Taxonomic identification of algae (morphological and molecular): species concepts, methodologies, and their implications for ecological bioassessment
Abstract
Algal taxonomy is a key discipline in phycology and is critical for algal genetics, physiology, ecology, applied phycology, and particularly bioassessment. Taxonomic identification is the most common analysis and hypothesis-testing endeavor in science. Errors of identification are often related to the inherent problem of small organisms with morphologies that are difficult to distinguish without research-grade microscopes and taxonomic expertise in phycology. Proposed molecular approaches for taxonomic identification from environmental samples promise rapid, potentially inexpensive, and more thorough culture-independent identification of all algal species present in a sample of interest. Molecular identification has been used in biodiversity and conservation, but it also has great potential for applications in bioassessment. Comparisons of morphological and molecular identification of benthic algal communities are improved by the identification of more taxa; however, automated identification technology does not allow for the simultaneous analysis of thousands of samples. Currently, morphological identification is used to verify molecular taxonomic identities, but with the increased number of taxa verified in algal gene libraries, molecular identification will become a universal tool in biological studies. Thus, in this report, successful application of molecular techniques related to algal bioassessment is discussed.
Abbreviations
-
- ITIS
-
- Integrated Taxonomic Information System
-
- NAWQA
-
- North American Water Quality Assessment
-
- NGS
-
- Next-generation sequencing
-
- USEPA
-
- United States Environmental Protection Agency
-
- USGS
-
- United States Geological Survey
Algae are important aquatic organisms for understanding ecosystem processes, conservation, and water quality. Accurate ecological bioassessments of aquatic habitats require algal identification at a meaningful taxonomic level for correct interpretation of the conditions algae grew. The type of taxonomic identification is driven by the goals of the study, and a variety of identifications to the species, genus, or higher taxonomic level can be useful (Rimet 2012). Taxonomic identification of algae presents opportunities to understand systematic entities such as species (Mayr 1942) and relate them to evolutionary and ecological processes. Taxonomy captures tremendous information about each species because the evolution of traits confers physiological and morphological adaptations to a wide diversity of environmental conditions, which are then summarized in a Latin binomial. Algae have predictable and sensitive species-specific responses to many chemical, physical, and biological changes in aquatic environments (Stevenson 2006, Bellinger and Sigee 2010, Schneider et al. 2011). Taxonomic entities have characteristics that can be used in bioassessment, like their relative abundance and relationship with environmental optima (ter Braak and van Dam 1989, Stevenson et al. 2008), cell size (Lavoie et al. 2006, 2010), and changes in shape (Wang et al. 2005). Correct taxonomic identification provides consistency and the transferability of ecological inferences (Kociolek and Stoermer 2001, Stevenson 2006), which can be translated across different geographic areas (Kahlert et al. 2008).
Cyanobacteria and eukaryotic algae are an evolutionarily diverse assemblage (De Clerck et al. 2013) that can adapt to microhabitats through varying growth rates (Manoylov 2005) and variable genomes (Kapraun 2007, Janouškovec et al. 2013) that can potentially express different genes under different environmental conditions (Zani et al. 2000). Two basic methods of algal species identification are currently employed: morphological (for both cleaned and un-cleaned algal material), which uses various features observed under a microscope, and molecular, which uses a variety of gene regions (Fig. 1). In recent years, many natural microbial communities have been studied with either morphological (Charles et al. 2002, Prygiel et al. 2002) or molecular (Marsh et al. 1998, Raviraja et al. 2005, Zwart et al. 2005) approaches or a combination of the two (Drummond et al. 2005, Sakayama et al. 2005, Manoylov et al. 2009, Kermarrec et al. 2013). Molecular identification has the potential to provide revolutionary discoveries in taxonomy that may have great benefits for bioassessment. Recently published (Kermarrec et al. 2013) methods and improved DNA reference libraries (vs. books of morphology) show great promise for bioassessment. Reference sequences of positive control taxa are being created and will be used in future libraries for the accurate assessment of algal biodiversity (De Clerck et al. 2013, Sluys 2013).
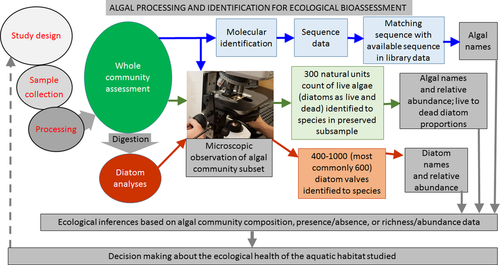
The goals of this review are to summarize the current processes and challenges of taxonomic analysis of algal communities used in bioassessments. Second, I will review the use of algal taxonomy in bioassessment, the methods of generating taxonomic data, and required training for taxonomists. Then, I will discuss the challenges in morphological taxonomy, such as the consistency and accuracy of identifications (Kociolek and Stoermer 2001), and improvements to bioassessment through advanced technology. Finally, I will review the molecular methods of taxonomic analysis of algal samples and their potential for enhancing bioassessments.
Application of Algal Taxonomy in Bioassessment
Many reviews have been written concerning the use of algae in bioassessment (Stevenson and Lowe 1986, McCormick and Cairns 1994, Lowe and Pan 1996, Bellinger and Sigee 2010, Smol and Stoermer 2010, Stevenson 2014), which indicates the long history and widespread use of algae in ecological assessment. Species lists from samples collected and the traits of taxa are used to calculate metrics for which Stevenson (2006, 2014) distinguished two main applications: characterizing stressors and assessing biological conditions. Algae are used to characterize water chemistry using the environmental preferences of a taxon, such as its environmental optima and tolerances (ter Braak and van Dam 1989). Algae are used to infer historical pH conditions in lakes, total phosphorus concentrations in streams, or complex stressors such as acid mine drainage (Kelly and Whitton 1995, Charles et al. 2002, Smucker et al. 2014). Biological condition is a measure of the deviation in structure and function of communities from natural conditions, that is, our best estimates of natural, minimally disturbed conditions. Nuisance and harmful algal abundances and multimetric indices could be considered elements of biological condition (Stevenson 2014). Assessments of stressors and biological conditions have important resource management applications related to laws such as the Clean Water Act in the United States and the Water Framework Directive in Europe (EU 2000).
