Molecular and morphological evidence for Sheathia gen. nov. (Batrachospermales, Rhodophyta) and three new species
Abstract
The freshwater red algal genus Batrachospermum has been shown to be paraphyletic since the first molecular studies of the Batrachospermales. Previous research, along with this study, provides strong support for the clade Batrachospermum section Helminthoidea. This study has found that heterocortication, the presence of both cylindrical and bulbous cells on the main axis, is an underlying synapomorphy of this clade. Based on support from DNA sequences of the rbcL gene, the COI barcode region and the rDNA ITS 1 and 2, along with morphological studies, the new genus Sheathia is proposed. Seven heterocorticate species were recognized from the molecular clades. Sheathia boryana and S. exigua sp. nov. appear to be restricted to Europe, whereas S. confusa occurs in Europe and New Zealand. Sheathia involuta is widespread in the USA and reported for the first time from Europe. Sheathia americana sp. nov., has been collected in the USA and Canada, and S. heterocortica and S. grandis sp. nov. have been collected only in the USA. Sheathia confusa and S. grandis can be distinguished based on morphological characters, whereas DNA sequence data are required to conclusively distinguish the other species. Sheathia fluitans and S. carpoinvolucra also are placed within this genus based on the presence of heterocortication. These data also hint at greater diversity among non-heterocorticate Sheathia than is recognized by the single species name S. arcuata.
The Batrachospermales is an order of freshwater red algae (Rhodophyta) that encompasses ~2/3 of all freshwater red algal diversity, and the genus Batrachospermum is the most species rich (100–150 species; Sheath 1984, Kumano 2002). Simon Sirodot was the first researcher to study Batrachospermum in depth, dividing it into six sections based on morphological features (Sirodot 1884). Over the past century, these sections have been reclassified and expanded upon, increasing to eight, as new taxonomically important characters have been proposed and new regions of the world explored (Kumano 2002). From the earliest studies using molecular data to infer phylogenetic relationships within Batrachospermales, Batrachospermum has been shown to be polyphyletic and clearly in need of a major taxonomic revision (Vis et al. 1998, Entwisle et al. 2009). Current systematics research is focused on resolving the polyphyly of Batrachospermum, elevating sections to segregate genera, and elucidating the true species diversity. For example, the two sections Contorta and Hybrida were combined to form the genus Kumanoa, and a subsequent study determined phylogenetic relationships among species and description of new species (Entwisle et al. 2009, Vis et al. 2012).
Batrachospermum section Batrachospermum sensu lato and the taxa therein have been the subject of numerous revisions in the past 20 years. Vis et al. (1995) conducted an extensive morphological study of 48 type and historically important specimens from this section. These researchers concluded that numerous morphometric characters, as well as qualitative traits such as monoecy, dioecy, and placement of spermatangia were taxonomically informative (Vis et al. 1995). As a result of that study, Batrachospermum section Batrachospermum was revised to contain 15 well-defined species based on both quantitative and qualitative characters. The first molecular phylogeny of the Batrachospermales showed that species from Batrachospermum section Batrachospermum formed two distantly related clades (Vis et al. 1998). One of these clades contained B. gelatinosum (L.) DC., the nominate species for the genus and section Batrachospermum. The other clade included B. boryanum Sirodot and B. involutum M. L. Vis et Sheath. After further research, section Helminthoidea Sirodot ex De Toni was reinstated for the clade containing B. boryanum and B. involutum, and section Batrachospermum was retained for B. gelatinosum (Entwisle et al. 2009).
