3D printing in zoological systematics: Integrative taxonomy of Labrys chinensis gen. nov., sp. nov. (Nematoda: Tylenchomorpha)
Abstract
Three-dimensional printing technology has shown its importance in many fields. In this study, the potential of this technique in zoological systematics was assessed. For the first time, 3D printed models were incorporated in the description of a new genus as a complement to pictures and drawings to illustrate complex 3D structures and to be used in education. Hereby, we also tested the performances of different printing materials and suggest resin as the most suitable option for the zoological field. As a case study, Labrys chinensis gen. nov., sp. nov. was described using an integrative approach: detailed morphology based on light- and electron microscopy, phylogenetic position as revealed from two ribosomal RNA genes, generic traits were tested for homoplasy, and the intra- and interpopulation variations of four sampled populations were analyzed. The new genus belongs to the subfamily Tylenchinae, family Tylenchidae in the infraorder Tylenchomorpha. It is characterized by a unique labial plate that has four narrow lobes with tips detached from the adjacent cuticle, laterally broad elongated amphidial apertures, a strong sclerotized excretory duct, a round spacious postvulval uterine sac, and a spicule with a sharp protrusion at the blade.
1 INTRODUCTION
Integrative taxonomy was introduced as a comprehensive framework to delimit and describe taxa by gathering together information from different types of data and methodologies (Dayrat, 2005; Will, Mishler, & Wheeler, 2005) and has been considered as the most efficient and theoretically grounded approach to define robust species hypotheses (De Queiroz, 2007; Samadi & Barberousse, 2006). Commonly used complementary perspectives include phylogeography, comparative morphology, population genetics, ecology, development, and behavior. In this study, we introduce 3D printing to generic descriptions. Models are incorporated as a complement to pictures and drawings to illustrate complex 3D structures. Aside from taxonomy, we show its potential applications in linking research frontiers to education. We also compared the performance of printing materials and proposed the most suitable option.
Nematodes of the family Tylenchidae are abundant and diverse. Ecologically, they are important soil fauna which may constitute up to 30% of the nematodes in any given soil sample (Ferris & Bongers, 2006; Yeates & Bird, 1994). However, this group is taxonomically notoriously difficult as most species combine a low observational resolution with high intraspecific variability. As a result, many descriptions are ambiguous, especially if only based on light microscopy and/or based on a limited number of representatives, and several genera are polyphyletic (Qing, Decraemer, Claeys, & Bert, 2017). Here, we describe a new genus, Labrys chinensis gen. nov., sp. nov. Integrative approaches were applied to increase descriptive resolution: detailed morphology based on light microscopy (LM) and scanning electron microscopy (SEM); 3D models were generated and actually printed in 3D; selected generic traits were tested for phylogenetic homoplasy; four populations were sampled, and their intraspecific variation was analyzed. The results expanded our knowledge on Tylenchidae and provided an example for future species description in taxonomically difficult group.
2 MATERIALS AND METHODS
2.1 Sample collection and processing
A total of 38 individuals were collected in four locations in China (Table 1). Soil samples were incubated for 48 h on plastic trays lined with paper towels, and nematodes were subsequently concentrated using a sieve (25 μm opening). After removing water, nematodes were rinsed with DESS solution and transferred to glass vials for preservation and transportation. DESS-preserved specimens were rinsed several times with deionized water and then transferred to anhydrous glycerin for morphological analyses (Yoder et al., 2006).
| Populations | Individuals | GenBank accession number | Locations | |
|---|---|---|---|---|
| 28S | 18S | |||
| P1 | 13 | KY776621 KY776622 KY776623 KY776624 |
KY776632 | Taibai, China (34°03′40″N, 107°41′ 9.6″E) |
| P2 | 8 | KY776616 KY776617 KY776618 KY776619 KY776620 |
KY776633 | Meixian, China (34°05′18.5″N 107°47′26.6″E) |
| P3 | 8 | KY776611 KY776612 KY776613 KY776614 KY776615 |
KY776630 | Shimen, China (29°56′08.3″N 110°47′13.1″E, 30°01′55.2″N, 110°39′54.0″E) |
| P4 | 9 | KY776625 KY776626 KY776627 KY776628 KY776629 |
KY776631 | Zhouzhi, China (107°47′9.4″E, 33°54′6.5″N) |
2.2 Morphological analyses
Measurements and drawings were prepared manually with a drawing tube mounted on an Olympus BX51 DIC Microscope (Olympus Optical, Tokyo, Japan). The holotype of the new species was recorded as video clips mimicking LM multifocal observations (De Ley & Bert, 2002) and these are available online at http://nematodes.myspecies.info/gallery?f[0]=im_field_smg_galleries%3A734. Illustrations were prepared manually based on light microscope drawings and edited in Adobe Illustrator CS3 and Adobe Photoshop CS3. For SEM, specimens from DESS were gradually washed with water and postfixed with 2% PFA + 2.5% glutaraldehyde in 0.1M Sørensen buffer, then washed and dehydrated in ethanol solutions and subsequently critical point-dried with CO2. After mounting on stubs, the samples were coated with gold and observed with a JSM-840 EM (JEOL Ltd., Tokyo, Japan) at 12 kV.