Understanding the different goals of algal bioassessments is important for evaluating the significance of identification errors. The consistent identification of taxa is more important for assessing stressors than biological condition because biological condition characterizes the loss of naturally occurring and invasive taxa. The characterization of stressors should identify taxa to the same level as did the research in which the environmental optima were determined rather than matching them to type material or splitting morphologically distinct types into greater detail than occurred when the taxa traits were originally characterized. The same is true for biological conditions, which are most commonly assessed with metrics and taxa traits. However, the accurate identification of taxa is important for assessing biodiversity, high biodiversity is an element of good biological condition (Davies and Jackson 2006). Methods of routine bioassessment do not allow for a full accounting of all taxa in a habitat, but they do allow for a determination of whether the physical and chemical conditions are suitable for supporting native taxa and whether high proportions of the observed taxa in a sample are native taxa. Therefore, the accurate identification of taxa relative to their type instead of to their previous identifications has become more important than characterizing stressors in the short term for assessing biological conditions. Many problems with misidentification can be corrected in the process of harmonizing taxa lists (discussed later); however, the long-term goal should be more consistent and accurate taxonomy in bioassessments to provide better transferability of information among projects and assessments of trends in conditions and current status (Kociolek and Stoermer 2001). Ideally, scientists should also be able to use data from multiple projects to compile standardized lists of taxa and their traits that can be applied more broadly in future projects. Inaccurate identifications result in poor characterization of taxa traits and limit the harmonization of taxa lists across projects, which require more assumptions compared with harmonizations within a single project.
Under the Clean Water Act (1971), protection is extended to species, subspecies, reference communities, and natural communities for all organisms, including algae (Jackson and Davis 1994). Species-level identification is the most desirable identification level because that level is afforded protection under the law, and the population responses of algae at the species level (rather than at the genus or higher taxon level) carry specific adaptations to environmental conditions (Stevenson et al. 2009).
Sample Analyses for Bioassessment with Algae
Algal collection and sample analyses have relatively standard methods based on the region and habitat being assessed. Phytoplankton, periphyton, and sedimentary diatoms are three common communities of interest for assessment, but their inclusion in assessments varies among lakes, streams, wetlands, and coastal zones. Analyses of algae from different habitats and climates can vary; for example, assessments of phytoplankton and periphyton commonly emphasize the characteristics of all algae in the sample (Stevenson and Bahls 1999), whereas sediment analyses commonly focus on diatom identification. Because all the algae cannot be identified, a pre-determined number of 300–400 natural algal units documented per site is used as a “stopping rule” for periphyton and phytoplankton in the United States Environmental Protection Agency (USEPA) and United States Geological Survey (USGS) protocols. Samples for diatom analyses are usually cleaned with oxidants to remove organic material before the diatoms are mounted on slides in a highly refractive medium. The number of diatoms identified in counts varies from 200 to 600 valves for most bioassessment programs. The analysis of all algae in a sample is usually performed using a settling chamber with an inverted microscope or a Palmer-Maloney counting chamber with a regular microscope. Protocols for the taxonomic analysis of all algae in large-scale assessments by the USEPA and USGS (Fitzpatrick et al. 1998, Moulton et al. 2002) call for the identification of non-diatom algae to the lowest possible taxonomic level, which is usually the genus or species level. The level of identification of non-diatom algae varies among groups and depends primarily on sexual reproductive structures to identify species. The treatment of diatom identification varies in counts of all algae because their identification is challenging without cleaning. Routine SEM observation of diatoms for bioassessment is not practical, although SEM has been applied for the clarification of ecologically important yet unknown taxa (Morales et al. 2012). In many protocols, diatoms are identified as alive (having plastids evident in frustules) or dead (Moulton et al. 2002). In addition, assessments of the taxonomic composition of diatoms from cleaned samples (or samples with removed organic material as a result of an oxidation process, Stevenson and Bahls 1999) are used to characterize the taxonomic composition of the live diatom proportion of all algae. Because so many algae occur in colonies and filaments and because the cell density on slides is patchy, the number of cells identified and counted in a sample is determined by natural units, which are cells for unicellular algae, colonies, or filaments. Documenting the identical number of taxa for samples collected and processed under the same protocol allows for meaningful comparisons (Charles et al. 2002). Morphological identification is a process used by most scientists and follows standard protocols (Stevenson and Bahls 1999, APHA 2012). As in other sciences, taxonomic research requires hypothesis testing, analysis of data, and taxonomy-specific tools for algal identification that include research-quality microscopes, a set of taxonomic keys, initial training, and expertise.
The identification of algae for bioassessment requires standardized taxonomic literature and a list of taxa. The Susswasserflora von Mitteleuropa series (Krammer and Lange-Bertalot 1986–1991) is the most common taxonomic reference for diatom identification because it provides the most comprehensive coverage of diatom taxonomy compiled by a single, authoritative team of authors and includes illustrations of specimens for most taxa, which provides a resource for investigating the accuracy of taxonomy. LM and SEM images from European material that includes type localities and population series are also valuable. Krammer and Lange-Bertalot (1986–1991) utilize a taxonomic approach that is referred to as sensu lato, meaning a broad interpretation of the morphological variability in specimens that are grouped with a type specimen. This approach is contrasted with a sensu stricto definition of taxa, in which little variation from the type description is allowed for identification.
A wider diversity of literature is required for the identification of non-diatom algae because of the great diversity in types of algae. Few of the common identification keys for all algal groups were developed for specific regions (e.g., Tilden 1910, Geitler 1931, Prescott 1931, 1962, Forest 1954, Dillard 1989, 2007, Komárek and Anagnostidis 1989, John et al. 2011); however, because of the lack of comprehensive literature, algal literature from one region is applied to many others.
The taxonomic libraries in most algal identification laboratories include extensive lists of books and papers that contain the historical treatments of taxa (e.g., Ehrenberg 1843, Kützing 1844, Van Heurck 1880-1896, Hustedt 1930) and a limited number of recent and regional investigations of algal taxonomy (e.g., Van de Vijver et al. 2004, Levkov 2009, Furey et al. 2011). These libraries are used for a deeper taxonomic investigation and a more sensu stricto identification of the taxa found in samples, and they are assumed to provide better bioassessments with detailed and accurate taxonomy compared with sensu lato. Taxonomists who are involved with bioassessment are often highly interested in the accurate identification of taxa and find satisfaction in observing new taxa and refining distinctions among taxa. Taxonomic libraries should be developed online to allow for a thorough investigation of original species descriptions, more recent refinements in taxonomy, and widely accepted sensu lato identities of species. With these resources, specimens requiring identification that do not satisfy descriptions can be compared with the original descriptions of the taxa. In addition, more recent refinements of taxonomy can be explored to determine whether a mystery specimen can be matched with recent treatments of morphologically variable, endemic, or narrowly distributed taxa. Personal traits required for algal identification are attention to detail, willingness to document all taxa observed, patience, intellectual curiosity, and a vested interest in the process being ultimately used for improving environmental stability. In addition, diatom taxonomists should apply critical thinking in recognizing the complex tridimensional structure of diatoms regardless of the view observed on the permanent slide.