Presently, only taxa with DNA sequence data were transferred to Batrachospermum section Helminthoidea, namely, B. arcuatum Kylin, B. boryanum, B. confusum (Bory) Hassall, B. confusum forma anatinum (Bory) S. A. Stewart et M. L. Vis, B. heterocorticum Sheath et K. M. Cole, and B. involutum (Stewart and Vis 2007b, Entwisle et al. 2009). All recognized taxa within this section, except B. arcuatum, exhibit the characteristic of heterocortication, in which both cylindrical and bulbous cells are present on the main axis (Vis et al. 1998, Stewart and Vis 2007a). Molecular analyses and morphological observations have shown heterocortication to be a synapomorphy of a well-supported clade sister to B. arcuatum (Stewart and Vis 2007a). Within this “heterocorticate” clade, monoecy versus dioecy or the presence of spermatangia on the branches that subtend the carpogonium, as well as other vegetative and reproductive features, is used to delineate species (Kumano 2002). However, monoecy versus dioecy, which was historically a key characteristic in distinguishing species within Batrachospermales, has been repeatedly shown to be unreliable (Entwisle et al. 2004, Stewart and Vis 2007a, Ji et al. 2011).
Many of the recognized species within Batrachospermum section Helminthoidea have broad morphological circumscriptions and are, in some cases, distinguished only by monoecy versus dioecy (Vis et al. 1995, 1996, Stewart and Vis 2007a). In addition, most taxa have been commonly reported from a broad geographic range in temperate regions in both the Northern and Southern Hemispheres (Kumano 2002). However, sparse sequence data are available for most taxa, and the data available are from a restricted geographic range. For example, there are sequence data for B. boryanum from three locations in the eastern United States, and one in eastern Canada even though this species has been reported from throughout North America and Europe (Kumano 2002). Likewise, sequence data for B. confusum were from two locations in New Zealand, but there has been no molecular data from Europe, the continent of the type locality. Therefore, further investigation is warranted to better understand genetic variation within and among these widespread taxa.
Numerous molecular markers have been utilized to study the species diversity of red algae (Verbruggen et al. 2010). The plastid-encoded ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit gene (rbcL) has proven to be a consistent marker for elucidating the evolutionary relationships and interspecific diversity in dozens of studies of red algae (e.g., Vis et al. 1998, Maggs et al. 2007, Entwisle et al. 2009). The rbcL gene not only provides well-supported species relationships within the Batrachospermales, but also has a large number of published sequences available on GenBank (Benson et al. 2005). A 664 base pair (bp) fragment of the mitochondrial cytochrome oxidase 1 gene (COI) has been extensively used as a “barcode” for red algae (e.g., Saunders 2005, Robba et al. 2006, Clarkston and Saunders 2012). In addition to being a barcode, this region has been combined with other markers in phylogenetic studies to provide further sequence data to distinguish species (Saunders and Lindstrom 2011, Vis et al. 2012). The nuclear encoded ribosomal internal transcribed spacer region (ITS) has less often been used in studies of red algae, but may help elucidate the nuclear genome history. The ITS region has been shown to be effective in studies of closely related species as a highly variable marker with the potential to be a second barcode region in red algae (Vis and Sheath 1997, Hu et al. 2009).
It would appear that Batrachospermum section Helminthoidea is a good candidate for a taxonomic revision using both morphological and molecular tools. Previous phylogenetic analyses (Vis et al. 1998, Entwisle et al. 2009) have shown the clade of heterocorticate species and B. arcuatum to be well-supported and distinct from Batrachospermum gelatinosum, suggesting that a new genus needs to be described to continue to resolve the polyphyly of Batrachospermum. In addition, there has been no study focused on this section as a whole, especially combining morphological and molecular analyses. With so many broadly circumscribed species that have large geographic ranges, it is likely that cryptic diversity may be uncovered with broader taxonomic and geographic sampling. This research was conducted to establish a new genus for the Batrachospermum section Helminthoidea clade, to provide a robust phylogeny to elucidate the species diversity and to determine geographic ranges along with a revised taxonomy.