2.3 Molecular phylogenetic analyses
Genomic DNA was extracted from DESS-preserved specimens with worm lysis buffer (Yoder et al., 2006). The single nematode was transferred to 40 mL worm lysis buffer and frozen for 10 min at 20°C. 1 μl proteinase K with the concentration of 1.2 mg/ml was added to the samples before incubation, 1 h at 65°C followed by 10 min at 95°C. The D2-D3 domains of 28S ribosomal RNA (28S) were amplified with primers D2A (5′-ACAAGTACCGTGAGGGAAAGT-3′) and D3B (5′-TGCGAAGGAACCAGCTACTA-3′). The partial 18S ribosomal RNA (18S) was amplified with primers TylF1 (5′-GCCTGAGAAATGGCCACTACG-3′) and TylR2 (5′-TGRTGACTCRCACTTACTTGG-3′). The PCR conditions were 30 s at 94°C, 30 s at 54°C, and 2 min at 72°C for 40 cycles. Newly obtained sequences were deposited in GenBank (Table 1). Alignments of the different genes were made using the Q-INS-i algorithm implemented in MAFFT v. 7.205 (Katoh & Standley, 2013), and alignments are available at TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S21277). The best-fitting substitution model was estimated using AIC in jModelTest v. 2.1.2 (Darriba, Taboada, Doallo, & Posada, 2012), and GTR + I + G was selected as best scored model for both markers. A maximum likelihood (ML) analysis was performed with 1000 bootstrap (BS) replicates under the GTRCAT model using RAxML 8.1.11 (Stamatakis, Hoover, & Rougemont, 2008), and a Bayesian inference (BI) was carried out with the GTR + I + G model using MrBayes 3.2.3 (Ronquist et al., 2012). Analyses were run for 5 × 106 generations, and Markov chains were sampled every 100 generations. Burnin was arbitrarily chosen to be 25% of the results and evaluated using a generation/log-likelihood scatter plot. The ML and BI analyses were performed at the CIPRES Science Gateway (Miller, Pfeiffer, & Schwartz, 2010). Gaps were treated as missing data for all phylogenetic analysis. All trees were visualized with TreeView v. 1.6.6 (Page, 1996). ML BS values and Bayesian posterior probabilities (PP) were summarized on the consensus tree using Adobe Illustrator CS3.
2.4 Homoplasy test
To provide an objective estimation on evolutionary conservation of lip morphology and its robustness as a generic delimitation marker, we calculated the homoplasy indices, the retention index (RI), the consistency index (CI), the observed number of character transitions (obs.), and the permutation of character values (perm.) (Maddison & Slatkin, 1991) on the BI consensus tree. We consider high RI and CI values (≥0.80) or low obs./perm. ratio (≤0.45) to be indicative that the analyzed traits evolved slowly enough to retain phylogenetic information and low homoplasy. All analyses were performed in Mesquite 3.10 (Maddison & Maddison, 2016).
2.5 Analyses of population genetic structure
To visualize the genetic structure at the population level and to display conflicts in the data by taking into account incompatible phylogenetic signals, we generated phylogenetic networks by employing the NeighborNet algorithm (Bryant & Moulton, 2004) with uncorrected pairwise p-distances in the program SplitsTree v4.10 (Huson & Bryant, 2006). With 1000 pseudo-replicates (result only shown between populations), bootstrap analysis was conducted to assess the support for splits in the network. We also estimated the nucleotide diversity (θπ and θS) within population and genetic variation between the four populations by calculating the fixation index (Fst). All diversity and demographic analyses were performed using Arlequin 3.1 (Excoffier, Laval, & Schneider, 2005).