Web-based taxonomic resources offer exciting new solutions to the availability of up-to-date taxonomic information. For example, the Western Diatoms database (http://westerndiatoms.colorado.edu/) has more than 500 diatom species, includes illustrations of multiple specimens to characterize the morphological range in the United States, and provides comparisons with type specimens from original publications. Over the last 4 years, the website has received increasing numbers of visitors; hopefully, this free tool and “the era of cybertaxonomy” will promote taxonomic discussion and collaboration. Standardized taxonomic lists also provide another web-based taxonomic resource. Standardized lists provide, at a minimum, a list of taxon names that are recommended for use. For example, the Integrated Taxonomic Information System (ITIS; http://www.itis.gov update 7/13) is a taxon list that has been proposed for use in standardizing taxa names used by the USEPA and is harmonized with the taxon list developed as part of the USGS bioassessment program. As a reference of taxonomic names can also include higher-level taxonomy, as does AlgaeBase (Guiry and Guiry 2013); reference like that can also provide past names or names used in other projects associated with current names in use. The latter are often called harmonization lists because they provide a tool for making taxonomy consistent among projects. Such lists can take taxon lists maintained by multiple individual counters and group the split taxa (to variety and form for example) into “lumped” taxa levels (species level) that may provide more consistency among taxonomists and ease ecological interpretation.
There are several notable examples of harmonized taxon lists that have unified nomenclature (Dodd 1972 and North American Water Quality Assessment [NAWQA] Program of USGS work 1991–2010). Dodd (1972) reported using classic identification books (citations list available at http://westerndiatoms.colorado.edu/citations, Spaulding et al. 2010) for the 900 taxa identified in lakes in northern Iowa. Dr. Dodd and his students verified each name by cross-referencing all books and updated the taxonomy with the name and identification of the newest source for taxa without a documented discrepancy. If discrepancies between references were discovered, the authors went to the original publication and verified the description, spelling, authority, and distribution. Another example is the standardized taxon list developed for the USGS bioassessment by multiple laboratories. This list has been adopted by the USEPA and is being incorporated into ITIS. This list (http://diatom.ansp.org/nawqa/Taxalist.aspx) includes key references for each taxon and an image in the literature that was meant to typify the morphology of the taxon.
Taxonomic training is needed to assign an individual to a name. The basic training for morphological taxonomists in bioassessment should include a basic phycology class, advanced training in species-level identification, and experience with the taxa in the samples being analyzed. A phycology class is the basis of a broad foundation in the taxonomy of all algal groups and can be upgraded to a deeper understanding of algal biology, physiology, and ecology. Training in most algal classes provides a good foundation for identifying genera, but bioassessment routinely calls for species-level and even variety-level identifications. This training can be acquired by practice and access to several general taxonomic keys (e.g., Prescott 1962, Patrick and Reimer 1966, 1975, Krammer and Lange-Bertalot 1986–1991). During the initial stages of a project, each sample should be reviewed by scanning and documenting the dominant taxa. Photographic images should be taken and shared within the project to provide taxonomic consistency. Taxonomic accuracy, i.e., consistency with other taxonomists and projects, together with the best known name for a taxon, is developed during an exploration of the literature. Accuracy can be established during counting because identifying every specimen in a count is a good method of learning taxonomy. Because morphological identification and counts can be at the whole community level (soft counts) or diatom level, there is a need for species-level knowledge for all algal groups and at both a 400× and 1,000× magnification. Once a novice counter can perform soft and diatom counts in ~4 h, he/she should compare counts with more experienced taxonomists. The two independent counts should be more than 50% similar for soft counts and higher than 75% similar for diatom counts. Preliminary plans have been developed for a certification program for algal taxonomists (R.J. Stevenson, personal communications) that would also provide guidelines and training for satisfactory performance. When the trainee can consistently meet these goals for consistency with an experienced taxonomist, then he/she is ready to start sample analysis. An ongoing program to confirm consistency in taxonomy is recommended with routine counting and comparison of duplicate counts.
Issues in Taxonomy and Relationship to Bioassessment
Taxonomy is a science that is constantly improving its ability to identify and systematically relate taxa according to evolutionary relationships. Algal bioassessments use taxonomy as a tool; however, the extent to which advances in taxonomy improve ecological assessments of algae is unclear. Few studies have addressed this question, and the answers differ. Prygiel et al. (2002) suggested that 80% of the variance in a tested diatom index resulted from the diatomist, 10% resulted from the sampling, 5% resulted from preparation of the sample and diatom slides, and 5% resulted from the replicates on each slide. Lavoie et al. (2005) reported that field sampling and laboratory methods were not important contributors to the variation of diatom community analyses across stream sites but that the familiarity of a diatomist with the species present was important. Kahlert et al. (2008) found that years of experience were not as important as using harmonized taxonomic lists among a group of taxonomists from across Europe. Among these studies, one reason for the variability was the different schools of taxonomy in which the taxonomists trained. Another reason may be an apparent tradeoff between accuracy and consistency. If more experienced taxonomists distinguish more species than less experienced taxonomists because they split more morphologically variable groups, then there will be less similarity between their counts and those of others simply because the other taxonomists did not use the same names. This tradeoff between more names and coarser taxonomy was also evident in analyses of duplicate counts by different taxonomists in a national assessment program in the United States that showed the greatest similarity in taxonomic composition of counts among taxonomists using a sensu lato taxonomic approach (R.J. Stevenson and K.M. Manoylov, unpublished data).
The importance of algal misidentification to ecological metrics variability has not been sufficiently addressed. However, bioassessment has been very useful in measuring aquatic health, and the use of algae in bioassessment is regarded as a better strategy compared with nutrient-only or chl a analyses because algae respond rapidly to human impacts (Griffith et al. 2005, Carliste et al. 2008). In a review of data presented by Kahlert et al. (2008), algal metrics distinguished ecological conditions among the sites relatively well given the great dissimilarity in species composition among the counts. Stevenson et al. (2010), R.J. Stevenson (personal communication) argued that algal metrics work because of the law of large numbers and random, unbiased effects of misidentifications. Random, unbiased errors in identifications should increase the variability of metric values without changing the mean value (or central tendency) because when calculating the metrics, the identity of a species is translated into a value related to environment preference or optima; thus, there is no reason why a misidentification would consistently make that value either higher or lower. Variation in calculated metrics should also decrease with the number of taxa observed in samples, just as the ratio of heads:tails in successive flips of a coin should approach 1:1. Of course, all taxa cannot be misidentified, and diatom metrics perform relatively well compared with other organisms because there are relatively large numbers of taxa in samples and because most taxa can be identified to the species level, whereas few other groups of organisms satisfy both of these characteristics.