Materials and Methods
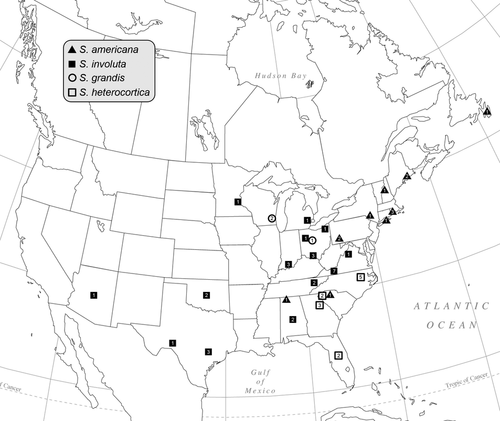
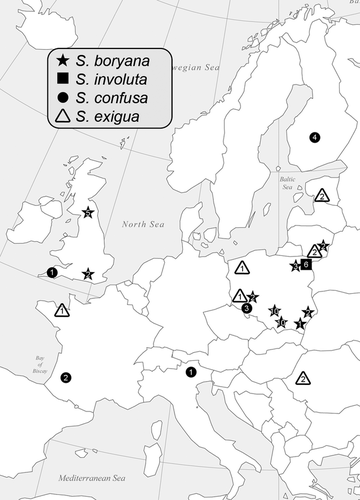
Specimens exhibiting heterocortication were collected from locations in North America, New Zealand and Europe for morphological and molecular analyses (Figs. 1 and 2; Table S1 in the Supporting Information). Locations for type and historically important specimens were visited, when possible, in an effort to collect material to tie molecular clades to historical names (Sheath and Cole 1990, Vis et al. 1995, Vis and Sheath 1996). For molecular analyses, one to many specimens from each location were cleaned of visible epiphytes, blotted dry, and immediately placed in silica desiccant for rapid dehydration and preservation of the DNA. For morphological analyses, specimens from each location were preserved in a 2.5% calcium carbonate buffered glutaraldehyde solution. When possible, a portion of the same specimen as was placed in silica desiccant was also dried on herbarium paper to serve as the morphological voucher; otherwise, specimens from the same location were used as vouchers. The dried voucher specimens for all new specimens, including isotypes, have been deposited at the Bartley Herbarium at Ohio University (BHO). The holotype specimens, both dried herbarium sheets and DNA vouchers, have been deposited at the New York Botanical Gardens (NY) and isotype specimens at the University of Michigan Herbarium (MICH).
Samples for DNA analyses were either ground by hand in liquid nitrogen using a mortar and pestle, or in the Tissuelyser LT (Qiagen Inc., Valencia, CA, USA) set at 20 oscillations per second for 40 s using 3 mm tungsten beads. DNA was extracted with the NucleoSpin® Plant II (Macherey-Nagel, Düren, Germany) kit according to the manufacturer's protocol. Three DNA markers, the rbcL gene, COI barcode, and rDNA ITS region (partial 18S, ITS1, 5.8S, ITS2, and partial 28S) were PCR amplified using either a MJ Research Minicycler™ (Bio-Rad, Hercules, CA, USA) or the Applied Biosystems 2720 Thermocycler™ version 2.08 (Applied Biosystems, Foster City, CA, USA). For the rbcL gene, a 1,282 bp fragment was amplified using the F160 and rbcLR primers (Vis et al. 1998). The PCR cocktail consisted of 19 μL dH20, 25 μL AmpliTaqGold master mix (Applied Biosystems, Carlsbad, CA, USA), 2.5 μL each of the amplification primers, and 1 μL extracted DNA. The PCR parameters were as follows: an initial denaturing at 95°C for 1:00; 35 repeated cycles of 93°C for 0:30, 50°C for 0:30 and 68°C for 1:00; and a final elongation period at 72°C for 10:00. For the COI barcode (664 bp), the PCR amplification cocktail consisted of 19 μL dH20, 25 μL AmpliTaqGold, 2.5 μL each of the GazF1 and GazR1 primers (Saunders 2005), and 1 μL of the extracted DNA. The PCR parameters were as follows: 94°C for 1:00; 40 repeated cycles of 94°C for 1:00, 50°C for 1:30, and 72°C for 1:00; and a final extension at 72°C for 5:00 (Le Gall and Saunders 2010). Unfortunately, these primers and PCR cycle were effective at amplifying the cox1 gene in a wide range of organisms, and this protocol would often result in the sequencing of invertebrate or other algal contaminants. Also, occasionally the concentration of PCR product was not enough for sequencing. For specimens that were not successfully amplified using the previous method, a semi-nested PCR method was utilized (Puławska and Sobiczewski 2005). A larger fragment (>1,400 bp) was first amplified using the forward GazF1 primer or GWSFn primer (Le Gall and Saunders 2010) and either the cox2R primer (5′-ATC ACA TTT HMY YCY HAA AGA WGG TAC-3′) or the cox3R primer (Zuccarello et al. 1999) as the reverse amplification primer. For this reaction, Ex Taq® DNA Polymerase (TaKaRa Bio Company, Tokyo, Japan) was utilized in a 50 μL PCR cocktail consisted of 34.25 μL dH20, 2.5 μL of a forward primer and 2.5 μL of a reverse primer, 5 μL Ex Taq Buffer, 4 μL Ex Taq dNTPs, 0.25 μL Ex Taq Polymerase, and 1.5 μL of the DNA extraction. The PCR parameters for this cocktail consisted of an initial denaturing at 94°C for 4:00; 38 cycles of 94°C for 1:00, 45°C for 0:45, and 72°C for 3:00; and a final extension at 72°C for 7:00. The resulting PCR product was then used as the template for PCR amplification using the COI PCR cycle with the GazF1 and GazR1 primers as described previously. The PCR cocktail to amplify the ~900 bp ITS region consisted of 19 μL dH20, 25 μL AmpliTaqGold, 2.5 μL each of the ITS5.1 (Vis et al. 2001) and AB28 primers (Vis and Sheath 1997), and 1 μL of the extracted DNA. The amplification cycle consisted of an initial denaturing at 95°C for 5:00; five cycles of 90°C for 1:00 and 50°C for 2:00; 30 cycles of 90°C for 1:00, 60°C for 1:00, and 72°C for 1:00; and a final extension at 72°C for 5:00 (Hu et al. 2009).
The rbcL and COI PCR products were purified using the UltraClean™ PCR Clean-up DNA purification kit (Mo Bio, Carlsbad, CA, USA) according to manufacturer's protocols. The ITS PCR resulted in multiple products of different lengths. Gel electrophoresis was used to separate the PCR products, and, at first, multiple bands were excised from the gel, purified and sequenced. One strong band (~900 bp) produced sequences that partially aligned with published 18S nrDNA sequences of Batrachospermum, while other bands would not produce clean sequence. This ~900 bp band was excised and purified using the GelElute Extraction Kit (5Prime, Hamburg, Germany) according to the manufacturer's protocol for all specimens.
The purified PCR products were sequenced using the PCR amplification primers. For the rbcL, the additional internal primers F650 (5′-ATT AAC TCT CAA CCA TTT ATG CG-3′) and R897.1 (5′-CGT GAG TAT GTT GAA TTA CCA GC-3′) were used to ensure that the 1,282 bp fragment was fully sequenced in both directions. For the ITS, a combination of the additional internal ITS3, ITS10 (Vis and Sheath 1997) and ITSF670bory (5′-YGC GTG GYG CTT YGT TGC GT-3′) were used to obtain the full ~900 bp sequence. No internal primers were needed for the 664 bp COI. All DNA sequences were assembled and edited using Sequencher™ version 4.10.1 (GeneCodes Corp, Ann Arbor, MI, USA).
In an effort to apply appropriate historical names to modern molecular clades, material was sampled from type and other historically important specimens from the herbarium at the Muséum National d'Histoire Naturelle in Paris, France (PC). Small fragments were removed with a clean razor blade after wetting with dH2O and placed immediately in silica desiccant. The DNA extraction and PCR preparation were performed using the previously described methods in a sterilized laminar flow hood to reduce the risk of contamination. In addition to the regular PCR protocols for the rbcL, COI, and ITS regions, semi-nested PCR for the COI (as previously described) and rbcL region was attempted. For the rbcL region, first the PCR was completed using the F160 and rbcLR primers, and then the resulting PCR product was amplified using either the F160 and R897.1 primer pair, the F160 and R472 (Stewart and Vis 2007a) primer pair, or the F650 and rbcLR primer pair.