2.6 3D modeling and printing
To visualize important morphological characters and facilitate zoological education, 3D models were reconstructed in Autodesk Maya following the procedure of Qing, Sánchez-Monge, and Bert (2015). Next to the new genus, three other tylenchid genera (Tylodorus sp., Cephalenchus sp., and Cucullitylenchus sp.) were modeled to visualize intrafamily variations of the morphology of lip regions and to test printing performance across a variety of nematode taxa. The constructed models were converted to the stl format, and MiniMagics 3.0 was used to optimize the models. Each model was printed using three commercial materials: Polylactic acid (PLA) and acrylonitrile butadiene styrene (ABS) were printed by MakerBot Replicator 2 using fused deposition modeling (FDM) method, while resin was printed by RSPro 450 Industrial 3D printer using the stereolithography method. All the model files and the printing video/pictures are freely available at worldwide 3D designer community Thingiverse (www.thingiverse.com/thing:2418910), allowing our designs to be discussed and improved by the scientific community.
3 RESULTS
3.1 Phylogenetic analysis and homoplasy tests
In both analyses, the tree topologies regarding the major clades of Tylenchidae are congruent with recently published studies (Atighi et al., 2013; Qing et al., 2015, 2017). The monophyly of all Labrys chinensis gen. nov., sp. nov. populations is fully supported (BI = 1, BS = 100) across the two genes. The new genus is sister to a clade containing Filenchus + Malenchus based on 28S (BI = 1, BS = 98) or to Filenchus misellus (Andrássy, 1958) Raski & Geraert, 1987 based on 18S (BI = 1, BS = 91) (Figures 1 and 2). As already noted by Qing et al. (2017), the genus Filenchus is polyphyletic and comprises at least three clades. Among these clades, F. misellus and F. chilensis Raski & Geraert, 1987 formed a separate clade, separated from the type species of the genus (F. vulgaris (Brzeski, 1963) Lownsbery & Lownsbery, 1985). This separation is also morphologically supported by their unique amphidial aperture pattern (Brzeski & Sauer, 1982; Karegar & Geraert, 1998; Torres & Geraert, 1996). Therefore, F. misellus and F. chilensis can be designated as one or even two separate genus/genera. However, this phylogenetic grouping is based on only few GenBank sequences without morphological vouchers, limiting the validity of further taxonomic actions.
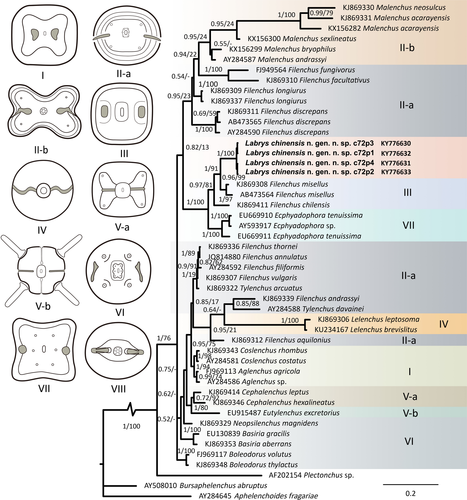
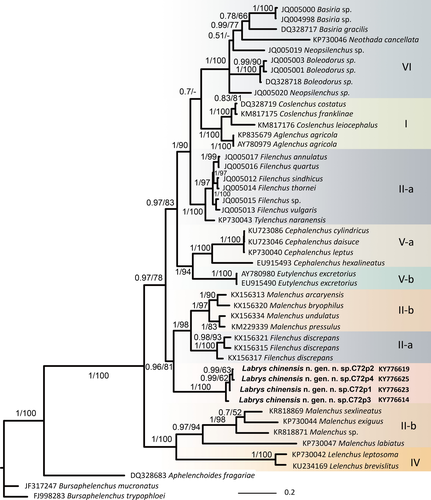
Tylenchidae taxonomy is controversial and problematic with several invalid or homoplastic generic characters (Qing et al., 2017) implying that an objective selection for morphological characters that define phylogenetic clades is necessary. In this study, homoplasy tests of lip region pattern morphology mapped on phylogenetic trees based on the in 18S and 28S sequences indicated strong phylogenetic signals (RI > 0.85, CI ≥ 0.80, obs/permu < 0.45) (Table 2), indicating that lip morphology is a relatively conserved character that can be used as a generic character.
| 18S | 28S | |
|---|---|---|
| RI | 0.91 | 0.96 |
| CI | 0.80 | 0.87 |
| obs. | 10 | 8 |
| perm. | 28 | 29 |
| obs/perm. | 0.36 | 0.27 |
- RI, retention index; CI, consistency index; obs, observed number of character transitions; permu, permutation number of character transitions.