Although diatom metrics work well, the question of whether an improved taxonomy will improve algal bioassessments remains unclear. To address this question, I will identify the sources of variability in morphological taxonomy, and I propose this question be addressed more rigorously in future research. Additional resources continually become available, and the ways to obtain taxonomy for bioassessment can vary. Assessing “soft algae” or “non-diatomaceous algae” is even more complicated because the diversity of the groups precludes anyone having all required literature for species-level identification. Standard operating procedures require “soft” counts (Charles et al. 2002) because it is possible for diatoms to be in smaller abundances compared with other groups of algae; moreover, soft counts provide evidence if the diatoms present at a site were alive at the time of analysis. Live and dead diatoms are recorded based on the presence of an intact chloroplast. Identification of live preserved material can be particularly difficult and uncertain, but Gillett et al. (2011) found no difference in the ecological inferences from counts of live diatoms (frustules with visible chloroplasts) and cleaned diatoms (acid-cleaned frustules) from the same sample.
Consistency in algal taxonomic identifications for bioassessment is an important goal. When revisiting the call for the “marriage of taxonomy and ecology” (Kociolek and Stoermer 2001), phycologists were asked to understand both disciplines. More than a decade ago, Kociolek and Stoermer (2001) argued that there was a historical discrepancy in the approaches of algal taxonomy and ecology. Algal taxonomists are describing many new species on the basis of significantly different morphological characteristics that are sometimes only visible with SEM and report on algal biodiversity from different geographic areas. Ecologists, however, analyze randomly collected samples and use somewhat older taxonomic floras without necessarily the need to integrate current algal taxonomy. Over the past 12 years, the conceptual and practical merging of the two disciplines can be seen in many examples of satisfactory integration at small experimental scales (Manoylov 2009) and regional scales from the United States and around the world (Rimet et al. 2004, Wang et al. 2005, Lavoie et al. 2009, Danielson et al. 2012). Recently, even larger national scales of bioassessment have been undertaken by linking accurate taxonomy in large-scale ecological surveys with thousands of samples (Potapova and Charles 2007, Stevenson et al. 2008, 2013). Stevenson et al. (2008, 2013) collected data across several algal labs and more than five taxonomists, and this process required communication between the taxonomists and successful agreement on the most current name. In algal taxonomy, there are constant name updates, but the use of an older name is not necessarily a wrong identification. Translation tables (for older and updated taxonomy) are required if more than one taxonomist is involved in algal identification for bioassessment. Intercalibration exercises between taxonomists have shown that identification mistakes can be improved between counters when there is an exchange of images and morphological descriptors (Prygiel et al. 2002) and that the most comparable results (in terms of diatom listings) are achieved among people who make an effort to harmonize the taxonomy rather than among people who have the longest experience with diatoms (Kahlert et al. 2008). Kahlert et al. (2008) reported that for identifying diatoms, access to literature is more important than experience of independent counts. It is difficult to create a usable taxonomic library without communicating with individuals experienced in phycology.
One common issue for discussion in algal bioassessment is whether genus-level taxonomy is sufficient. The need for accurate species-level identification has been well established (Patrick and Palavage 1994, Potapova and Ponader 2004, Kociolek 2005) because most of the algal bioassessment tools are based on species resolution. Species-based nutrient optima perform better than genus-level responses using multiple methods of defining concentration-based nutrient criteria in streams and their ecological relevance (Smucker et al. 2013). In a multi-assemblage case study to assess agricultural and coal mining impacts on streams, Smucker and Vis (2009) found that using diatom species provided a finer resolution of stressors than did using genera. Insights on diatom species' characteristics, communities, and population interactions under low-nutrient conditions have resulted from the study of diatom assemblages in experimental ecosystems with minimal human alteration (Manoylov and Stevenson 2004, 2006, Manoylov 2005).
However, considerable literature supports many metrics at the genus level, such as the diatom genera Cymbella or Nitzschia discussed in Wang et al. (2005). Groups of genera and even higher classification levels, such as families and orders in diatoms, provide characteristics for classification and some ecology, such as members of Surirellales indicate presence of raphe and motility necessary for living in sediments. (Potapova and Charles 2002, Olds et al. 2012). Rimet and Bouchez (2012) compared taxonomically tiered data for nearly 2,000 samples from France and show that the finer the taxonomic resolutions produced diatom assemblages were more consistent with ecoregion classifications, which resulted from diatom endemism and responses to environmental conditions that are mostly observed at the species level. However, coarse identification could be meaningful because (i) many species are too rare to describe their ecological requirements with certainty; (ii) additional environmental descriptors are required to explain the presence of certain species and (iii) the dataset includes identification errors, particularly at the species level (Rimet and Bouchez 2012). Excluding diatom taxa that have been identified as species with low relative abundance in bioassessment analyses is useful (Lavoie et al. 2009). Broad taxonomic resolution appears to be well adapted for use in some diatom studies (e.g., Hill et al. 2001). Raunio and Soininen (2007) found that generic level analyses must be combined with the identification of the dominant species for practical and reliable biomonitoring in rivers. Unfortunately, the processing methods (digestion methods and units and cells enumerated) and taxonomic literature used in species identification for soft counts and diatom counts are often hard to repeat based on the published materials and methods. For example, in the diatom species index for the bioassessment of Australian rivers (Chessman et al. 2007), the taxonomic keys for the whole community (soft algae) pollution-tolerance indices for Nordic Rivers (Schneider and Lindstrøm 2011), soft algae counts, and diatom species-level indices for Israel (Barinova et al. 2006) and California (Nelson et al. 2013) are incomplete or not reported.
It is not clear whether species-level identification and ecological inferences are better when performed on cleaned or uncleaned material in diatom analyses. Correlations between environmental characteristics and stream diatom assemblages that used species were stronger than those that used the genus level (Hill et al. 2001). Coarser aggregations to genus, family, order, class, and subdivision are reported according to available taxonomy in AlgaeBase (Guiry and Guiry 2013), ITIS (http://www.itis.gov update 12/13), or other naming conventions as master lists. Future online master lists will be a convenient site for updated taxonomy and will allow for wider participation by scientists in the process of preventing old and new names from being used concurrently in a dataset.