In addition to the new DNA sequences generated for this research, GenBank was searched for sequences within Batrachospermum section Helminthoidea. Thirty-seven rbcL sequences and one COI sequence were obtained; there were no ITS sequences for these taxa on GenBank (database accessed May 3, 2013). These sequences were downloaded, checked for taxonomic accuracy and when long enough to be used in phylogenetic analyses were included in the final data set (Table S1). Outgroup sequences for the rbcL data set were obtained from GenBank (AF029150; AF029141), and COI and ITS sequences from B. gelatinosum were newly generated for outgroup rooting. DNA sequences for the rbcL and COI markers were aligned using Sequencher™. For the ITS region, sequences were aligned with MAFFT (Katoh et al. 2002) under the 20PAM/K = 2 scoring matrix with a gap penalty of 1.53 and an offset value of 0.0. These alignments were checked by eye for errors.
Prior to starting the phylogenetic analyses, the best model of evolution for each of the three data sets (rbcL, COI, and ITS) was determined using the Bayesian information criterion (BIC) as implemented in jModelTest v0.1.1 (Posada 2008). For the rbcL data set, the model was: GTR substitution model with a gamma distribution = 0.1550; base frequencies A = 0.3154, C = 0.1451, G = 0.2050, T = 0.3345; and rate matrix A–C = 4.1874, A–G = 7.4965, A–T = 1.4359, C–G = 0.8379, C–T = 18.1663, and G–T = 1.0000. For the COI data set, the model was: TPM1uf substitution model with a gamma distribution = 0.1370; base frequencies A = 0.2833, C = 0.1553, G = 0.1381, T = 0.4233; and rate matrix A–C = 1.0000, A–G = 8.8519, A–T = 0.2084, C–G = 0.2084, C–T = 8.8519, and G–T = 1.0000. For the ITS data set, the model was: TPM3 substitution model with a gamma distribution = 0.2860; and rate matrix A–C = 0.4181, A–G = 1.4484, A–T = 1.0000, C–G = 0.4181, C–T = 1.4484, and G–T = 1.0000. A concatenated data set was made from the rbcL, COI, and ITS alignments using Sequence Matrix v1.7.8 (Vaidya et al. 2011). The concatenated data set was partitioned by gene for the phylogenetic analyses, and the model of evolution for each gene was set accordingly. Relative substitution rates, base frequencies, and gamma shape were unlinked across data subsets, while topology was linked.
Each gene individually and the concatenated data set were subjected to Bayesian Inference (BI) analysis using MrBayes v3.2 (Huelsenbeck et al. 2001, Ronquist and Huelsenbeck 2003, Ronquist et al. 2012). For the BI analyses, two Metropolis-coupled Markov chain Monte Carlo (MCMCMC) runs consisted of one cold chain and three hot chains. Each run was sampled every 100 generations for 5,050,000 generations. After confirming that the runs converged by checking to ensure that the average standard deviation of split frequencies was below 0.01, the trees were merged following the removal the first 500 trees from each run as burn-in. The resulting tree and posterior probabilities were calculated from the remaining 100,000 trees generated for all data sets. All data sets were subjected to maximum likelihood (ML) analysis using RAxML (Stamatakis 2006). The model parameters for the ML analyses were the same as those for the BI. ML bootstrap support values were calculated using 1,000 bootstrap replicates.