3.2 Population structure
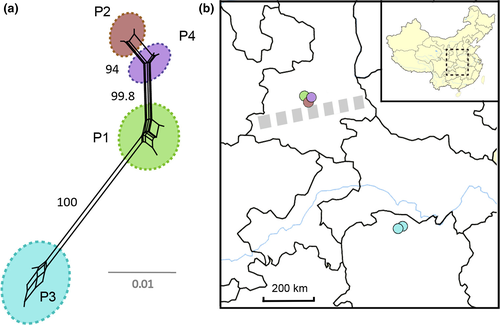
The NeighborNet analysis based on the alignment of the 28S gene revealed four major clades (Figures 3a), supported by high interpopulation genetic divergence (Fst > 0.8) (Table 3) and concordant with the geographic origin of the respective samples: Population No. 1 (P1), Population No. 2 (P2), and Population No. 4 (P4) that were sampled are ca. 10–20 km apart from each other, and Population No. 3 (P3) is distantly separated from the other populations with around 600 km (Figures 3b) between them. The most interesting general result is the presence of multiple lineages (P1, P2, P4) occurring in a relatively small geographic region. Although the divergence is relatively low, and network analyses show that historical admixture across the range may exist, all lineages are well separated. The intrapopulation nucleotide analyses also shows diversity, the lowest divergence in P1 (θπ = 0.50, θS = 0.54) and the highest in P3 (θπ = 9.8, θS = 2.9). However, all populations do not show morphological or morphometrical inter- or intrapopulation differences.
| P1 | P2 | P3 | P4 | |
|---|---|---|---|---|
| P1 | 0.50/0.54 | |||
| P2 | 0.89 | 1.4/0.96 | ||
| P3 | 0.92 | 0.93 | 9.8/2.9 | |
| P4 | 0.94 | 0.94 | 0.93 | 2.8/1.9 |
3.3 Taxonomy
3.3.1 Labrys gen. nov
Description
Same as species description.
Diagnosis and relationship
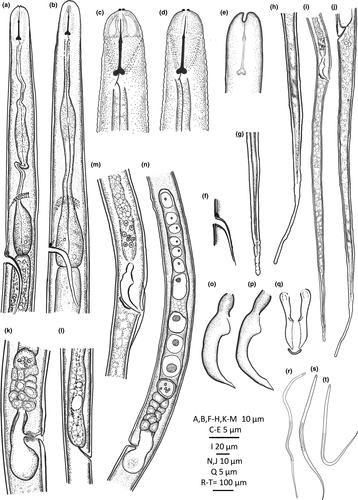
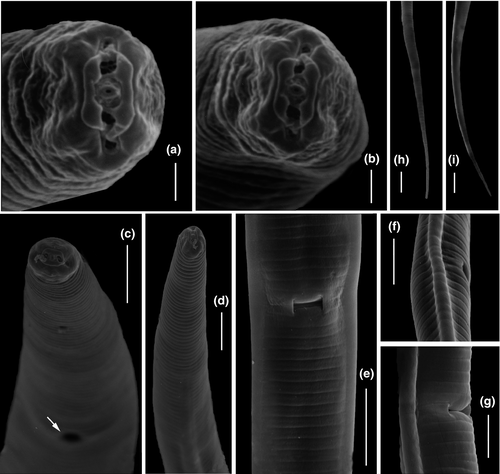
The new genus belongs to the subfamily Tylenchinae, family Tylenchidae. It is characterized by a unique labial plate that has four narrow lobes with tips detached from the adjacent cuticle, visible in LM as two small protruding lips at the anterior end (Figures 4a, b and 5c, d). Geraert and Raski (1987) distinguished seven patterns of the morphology of the lip region in Tylenchidae. The pattern in Labrys gen. nov. differs from all known lip patterns in Tylenchidae and is here considered as an eighth unique pattern. Beside, the wide, laterally broad, and elongated amphidial aperture (Figure 4d), the spicule with a sharp protrusion at the blade (Figure 4m), and a round spacious postvulval uterine sac (PUS) are also very rare in Tylenchidae. The new genus resembles the genera Allotylenchus, Lelenchus, Filenchus, and Polenchus in general appearances, for a detailed comparison see Table 4. The genus Sakia is also similar to the presented new genus (broad cap-like cephalic region and a sclerotized excretory duct), although its validity is still in discussions (Fortuner & Raski, 1987; Geraert, 2008; Husain, 1972; Siddiqi, 1986, 2000). The type species S. typica Khan (1964) was described without drawings but still shows several differences: amphidial aperture small, oval slit (vs broad and elongated aperture, obvious in laterally view), stylet with cone equal in length to shaft (vs shaft two times longer than cone), spermatheca not set-off (vs spermatheca set-off), and a reduced PUS (vs a round spacious PUS). Other species of Sakia all have the anterior end without protruding lips, spicule without sharp protrusion and are different in lateral incisures (absent/four in Sakia vs two in Labrys gen. nov.).