Bioassessment with algae combines an understanding of algal ecology with expertise in phycology, but it also requires skills for data management. The current taxonomy list (http://diatom.ansp.org/nawqa/Taxalist.aspx) combines data from several current surveys in the US and is dominated by diatoms. Many diatom taxa, even the most common, are difficult to distinguish using light microscopy because diatoms in the microscope may occur in valve, girdle, or tilted views. In addition, electron microscopy has revealed inconsistencies in light microscopy identifications (Morales 2001). The relative ease of identifying diatoms and the ability of diatom-based analyses to reflect physical and chemical impacts are demonstrated by Desrosiers et al. (2013) for marine ecosystems and by Stevenson et al. (2008) for freshwater ecosystems. Knowing little about the functional role or true physical abundance of the “rare” organisms in situ makes it difficult to account for these organisms statistically, so rare algae are often ignored in ecological analyses (Lavoie et al. 2009).
Another issue in algal bioassessment is whether non-diatom algae should be assessed in habitats in which diatoms are abundant. Non-diatomaceous benthic algae show a good relationship with ecological conditions in Nordic streams if analyses are conducted at the species level, but not when the analyses are conducted at the genus level (Schneider and Lindstrøm 2009, 2011). Problems in identifying non-diatom algae with standard counting methods are caused by poor preservation, lack of distinguishing features, and the relative rareness of non-diatom algae in certain habitats. Sample treatment with common preservatives such as formaldehyde and Lugol's solution as preservatives (APHA 2012) can discolor cells or alter the morphology of colonial and filamentous forms, which are key characteristics for identification. For example, Lugol's stains starches, which can obscure organelle structures. Filamentous fragments of cyanobacteria can be confused at the genus level because the sheath and heterocysts are variably present. Filamentous green algae represented by Zygnematales have been traditionally identified to the genus level (for a notable exception, see Stancheva et al. 2013) because reproductive states are rarely observed in samples collected using standard bioassessment sampling protocols. In a recent study, Stancheva et al. (2012) proposed a solution for this problem for future bioassessment by implementing a new protocol that allows the identification of all algae at 1,000× magnification. Many filamentous species were distinguished morphologically and identified as different taxa but not named during counting. After they were grown and maintained in culture, morphological changes were documented continuously, and each culture was assessed molecularly. As a result, 12 new species were described morphologically, molecularly, and ecologically (Stancheva et al. 2013). An additional issue for many samples is that non-diatom algae are so rare that little information can be gained by their identification using standard counting protocols. Of course, having information about non-diatom algae would make algal bioassessments more complete, especially for assessments of biodiversity and potential nuisance and harmful conditions, but the costs and benefits must be evaluated more thoroughly to determine when and where non-diatom algae should be included in assessments.
Misidentifying and combining closely related species or cryptic species are also problems in algal bioassessment. Many closely related species have ecoregionally related distributions in European rivers and include Achnanthidium bisolettianum (Grunow in Cleve & Grunow) Lange-Bertalot, which is present in pristine rivers on limestone geology, whereas A. subatomus (Hustedt) Lange-Bertalot is also present in pristine rivers but only those overlying crystalline geology (European Committee for Standardization 2004, Rimet et al. 2004). Similarly, two varieties were lumped together at the species level as Cocconeis Ehrenberg (Potapova and Charles 2007) because of the common inability to separate taxa that do not have a raphe-less valve attached to the raphe-bearing valve. These varieties were recently separated morphologically (Romero and Jahn 2013), which resulted in the recognition of C. euglypta Ehrenberg in rivers with limestone geology and mesotrophic conditions and C. lineata Ehrenberg in rivers with crystalline geology (Rimet et al. 2004) and oligotrophic conditions (Monnier et al. 2007). The inclusion of taxa such as Gomphonema parvulum (Kützing) Kützing, Navicula cryptocephala Kützing, Nitzschia palea (Kützing) Smith, and Encyonema minutum (Hilse) Mann, which are commonly associated with high-nutrient conditions, in reference streams in Portugal, was attributed to potential misidentification. In contrast, the high abundance and commonality of A. minutissimum in those streams was attributed to initial colonizer abilities and not nutrient-related ecological preferences (Almeida and Feio 2012). N. palea (Trobajo et al. 2009, 2013) and G. parvulum (Moseley and Manoylov 2012) can be considered species complex based on morphological and size variation. Other taxa like N. cryptocephala and E. minutum should allow precise identification based on precise original descriptions and little size variation. N. palea may be a widely distributed diatom and is common in various lotic and lentic freshwater habitats. However, it is also taxonomically problematic. As part of a multidisciplinary study of this diatom, 25 clones that were identified morphologically as N. palea were isolated from different freshwater habitats around the world (Belgium, Brazil, Egypt, India, Japan, Paraguay, Spain, Sri Lanka, and the United Kingdom). Their morphological and genetic diversity (using the hypervariable D1–D2 domains of LSU rDNA) were investigated, and an almost complete set of interclonal crossing experiments was performed. The results indicated that N. palea is not a simple, homogeneous taxon and will most likely be separated into three or more species. Molecular and mating groups do not separate along the traditional morphological boundaries of N. palea varieties, in particular, between varieties palea and debilis, which are two taxa that are commonly used to discriminate degrees of water pollution. At least two of the putative species within the N. palea complex appear to be geographically widespread. Because of the complexity of the variation revealed by the LSU, mating, and morphometric data, additional work using other genetic markers and new isolates is required to determine the full extent of cryptic and pseudocryptic speciation in N. palea and to investigate whether the segregated species are ecologically differentiated and have value as indicators (Trobajo et al. 2013).
The Specific Example of Achnanthidium minutissimum (Kützing) Czarnecki
Applying names to different morphological variations of small and morphologically variable taxon is challenging. This problem is exacerbated by historical differences in the goals of taxonomic research. Originally, species were collected and documented to describe regional floras within species inventories (Ehrenberg 1843, Hustedt 1930, Patrick and Reimer 1966, 1975) rather than population studies where ecological optima can be differentiated and linked to a morphological entity. As a result, part of the uncertainty in identifying and correctly assigning names to morphologically distinct taxa is the original description.
