ realized climatic niche were more different than expected by chance because the regions they occupy are environmentally different. This result suggests that the observed realized climatic niche differentiation between these pairs is partly due to different patterns of habitat selection within the heterogeneous environmental background. Ecological differentiation between these forms may reflect adaptation to different conditions driven by the differences in the heterogeneity within each environmental background (Warren et al., 2008; Warren et al. 2014). Parapatry between P. bocagei and P. liolepis in Northern Spain, for example, may thus be maintained by distinct environmental conditions. However, an important proportion of the models (42%) were no more similar or different than expected by chance.
Overall, our results are compatible with the hypothesis that genetic divergence across this group of lizards was likely built in allopatric conditions. In such a scenario, the almost continuous distributions (either in parapatry or partial sympatry) observed at present across this clade would have developed after secondary geographic contact between lineages. Although the analyses presented here cannot fully disclose whether realized climatic niche changes directly caused differentiation or speciation events, or whether alternatively are a consequence of lineage divergence, it is clear that realized climatic niche divergence is poorly related to genetic divergence as even closely related lineages have restricted realized climatic niche overlap.
4.3 Climatic niche divergence and geographic distribution
Identity tests reveal that most pairs of lineages (85%) are significantly different in the environmental space. Most of these pairs are formed at least by one lineage with low tolerance but high marginality (e.g., P. hispanicus Jbel Siroua, P. bocagei, P. carbonelli, P. g. lusitanicus), that is, lineages with high specialization, occurring in marginal environmental conditions in the study area, and with restricted geographic distributions. Because these lineages have no significant realized climatic niche similarities with most other lineages (see Table 2), we can infer that their current distinct distribution ranges (Figure 6b) may be mostly shaped by environmental conditions.
According to prediction maps of suitability (Figure 5) and based also on the results of equivalence tests (Table 2), considerable realized climatic niche overlap would be expected between some lineages (15%), which should exhibit extensive areas of overlap. Some of these lineages have high tolerance (e.g., P. vaucheri Morocco/Algeria, P. virescens), reflecting large distribution ranges relatively to the study area, but most are lineages with low tolerance and marginality (e.g., P. hispanicus Albacete/Murcia, Valencia, P. hispanicus Galera, P. g. guadarramae), indicating that they inhabit relatively small areas close to average conditions in this region. This last group of lineages tends to have the highest realized climatic niche similarities with several other lineages (see Table 2). However, extensive areas of sympatry do not exist (Figure 6b). On the other hand, P. bocagei and P. g. lusitanicus co-occur in extensive sympatric areas across their distributions, and P. carbonelli is everywhere sympatric with other Podarcis species.
Assuming that most Podarcis lineages have been well sampled, at least in the Iberian Peninsula and in Morocco, the lack of extensive overlap shows that factors other than bioclimatic and topographic variables are important in shaping range limits in this species complex. This is particularly evident for lineages with low tolerance and marginality, as they present restricted distributions but niche models tend to occupy relatively large geographic areas. Such results show that generally lineages’ distributions may be primarily limited by physiological tolerances (Kellermann, van Heerwaarden, Sgrò, & Hoffmann, 2009), but a role for competition after secondary contact may be suggested, for example, between pairs like P. virescens and P. g. guadarramae or P. hispanicus Galera and P. hispanicus Albacete/Murcia. Dispersal constraints and/or historical factors may also be important limitations for their presence and the occurrence of extensive contact zones (Barbosa et al., 2012; Peterson, 2011). This may be illustrated by some cases where important geographic barriers are present or where geographic segregation is a consequence of the presence of other Podarcis lineages which diverged largely in allopatry (Diamond, 1973; Price, 2008). The first scenario includes P. vaucheri Southern Spain plus P. vaucheri Morocco/Algeria or P. vaucheri Southern Spain plus Tunisia/Algeria group disjointed by the Strait of Gibraltar. Actually, such allopatric lineages separated by the Strait of Gibraltar does not have significant realized climatic niche differences or significant distinct backgrounds as similar ecological conditions can be found in the Iberian Peninsula and North of Africa. In the second case, P. virescens occurrence between P. hispanicus Galera and P. g. guadarramae at central and southeastern Iberia may contribute to allopatry between these two latter lineages according to the distribution and overlap of suitable climatic areas for their occurrence. Jbel Siroua is in a similar situation with Tunisia/Algeria group and P. vaucheri Morocco/Algeria, occupying most of the available North African habitats. In the last case, these forms are generally allopatric, but the presence of P. vaucheri Morocco/Algeria may have contributed to a restricted geographic distribution as well as a more specialized realized climatic niche of the other two Podarcis lineages (Lima et al., 2009).