| Genera | Characters | |||||
|---|---|---|---|---|---|---|
| Anterior end | Amphideal fovea | Median bulb | Flap in Vulva | Excretory duct | Spicule blade | |
| Labrys gen. nov. | Two lips protruding | Indistinct | Elongated fusiform | Indistinct | Sclerotized | Sharply protruding |
| Allotylenchus | Continuous | Indistinct | Well developed | Large | Sclerotized | Less protruding |
| Polenchus | Continuous | Indistinct | Well developed | Indistinct | Weak | Less protruding |
| Filenchus | Continuous | Indistinct | Well developed to elongated fusiform | Indistinct | Weak | Less protruding |
| Lelenchus | Continuous | Pouch-like | Elongated fusiform | Indistinct | Weak | Less protruding |
| Sakia | Continuous | Indistinct | Elongated fusiform | Indistinct | Sclerotized | Less protruding |
Etymology
The selected genus name is derived from the shape of the labial plate, which resembles a symmetrical double-bitted axe, one of the famous symbols of Greek civilization.
Type and only species
Labrys chinensis gen. nov., sp. nov.
3.3.2 Labrys chinensis gen. nov., sp. nov. (Figures 4-6, Table 5)
| Characters | Female | Male | |
|---|---|---|---|
| Holotype | Paratypes | Paratypes | |
| n | – | 9 | 4 |
| L | 636 | 616 ± 24 (580–637) | 611 ± 12 (602–627) |
| a | 41.6 | 44.1 ± 3.9 (37.7–48.7) | 35.1 ± 1.6 (33.7–37.4) |
| b | 6.9 | 6.6 ± 0.6 (5.4–7.3) | 6.2 ± 0.38 (5.6–6.5) |
| c | 3.8 | 3.8 ± 0.23 (3.5–4.4) | 3.6 ± 0.20 (3.4–3.8) |
| c' | 17.3 | 18.0 ± 2.1 (13.3–20.3) | 17.6 ± 2.9 (14.8–21.5) |
| V | 56.5 | 56.6 ± 1.5 (53.9–59.8) | – |
| V' | 76.5 | 77.1 ± 1.0 (75.0–78.1) | – |
| Tail length/vulva-anus distance | 1.5 | 1.6 ± 0.12 (1.3–1.7) | – |
| Body diam. | 15 | 14 ± 1.3 (12–16) | 17 ± 0.91 (16–19) |
| Stylet | 8.5 | 9.3 ± 0.42 (8.9–10) | 9.4 ± 0.54 (8.7–10) |
| MB | 49 | 43 ± 4.2 (38–51) | 44 ± 1.2 (44–46) |
| Excretory pore to anterior end | 90 | 84 ± 4.0 (76–87) | 80 ± 3.8 (76–84) |
| Excretory duct length | 19 | 20 ± 4.1 (15–25) | 22 ± 3.3 (18–26) |
| Pharynx | 92 | 95 ± 9.0 (82–108) | 99 ± 5.0 (96–106) |
| Nerve ring | 67 | 69 ± 8.8 (58–89) | 70 ± 4.8 (64–76) |
| Anus width/cloacal width | 9.6 | 9.2 ± 0.83 (8.3–11) | 9.7 ± 0.87 (8.6–11) |
| Spicule | – | – | 16 ± 1.3 (14–17) |
| Postuterine sac/gubernaculum | 15 | 15 ± 1.2 (13–17) | 4.8 ± 0.45 (4.4–5.4) |
| Tail | 166 | 164 ± 12 (132–171) | 169 ± 13 (159–186) |
ZooBank (zoobank.org) identifier: CE6004B6-D242-4989-9ABE-1F000FA2AEFE.