As an example of the significance of this problem, Achnanthes minutissima is likely the most commonly identified diatom species in the world. A. minutissimum has been reported as widespread and abundant in North America (Patrick and Reimer 1966, Potapova and Charles 2007). In the General Collection at the Academy of Natural Sciences in Philadelphia, there are records from more than 2,500 localities in the United States (K.M. Manoylov, personal observation) in which A. minutissima was found at an abundance of more than 10% at the time of collection. The type of water body, habitat, environmental conditions, and time of collection varied, but all the slides contained what current taxonomy accepts as A. minutissimum. A similarly wide distribution across variable environments was reported in a study of 1,109 rivers in the US (USGS, NWQAP) in which A. minutissimum was reported in 79% of the 2,674 samples counted (Potapova and Charles 2002).
Achnanthes minutissima was described by Kützing (1833-36, pl. 16, fig. 54) as only four times smaller than A. exilis Kützing. In the line drawing, Kützing only illustrated the girdle views of curved frustules on stalks. Based on later observations of the type material, Krammer and Lange-Bertalot (1986–1991) added characteristics to the vague original description, including rounded ends, a straight distal raphe, and striae that become finer toward the ends. Adding more confusion, Lange-Bertalot stated that the lectotypes he had chosen for A. minutissima and A. microcephala Kützing (a completely different diatom until that point) were the same. As a result, the routine distinction between A. minutissima and A. microcephala could not be achieved with morphometric data when they were included in the same species complex (A. minutissima “Sippen”-complex in Krammer and Lange-Bertalot 1991 with a length of 5–25 μm, usually less than 20, and a width of 2.5–4 μm). In addition, other authors such as Hustedt (1930) and Patrick and Reimer (1966) presented the two entities as having overlapping size ranges, with a given length of 5–40 μm.
Along with the significant morphological variability in this taxon is confusion about its ecology. This taxon has been reported as being indifferent to nutrient concentrations (Van Dam et al. 1994); however, experimental work suggests that A. minutissimum is tolerant of low-nutrient concentrations and relatively slow growing in high-nutrient concentrations compared with other taxa (Manoylov 2005). Indeed, other authors have since found that A. minutissimum is a good competitor for nutrients when they are in low supply in natural communities (DeNicola et al. 2006, Potapova and Hamilton 2007, Almeida and Feio 2012). Questions still remain about whether the different findings are incompatible or if a broader explanation can be found for the differences, and there even questions about whether different ecophenes might be hidden in the species complex. Using genetically identical populations of this taxon (culture obtained from Dave Czarnecki), responses of one genotype were characterized under different light and nutrient conditions (Manoylov 2009). Periphyton communities of genetically identical populations were created to evaluate the ecological effects of other diatoms on clonal populations of A. minutissimum. Nutrients limited the growth of A. minutissimum, and its growth rate was further decreased with an increase in diatom density. Light had a strong negative effect when nutrients were limited but had a positive effect when nutrients were available. A. minutissimum and C. placentula var. lineata are both monoraphid diatoms, but the latter species has a slower growth rate compared to A. minutissimum. Recognizing the importance of intraspecific competition in addition to interspecific completion that could be regulating A. minutissimum, the most common diatom in North American streams, is an important contribution to the knowledge about competition regulating the composition of stream periphyton (Manoylov 2005, Manoylov et al. 2013). A. minutissimum is merely one example that shows the need for interdisciplinary research that combines natural history knowledge, observations, and experiments. Therefore, the algal taxonomic literature is currently evolving and changing rapidly as types and original descriptions are tested with currently existing populations from North America (Morales 2001, 2002, Manoylov et al. 2003, Potapova and Hamilton 2007, Stancheva et al. 2013).
Molecular Taxonomy and Identification for Bioassessment
Molecular approaches for taxonomic identification of algae are being tested to augment or even replace morphological identification. Most molecular work has been related to characterizing biodiversity and systematics (Deans et al. 2012), but its uses in bioassessments are being advanced. Detecting species using nucleic, mitochondrial, and/or chloroplast DNA has become commonplace in recent years. DNA barcoding (Hebert et al. 2003) was proposed as a new system of species identification and discovery using a short section of DNA from a standardized region of the genome. This technique bypasses the need for laboratory cultivation and/or isolation of individual specimens. Molecular assessments of algal biodiversity have the potential for more automated, complete, and accurate characterization of taxa in natural communities.
Early comparisons of molecular and morphological identification of taxa in stream periphyton analyses illustrate some of the opportunities and challenges of using molecular approaches. In a stream benthic community, gene sequences were matched with sequences from the database and then compared with taxa identified with morphological characters (Manoylov et al. 2009). Both bacterial and eukaryotic sequences were targeted with domain-specific primers for 16S and 18S rRNA genes, and 90% of the sequenced data was potentially identifiable. The 16S library revealed that 23% of the 46 prokaryotic isolates were classified as representatives of Cyanophyta. Two genera of cyanobacteria were observed microscopically in the natural sample, but they were rare. No cyanobacteria were reliably identified at the genus level using molecular techniques. For the eukaryotic community, 13% of the 98 sequences corresponded with the phototrophic taxa using 18S sequences. All eukaryotic phototrophic taxa were identified reliably at greater than or equal to a 98% similarity with the sequence data. Only three of the 53 algal species identified with morphological characteristics were also identified molecularly. There was no correlation between the dominant taxa identified morphologically and molecularly. While species-level matching with the two techniques was not as high as expected, the data suggested that with more comprehensive databases, there may be a great potential for applications in bioassessment. Considerable work is required to document RNA sequences for more taxa and to evaluate variability in sequences within and among taxa to improve both the molecular and morphological identification of algae.
New methods and improved DNA reference libraries show great promise for bioassessment (Kermarrec et al. 2013). A growing number of investigations have focused on the composition and structure of the protistan community within marine and freshwater systems, including analyses performed by the 454 pyrosequencing method (Zimmermann et al. 2011). Often, morphological assessment is considered inadequate for the smallest members of algal communities, so the community structure of small freshwater eukaryotes (0.2–5 μm) was investigated in a mesotrophic lake every 2–3 d over one summer by coupling three molecular methods: 454 amplicon pyrosequencing, qPCR, and TSA-FISH (Mangot et al. 2013). In the later study algal communities did not change as much as fungal and heterotrophic Protista. Aquatic communities are composed of a vast majority of taxa that are always rare (99.8%), i.e., never detected as abundant in any of the samples analyzed. This finding is consistent with Galand et al. (2009) and provided further information about the taxonomic composition and putative ecological role of the rare biosphere in this system. Indeed, the rare biosphere was mainly composed of fungal sequences and, to a lesser extent, photosynthetic organisms (Chlorophyta, Bacillariophyta, and Dinophyceae).