The almost continuous distribution of Podarcis’ lineages studied here results from a wide range of realized climatic niche divergence across the group. Furthermore, realized climatic niche overlap between several lineages suggests the potential for the existence of extensive areas of sympatry between several lineages that are, however, currently allopatric or parapatric. Hence, biotic interactions and historical factors seem to play an important role in shaping current patterns of distribution. Different realized climatic niches and competition both seem to interact to shape lineage distribution and limit overlap.
4.4 Some methodological remarks
Because it is still unsafe to identify all lineages of the P. hispanicus species complex on the basis of morphology (i.e., Kaliontzopoulou, Carretero, & Llorente, 2012), the use of mtDNA for identification guarantees numerous genetically confirmed presence data. However, a certain level of mismatch with evolutionary relationships has to be expected, especially in the southeast of the Iberian Peninsula, where the Valencia lineage has been identified only as an introgressing mtDNA lineage in populations of P. liolepis or P. hispanicus Galera (Renoult et al., 2009).
Characterizing only a subset of the species’ fundamental niches leads to the limited representation of the full dimensions of these niches, that is, the realized niche (Peterson et al., 2011). Bioclimatic models can reconstruct environmental correlates of species’ geographic distributions (see Araújo & Peterson, 2012). Because all organisms require a stable input of energy for successful growth, survival, and reproduction (Porter and Gates, 1969; Kearney & Porter, 2004), the fundamental niche includes those environmental variables that influence organisms’ energy. Particularly for terrestrial ectotherms, like Podarcis hispanicus lineages, this “climate-space” is important. Since in this study, rather than predicting limits and detailed geographic distributions, we wish to understand environmental correlates that explain species’ distributions, and given the availability of spatiotemporal data for climatic conditions, the representation of the realized climatic niche obtained thus explains much about Podarcis hispanicus lineage distributions as well as provides a solid basis from which to consider the effects of other environmental variables such as biotic interactions (Kearney & Porter, 2004).
An important issue was whether our dataset was geographically biased. To overcome this possible problem, we used a “restricted background” correction method proposed by Phillips (2008) and discussed by Fourcade et al. (2014). We are aware that this correction method may not be the most efficient in some cases (Fourcade et al., 2014), but after model calculations, the AUC values were the same for all lineages as the models without correction for sampling bias. So the models with this correction method did not perform worse or better as the uncorrected dataset. These results together with the fact that the total area where different lineages of P. hispanicus complex occur is well covered with samples, the sampling bias should not be a major concern and that general conclusions are identical if we use the uncorrected dataset.
Furthermore, despite the high resolution of the GIS data (1 km2), the scale used in this study limits analyses to regions, rather than specific localities. We can only investigate environmental determinants of regional co-occurrence or allopatry, rather than strict sympatry or allopatric distribution at the local scale as differential habitat selection patterns may be responsible for the allopatric distribution at such scale. The next step should be the integration of fine-scale niche characteristics to elucidate the determinants of local distributions and habitat use, especially factors contributing to the formation and location of hybrid zones, which is of cornerstone importance for our understanding of the ecological and evolutionary processes taking place when different, closely related species, meet.
ACKNOWLEDGEMENTS
We thanks P. Bacquet, O. Chaline, A. Cluchier, V. Fradet, M. Geniez, P. Geniez, C. P. Guillaume, G. Guillaume, J. Magraner, U. Mathis, V. Rufray, M. Siol, I. Rocha, D. Salvi, M. A. Carretero, C. Rato, A. Perera, M. Ribeiro, D. Donaire, F. Jorge, G. Perez-Lanuza, E. Ayllón, P. Hernández-Sastre, J. C. Barberá, M. Barata, F. Martínez-Freiría, D. J. Harris, J. Maia, B. Tomé, I. Damas and J. C. Brito for helping with sample collection, P. Bacquet, M. Beddek, P. Sourrouille and M. Ribeiro for sequencing some of the samples and P. Geniez for managing the tissue database in Montpellier and helping us in many other ways. GCD was supported by a PhD grant (SFRH/BD/89750/2012); CL by a research grant (PTDC/BIA-BEC/102179/2008); NS, CP, and AK by IF contracts (IF/01526/2013, IF/01597/2014/CP1256/CT0009, and IF/00641/2014/CP1256/CT0008, respectively), all under the Programa Operacional Potencial Humano—Quadro de Referência Estratégico Nacional funds from the European Social Fund and Portuguese Ministério da Educação e Ciência. Support was also provided by FCT project PTDC/BIA-BEC/102179/2008, under FEDER COMPETE funds (FCOMP-01-0124-FEDER-007062) and by the project “Biodiversity, Ecology and Global Change” co-financed by North Portugal Regional Operational Program 2007/2013 (ON.2—O Novo Norte), under the National Strategic Reference Framework (NSRF), through the European Regional Development Fund (ERDF).