Holotype
Female, from population P1, recovered from soil underneath Quercus aliena from Taibai (34°03′40″N, 107°41′ 9.6″E), China, at an altitude of 1963 m a.s.l. Deposited at the Ghent University Museum, Zoology Collections, collection number UGMD 104322.
Paratypes
Four females and one male paratypes collected from the same location and same sample of holotype. Deposited at the Ghent University Museum, Zoology Collection, collection number UGMD 104323, Ghent University, Belgium. Additional paratypes are available in the UGent Nematode Collection (slide UGnem-162) of the Nematology Research Unit, Department of Biology, Ghent University, Belgium.
Type habitat and locality
Type population P1 from soil underneath Quercus aliena from Taibai (34°03′40″N, 107°41′ 9.6″E), China, at an altitude of 1963 m.a.s.l. Three other populations were found in different locations in China (Table 1).
Description
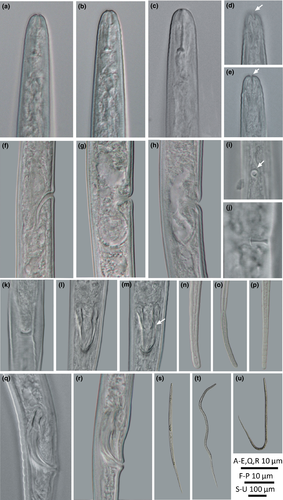
Female: body slender, straight to ventrally arcuate. Cuticle appearing as bright lining in stereomicroscopy, smooth in LM but finely striated in SEM. Lateral field distinct, an elevated ridge forming two incisures, starts at level of metacorpus. Cephalic region rounded, continuous, framework weak, not sclerotized. Labial plate offset and constricted dorso-ventrally, forming four lobes, tapering toward tip and detached from adjacent cuticle (Figure 6a, b; 1 type VIII). Labial papillae six, arranged as a circle in oral disk. Cephalic papillae invisible. Amphidial apertures broad slits, laterally elongated, confined in first annulation after labial plate, edge of apertures thicker than adjacent cuticle forming a elevated ring (Figure 6a, b). Stylet knobbed, shaft about two times longer than cone. Dorsal pharyngeal gland orifice close to stylet base. Excretory pore wide 2.0–2.5 μm, excretory duct long, heavily sclerotized, generally at the level of pharyngo-intestinal junction. Deirids at the level of basal bulb. Hemizonid just above excretory pore. Corpus cylindroid, metacorpus elongated fusiform, its cuticular valve weak, and gradually transiting to cylindrical isthmus. Basal bulb spindle-shaped. Vulva with small flap, one annulus wide. Vagina wall thin, perpendicular to body. Postvulval uterine sac (PUS) round, occupying full body width, one body diameter long. Female gonoduct monodelphic, prodelphic. Ovary outstretched, oocytes in single row. Spermatheca offset, filled with spherical sperm cells. Uterus quadricolumellate, probably with five or six cells in each row.
Male: Bursa ad-cloacal, slightly crenated. Spicules with velum, proximal part of blade sharply protruding, gubernaculum simple. Tails filiform, ending in a rounded terminus.
Etymology
Species name is given after China, where it was recovered.
4 DISCUSSION
Currently, 44 genera are recognized in the family Tylenchidae: 42 genera as listed by Geraert (2008), one new genus recently described by Yaghoubi et al. (2016), and one by the present study. In this study, integrative approaches were used to describe Labrys chinensis gen. nov., sp. nov. Although the phylogeny of Tylenchidae remains unresolved, current results extended its diversity and highlighted the importance of detailed morphological analyses in such taxonomically difficult group. Given that many of the generic characters are only obvious in SEM (e.g., Labial patterns), new species descriptions should not solely be based on LM.
The morphology- and molecular phylogeny-based homoplasy test supports lip morphology to be informative, concurring with previous morphological observations (Geraert & Raski, 1987; Sher & Bell, 1975). Therefore, more attentions need to be given on lip region (especially the shape and position of amphidial aperture) in future taxonomy studies. Nevertheless, a combination of different characters is necessary, as congruence may still occur in lip morphology (lip pattern II-a).