Next-generation sequencing (NGS) platforms have revolutionized the amount of sequence data returned from environmental sequencing runs. Recovery of DNA sequence data directly from environmental samples (e.g., Sogin et al. 2006) instead of a single cell (Ki and Han 2005) was possible with Sanger and other sequencers, but not applied to algae. NGS has been used in a variety of applications, including estimating the health of an ecosystem by analyzing invertebrate biodiversity from benthic communities in rivers (Hajibabaei et al. 2011) and the diversity of photosynthetic protists (Saunders and McDevit 2012) and studying algal DNA from permanent collections (Hughey and Gabrielson 2012). By comparing obtained sequences to a growing standard reference library of known organisms, the taxa present in an environmental sample can be identified with high confidence. A recent explosion in both the number and breadth of studies employing NGS platforms demonstrates the potential for applications in ecological research and high volumes of sequence data (Shokralla et al. 2012).
Taxonomic inventories have also been characterized using the complete DNA reference libraries and three genes as if a mock community was ‘unknown’ (Kermarrec et al. 2013) and as they would be performed for unknown environmental samples. False-positive species (i.e., species not actually included in the combination of taxa developed as a control) may also appear in the molecular inventories. For the rbcL and cox1 genes, almost all reads (>94%) displaying a perfect match with a sequence from reference libraries were associated with a single species. However, a high proportion of small subunit ribosomal DNA (SSU rDNA) reads displaying 100% sequence identity with sequences from reference libraries were associated with several genera, with top hits such as Gomphonema and Nitzschia. For algal specific isolates, ‘informative’ reads at different taxonomic levels only had high taxonomic resolution for cox1 (a mitochondrial gene); these reads had slightly lower resolution for rbcL and much poorer resolution with the SSU rDNA, which only provided a full taxonomic resolution at the class level with the sequences available in the databases (Kermarrec et al. 2013).
Molecular NGS techniques have been tested and will no doubt continue to be tested because of their ability to substitute for the laborious morphological identification of thousands of samples. Genetic source molecules are assumed to come from viable cells, but they can also come from dead cells, dormant cells, or molecular debris of rare taxa that can persistent in algal communities (active or non-active resting stages of microbial communities, Jones and Lennon 2010). Dormancy is a survival strategy used by a variety of algae to overcome unfavorable environmental conditions in eukaryotic microbial communities dominated by green algae (spores) and cyanobacteria (akinetes). How the presence or absence of those dormant stages will be recognized with molecular tools and used in bioassessments is not clear.
The reference library of SSU rDNA sequences is the largest reference library because it is the ribosomal region that has been the main marker used for diatom phylogenetic studies since Medlin et al. (1988). This gene sequence has been used so often because the presence of conserved regions facilitates the design of universal primers and because the presence of variable regions allows phylogenetic study. Despite the conserved regions, many investigators include SSU in their studies because of the large amount of available sequences and the presence of a highly polymorphic region that could be a potential barcode (Zimmermann et al. 2011, Stancheva et al. 2013). Similarly, the rbcL reference library comprises many sequences because this gene has been used for cryptic species evaluation (Evans et al. 2008) and phylogenetic studies (Bruder and Medlin 2007, Trobajo et al. 2010). Recently, rbcL has been proposed as the reference barcode for diatoms with LSU (Hamsher et al. 2011) or the ITS fragment (MacGillivary and Kaczmarska 2011) as a secondary barcode. As previously indicated by Moniz and Kaczmarska (2009), Trobajo et al. (2010), and Hamsher et al. (2011), little success has been found with cox1 fragment sequencing for diatoms, which is contrast with the findings of Kermarrec et al. (2013). The Shannon index values demonstrate the high variability distributed throughout the entire length of the rbcL fragment and explain why it is difficult to design universal primers for diatoms. Other markers studied for diatoms (ITS, LSU) were discarded from the study by Kermarrec et al. (2013) because of intraclonal variability and the lack of available data for building reference libraries. Primer design is still critical for the use of DNA barcoding; however, the available cox1 primers do not reach the 95% amplification success proposed by Hajibabaei et al. (2005). This problem has resulted in a small DNA reference library (only 266 sequences). The design of good primers could constitute an avenue of research that would improve the cox1 reference library and enhance the detection of diatom species, especially because the official barcode proposed by Hebert et al. (2003) was the cox1 gene.
According to Mann et al. (2010), new attempts to isolate and grow diatom strains are required to complete the DNA reference libraries and increase their taxonomic coverage, regardless of the marker. Other approaches are being developed to overcome the limits of incomplete reference libraries by assigning higher-level taxonomies when a reference is lacking at a lower level or even using functional genes rather than species (Burke et al. 2011). A consensus should also be reached to define the primers used to create the DNA reference libraries. Currently, many reference libraries are composed of different fragment lengths. Long amplicons for pyrosequencing are required for accurate matches on the whole-read size. Reads that overlap poorly with reference sequences are usually discarded and should be taken into consideration in future studies to optimize the design of the barcode used for NGS platforms. The process tested by Burke et al. (2011) makes it possible to identify diatom taxa at any taxonomic level.
With regard to the DNA reference libraries, resolving power, and methodological bias, 454 pyrosequencing could potentially be used for algal communities. Further efforts are required to optimize laboratory protocols and bioinformatics tools and to continue to develop the rbcL DNA reference library to improve its taxonomic coverage. However, the approach of using rbcL is promising for determining taxonomic inventories of diatoms in samples of natural assemblages, which could be valuable in the context of biomonitoring programs.
Relative Abundance Estimates of Species with Molecular Techniques
Questions remain about whether molecular data can be used to characterize relative abundances of taxa in samples. If richness is underestimated in morphological assessment, then molecular identification will improve the documentation of the number of species in a community. However, it is unclear how accurately pieces of DNA will relate to the relative abundance of actual organisms present in a community. Because sequence reads can come from the same individual (algae have variable genome sizes) (Cerutti et al. 2011), from different individuals within the population, or from different species, there is a potential to overestimate abundance. Amend et al. (2010) studied the relative abundance assessment in microbes as a general problem and found that repeated sequences within microbe genomes can be problematic for abundance estimates. The culture-independent analysis of sequences derived from environmental samples has revolutionized our understanding of microbial diversity, function, and processes (Rinke et al. 2013). Technological advances such as pyrosequencing enable rapid characterization of microbial communities that are faster and at a greater sequence depth than was deemed possible via cloning and Sanger sequencing (Sogin et al. 2006). Concurrent with these technological advances are new ideas and methods for statistical comparison of complex microbial communities (Schloss 2008), with considerations of “read abundance” being almost quantitative within species; however, between-species comparisons can be biased by the innate sequence structure. The results showed a trade-off between sequence quality stringency and quantification.