In order to verify the monophyly of Batrachospermum section Helminthoidea, eleven divergent rbcL sequences within the section were chosen, and sequence data for all other Batrachospermales genera were obtained from GenBank (Table S2 in the Supporting Information). Sequence alignment was completed using Sequencher™, and the best model for evolution was determined using the BIC as implemented in jModelTest v0.1.1. For this data set, the model was: GTR substitution model with a gamma distribution = 0.1550; proportion of invariable sites = 0.4870; base frequencies A = 0.3585, C = 0.1029, G = 0.1813, T = 0.3573; and rate matrix A–C = 8.8272, A–G = 8.1492, A–T = 2.7341, C–G = 4.9401, C–T = 48.5418, and G–T = 1.0000. Phylogenetic analysis of this data set followed the same methods as with previous data sets, and the data set was outgroup rooted with members of three closely related orders, Audouinella arcuata (Drew) Garbary, G. I. Hansen & Scagel, Ballia callitricha (C. Agardh) Kütz., and Thorea violacea Bory (Table S2).
Morphological observations were made using a BX40 Olympus microscope with an attached SC 20 camera system (Olympus American Inc., Center Valley, PA, USA). Specimens used for molecular analyses either had vouchers or other specimens from the same population for morphological analyses (Table 1, Table S3 in the Supporting Information). Distinctive qualitative morphological features and whether a specimen was monoecious or dioecious were noted. For morphometrics, fifteen measurements were taken for each morphological voucher of the following features: whorl diameter, carposporophyte diameter, fascicle cell number, carposporangium width and length. Seven measurements of the carpogonial branch cell number, carpogonium diameter and length were made from each specimen. These morphological characters were chosen because they have previously been shown to be useful in delineating taxa within the genus Batrachospermum (Vis et al. 1995, 1996). Measurements from the morphometric data were compiled into a data set. Specimens were grouped by molecular species, and each characteristic was tested for normality and transformed, if necessary. Morphometric data were examined using analysis of variance (ANOVA) with a Bonferroni correction in R (R Core Team 2012). There were two characteristics, whorl diameter and carpogonium diameter that did not meet normality assumptions after transformation; therefore, the non-parametric Kruskal–Wallis test with pair-wise comparisons using the Wilcoxon rank sum test was performed in R.
| Taxon | Number of specimensc | Whorl diameter (μm) | Carposporophyte diameter (μm) | Fascicle cell number | Carpogonium diameter (μm) | Carpogonium length (μm) | Carpogonial branch cell number | Carposporangium diameter (μm) | Carposporangium length (μm) | Qualitative characterd |
|---|---|---|---|---|---|---|---|---|---|---|
| S. arcuata type | 1 | 570 (428–727) | 106 (92–129) | – | 9 (6–12) | 34 (28–39) | 6.7 (5–9) | 9 (7–12) | 12 (10–15) | d, rc |