The implementation of molecular tools led to discovery unexpected diversity in a small spatial scale. This suggests that either the contemporary gene flow was interrupted for an unknown reason or that ancestral polymorphisms have been retained. Alternatively, these populations may represent several cryptic species that morphologically similar. Cryptic species have been identified in various nematodes: plant-parasitic, entomoparasitic, vertebrate-parasitic, and bacteria feeding (Jorge, Perera, Carretero, James Harris, & Roca, 2013; Kanzaki, Li, Lan, & Giblin-Davis, 2014; Palomares-Rius, Cantalapiedra-Navarrete, & Castillo, 2014; Sudhaus & Kiontke, 2007). These species only have very minute morphological differences which may otherwise be ignored without integrative approaches. In present study, detailed morphology and morphometric cannot recognize any interpopulation difference, but they can still be separated species, differing in invisible characters (e.g., mating behavior and ecological constraints) or morphology that cannot access by current techniques.
Although 3D printing technology has been used since the 1980s, it has only recently gained real momentum, as the technology matures and awareness grows. Driven by new applications, the “printable” category keeps expanding into many fields such as medicine, architecture, education, fashion, manufacturing, even food (Lombardi, Hicks, Thompson, & Marbach-Ad, 2014; Murphy & Atala, 2014; Petrick & Simpson, 2013; Qing et al., 2015; Sun, Peng, Yan, Fuh, & Hong, 2015; Thomas, Hiscox, Dixon, & Potgieter, 2016). Within zoology, it has already been showing great potential in functional morphology, pest detection, anatomy, and physiology (Domingue et al., 2015; Greco et al., 2014; Igic et al., 2015; Porter, Adriaens, Hatton, Meyers, & McKittrick, 2015; Thomas et al., 2016). Here, we extend the application of 3D printing to the field of taxonomy and describe for the first time a new taxon together with a printed model (Figure 7). Although the accuracy of our models is not comparable to 3D reconstructions based on serial transmission electron microscopy (TEM) sections or electron tomography techniques, the models are useful and time-efficient complements to pictures and drawings of species descriptions to illustrate complex 3D structures. Future taxonomical applications can also be extended to virtual reality approaches that allow observation and dissection without damaging precious specimens, which represents a promising direction for both taxonomy and education. Therefore, as we add 3D printing to the toolkit of taxonomical research, we also underline the relevance of its development as a synergistic disciplinary link of frontier zoology research and zoological education.
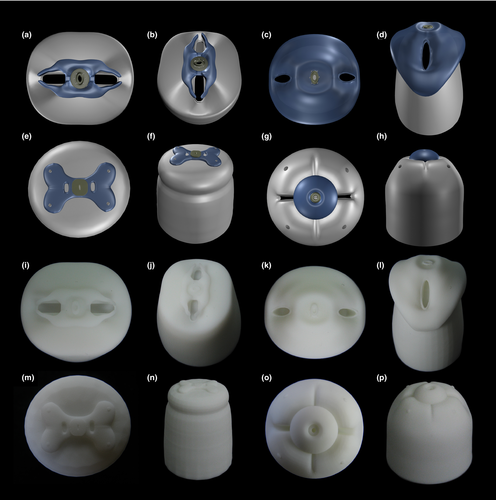
In this study, we also experienced the importance of selecting the optimal printing materials to achieve appropriate model quality. ABS and PLA are two effective commercial materials that combine mechanically desirable performance and low cost, whereas resin is considered a more advanced material that delivers the highest quality output but at a considerably higher price. Our tests based on four taxa reveal that the thermoplastic polymers ABS and PLA gave similarly acceptable coarse surfaces in 8-cm models, while all labial details were completely lost in 4-cm print (see http://nematodes.myspecies.info/gallery?f[0]=im_field_smg_galleries%3A734). Conversely, resin provides highly resolved details, that is, all papillae are clearly visible, even when model size is reduced to 4 cm (Figure 7i–p). Therefore, such high quality in small size print can compensate for the less competitive price of resin (usually 1.5 to two times that of PLA). Moreover, resin can be printed in light color, semi-opaque color, or even transparent which facilities the visibility of internal structures. In conclusion, resin is highly recommended for zoological anatomy education and research while PLA/ABS is also useful but only for larger print size (8 cm or more).
ACKNOWLEDGEMENTS
We thank Marjolein Couvreur for the SEM analysis. The first author thanks the China Scholarship Council (CSC) for providing a Ph.D. grant. This work was also supported by a special research fund UGent 01N02312.