A common assumption in targeted sequence analyses of environmental samples is that read abundance correlates with genic (gene based) and taxon abundances. Strictly quantitative measures in which absolute abundances are estimated (e.g., cells/sample) can be discounted in favor of current pyrosequencing technologies because upper (and effectively lower) constraints on the number of total reads produced are imposed. A looser definition of a “quantitative” measure that is analogous to relative abundance is the correlation between proportional read abundance and the proportional abundance of a given organism in relation to its neighbors in two subsamples from an original sample with assumed random species distribution within the original sample.
Intuitively, a dominant community taxon in a sample should dominate a pyrosequencing data set. In current algal studies, several genes are commonly used: SSU rDNA from the nuclear genome, rbcL from the chloroplast genome, and cox1 from the mitochondrial genome. The taxonomic coverage of the DNA reference libraries appeared to be the most crucial limitation, although marker polymorphism, which is essential for identifying taxa at the species level, was also a limitation. In Kermarrec et al. (2013), rbcL offered the highest power to correctly identify taxa, which may in part be a result of its large DNA reference library. Although pyrosequencing requires further optimization, it is potentially suitable for identifying diatom assemblages and may find applications in the field of freshwater biomonitoring. However, it may have to be combined with estimation of species abundance proportions with microscopy (Kermarrec et al. 2013). No applications of shotgun metagenomics to algal bioassessment have been found.
Concerns in Using Molecular Taxonomic Identification for Bioassessment
At this point, we cannot adopt molecular bioassessment of algal community composition for many reasons. The number of samples analyzed with molecular tools for bioassessment has been limited to between one (Manoylov et al. 2009) or five samples of natural habitats (Kermarrec et al. 2013). The identity of most taxa in the reference libraries of genetic sequences has not been rigorously evaluated, so the reference library taxonomy may not be accurate. Because the importance of correct identification for bioassessment has been established, it is necessary to mention that the complexity of algal shapes, adaptations, and survival cannot be simply translated into a DNA code because the expression of proteins depends on the unique set of environmental conditions in which algae live (Will and Rubinoff 2004, Sluys 2013). A large initial investment is required for equipment, or the price of analyses must be lowered. A low number of reads (<100) and reads with non-compatible tag combinations (Carlsen et al. 2012) are common and depend on the sequencing platform used. Manual inspection of BLAST searches (Altschul et al. 1997) and GenBank (Benson et al. 2012) to identify “contaminant sequences” is also required. Interpretations of sequences that have switched tags at both ends (see Carlsen et al. 2012 for microbial molecular identification) could produce problems. Descriptive statistics for molecular variations have not been developed and tested to include the number of reads, alignment sites, and haplotypes; the haplotype diversity; the nucleotide diversity; or the average number of nucleotide differences.
The high cost of reagents per megabase sequencing output and the reading of homopolymer regions (Claesson et al. 2010) are additional issues for 454 pyrosequencing. Problems with reading homopolymer regions result from the lack of a terminating moiety to stop the extension run and insertion–deletion rather than substitution errors. This issue has caused concerns for scientists employing this platform in the analysis of environmental DNA because sequence errors may be interpreted as unique haplotypes that represent rare biota (Sogin et al. 2006). However, this problem has been largely alleviated by using computational tools to distinguish and filter out erroneous sequences (Quince et al. 2009).
A novel fosmid clone-based metagenome isotope array approach termed the community isotope array (CIArray) was developed and validated for metabolically active microorganisms, that is, for the sensitive detection and identification of microorganisms that have assimilated a radio-labeled substrate within complex microbial communities (Tourlousse et al. 2013). This approach also has a great potential for algal bioassessment because it would solve the problem of DNA sequence assays including genes from resting spores (common in algae) and broken or damaged algal cells in freshwater environments. If an organism is in a dormant stage, it might be avoiding current environmental conditions.
Conclusions
For meaningful bioassessment, harmonization between taxonomists in large projects, consistency and accuracy of unknown taxa descriptions, and image documentation are required. Morphological analyses of algal communities will remain important as standalone options for bioassessment, documentation of biodiversity, or an initial stage of proof for the molecular taxonomic identification by expert taxonomists (Lücking 2008). Algae have variable (some large) genomes, repeated sequences, and a tremendous ability to survive environmental conditions, so there is a great potential for additional discoveries and documentation of biodiversity and for the efficient application of this knowledge to bioassessments.
It is unclear how we will describe new species in the future. Taxonomy will remain an active field (Wheeler et al. 2004, Stegen and Hurlbert 2011), but whether bioassessment will ever evolve past morphological observations, type specimens, original population size descriptions, and imaging for taxonomic evaluation is unclear. Reports have indicated that taxonomists will remain in high demand in the near future (Taylor and Harris 2012). The current need for explanatory types (epitypes) that combine micro-morphological traits as well as molecular data is important, and these epitypes will be used for reference barcodes for future molecular identification (Zimmermann et al. 2011, Pawlowski et al. 2012). A standard method for sequencing genes will most likely become routine in the future; however, applying that method to any “real” environmental communities to identify taxa to provide a suitable substitute for morphological identification in bioassessment is not imminent. For algal communities, microscopic identification makes it possible to compare inventories based on morphology to those obtained with molecular techniques; therefore, morphological identification will remain important, which has been documented in other organisms (Will and Rubinoff 2004). To study complex environmental samples, Kermarrec et al. (2013) are currently designing a bioinformatic tool capable of accounting for intraspecific variability by computing edit distances on local alignments, which were implemented by the Smith–Waterman algorithm, and filtering small distances (up to a few SNPs or short indels as in polyA and polyT). Such a tool will be able to detect intraspecies variability while maintaining accuracy, and associating this tool with NGS should lead to the generation of automatic taxonomic inventories of environmental diatom communities, which will be faster and more reliable. These types of approaches are the likely next steps in the development of next-next generation tools, which show great promise for characterizing biodiversity of eukaryotic communities and may meet the demands for the high rates of sample analysis, natural assemblage analysis, and accurate species identifications that are required for algal bioassessment.
Comments and ideas from Jan Stevenson, Morgan Vis, and Nate Smucker are greatly appreciated. This paper was presented as a part of the Algal Bioassessment Symposium invited talks at the 2012 Phycological Society of America Meeting in Charleston, South Carolina. I also thank the associate editor and an anonymous reviewer for critical comments and suggestions.