| S. americana | 11 | 702 (178–2172) | 95 (40–218) | 11.1 (6–23) | 7.3 (5.4–9.7) | 25.5 (16.8–43.9) | 6.5 (4–15) | 7 (5.0–9.5) | 10.4 (7.1–14.9) | m = 1, d = 10, hc |
| B. anatinum typea | 1 | 735 (479–994) | 96 (69–140) | – | 8 (6–10) | 28 (17–40) | 6.2 (3–8) | 9 (7–14) | 11 (8–16) | m, hc |
| S. boryana typea | 1 | 607 (337–1034) | 96 (68–139) | – | 8 (6–10) | 26 (16–35) | 6.3 (4–8) | 9 (7–14) | 12 (8–17) | d, hc |
| S. boryana | 6 | 561 (286–1190) | 119 (55–197) | 12.5 (7–18) | 7.07 (4.5–9.1) | 27.3 (17.1–42.3) | 8.7 (3–20) | 8.4 (5.9–10.2) | 11.2 (7.3–15.4) | m = 5, d = 1, hc |
| S. confusa typea | 1 | 658 (412–955) | 80 (56–112) | – | 7 (5–10) | 20 (14–28) | 6.1 (4–9) | 8 (6–10) | 10 (8–12) | m, s, hc |
| S. confusa | 9 | 670 (164–1510) | 92 (36–185) | 12.1 (8–19) | 8.3 (6.2–11.0) | 25.6 (14.8–38) | 7.6 (4–13) | 7.7 (4.7–11.6) | 10.3 (6.9–14.7) | s, hc |
| S. exigua | 6 | 615 (301–1236) | 115 (67–187) | 12.2 (8–16) | 8.4 (5.6–12.2) | 35 (22.1–49.4) | 8 (5–13) | 10.2 (8.0–13.9) | 14 (11.2–18.1) | d = 6, hc |
| S. grandis | 8 | 757 (292–2105) | 148 (74–239) | 13.8 (8–24) | 8.6 (6.2–12.8) | 42.7 (30.7–80.9) | 8.4 (5–17) | 11 (7.8–17.5) | 16.2 (11.1–23.5) | m = 4, d = 4, hc |
| S. involuta typeb | 1 | 999 (627–1405) | 161 (112–237) | 10.1 (8–12) | 8.4 (7–10) | 40.9 (30–54) | 5.1 (3–8) | 9.1 (7–11) | 14.9 (12–18) | m, s, hc |
| S. involuta | 15 | 629 (245–2635) | 117 (58–254) | 11.8 (6–19) | 8.6 (6.3–16.4) | 28.3 (17.4–46) | 6.9 (2–13) | 8.5 (5.9–10.9) | 11.9 (9–16.5) | m = 2, d = 13, hc |
| S. heterocortica typea | 1 | 617 (492–876) | 99 (71–126) | – | 8 (6–10) | 34 (27–40) | 4.9 (3–9) | 10 (8–11) | 12 (10–15) | d, hc |
| S. heterocortica | 9 | 478 (200–973) | 125 (53–258) | 10.8 (6–16) | 7.7 (5–11.1) | 31.3 (20.1–50.2) | 5.3 (3–9) | 8.5 (5.9–10.4) | 12.3 (9.1–15.6) | m = 2, d = 7, hc |
- a Data from Vis et al. (1995).
- b Data from Vis and Sheath (1996).
- c Specimens from the current study used in morphometric analysis marked with an “*” in Table S1 as well as specimens in Table S3.
- d d, dioecious; m, monoecious; s, spermatangia on involucral filaments, hc, heterocortication; rc, regular cortication.
Results
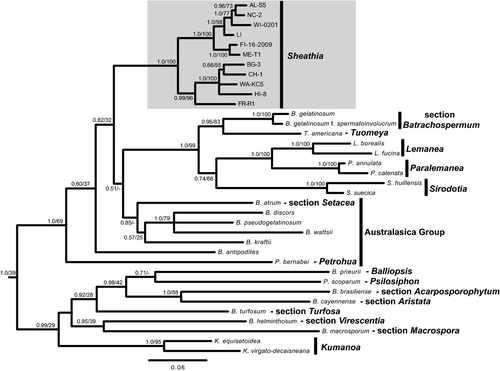
Eleven specimens with divergent rbcL sequences within Batrachospermum section Helminthoidea were combined with 25 sequences representing the known diversity of the Batrachospermales and three sequences from closely related orders as outgroups to examine the monophyly of the section. Phylogenetic analyses of this data set using both BI and ML produced similar topologies and only the BI tree is shown (Fig. 3). There was strong support (1.0 posterior probability [pp], 100% bootstrap) for a monophyletic section Helminthoidea within the paraphyletic genus Batrachospermum (Fig. 3). Therefore, a new genus, Sheathia, is proposed (see taxonomic proposals), and this clade will be referred to as the new genus throughout the remainder of results and discussion.
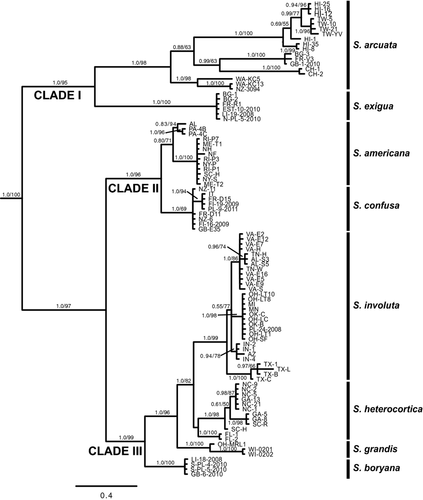
Sixty-six new rbcL sequences (1,282 bp) were combined with 36 sequences available on GenBank (Table S1). The sequence (DQ523252) attributed to the species B. antipodites Entwisle was excluded from the analysis because the identification could not be verified, and this species has been shown to belong to another Batrachospermum clade (Entwisle et al. 2009). Alignment of the sequences had no gaps as expected for the protein encoding rbcL gene. Phylogenetic analyses of three outgroup taxa and 95 Sheathia specimens using both BI and ML produced trees with similar topologies, such that only the BI tree is shown (Fig. 4). The genus Sheathia was divided into three main clades with strong support values (Clades I, II, and III; Fig. 4). Clade I was composed of specimens identified previously as B. arcuatum (Type locality: Hör, Skåne, Sweden) and recognized in this study as S. arcuata and S. exigua (Fig. 4). Sheathia exigua was a well-supported lineage containing specimens from Bulgaria, Estonia, France, Lithuania, and Poland. Clade II contained a lineage of specimens morphologically identified as B. confusum (S. confusa, Type locality: near Fougères, Brittany, France) from several locations in Europe and New Zealand, and a second lineage (S. americana) was composed predominately of specimens morphologically identified as B. boryanum (Type locality: near Bourg-des-Comptes, south of Rennes, France) from Maine, New Hampshire, New York, Pennsylvania, South Carolina (all USA), Newfoundland (Canada) and one specimen as B. confusum f. anatinum from Alabama (USA). In this analysis, S. confusa had high pp support, but the support values for the S. americana lineage were 0.80 pp and 69% bootstrap. Clade III contained four distinct lineages, three with high support values (Fig. 4). The first lineage, S. boryana, was comprised of specimens morphologically identified as both B. boryanum and B. confusum f. anatinum from Europe (Great Britain, Lithuania and Poland). The second, S. grandis, had specimens from Ohio and Wisconsin (USA) previously identified as B. boryanum. The third lineage, S. heterocortica, contained specimens of B. heterocorticum from the type locality (Mormon Creek, Marion County, FL, USA) and specimens previously identified as B. boryanum from North Carolina, South Carolina, and Georgia (USA). The S. heterocortica lineage did not receive strong support (0.59 pp/82% bootstrap). The last lineage, S. involuta, was geographically widespread in the USA and contained the type specimen of B. involutum (AF029143) from San Marcos, Texas (USA), B. boryanum from Virginia (USA) (GU476569), and newly collected B. boryanum specimens from Alabama, Arizona, Indiana, Michigan, Minnesota, Ohio, Oklahoma, Tennessee, Texas, and Virginia (USA) along with one specimen from Poland.
The COI data set had fewer specimens than the rbcL data set, primarily due to the lack of previously published sequences. Additionally, this region could not be amplified for five of the newly collected specimens despite numerous attempts; other new specimens were not sequenced because data had already been obtained from specimens of the same locations (VA-E2, IN-2, ME-T2; see Table S1). The only previously published sequence available was for a B. heterocorticum specimen (EU636740.1) collected at the type locality. This sequence was identical to a newly generated sequence for another specimen from the type locality collected on a different date (Table S1). There were fewer specimens in the ITS data set because there were no ITS sequences in GenBank for specimens attributable to Sheathia (database accessed May 3, 2013).
All specimens were within the same lineage in all single gene analyses, except FL-2. In both the rbcL and COI single marker analyses, FL-2 was within the S. heterocortica clade (Fig. 4 for rbcL; COI tree not shown). However, in the ITS analyses, FL-2 was on a separate branch and its inclusion in S. heterocortica was equivocal with no support for its placement with either this taxon or S. involuta (tree not shown).
The concatenated data set contained 54 specimens, for which all three regions were obtained. Other specimens included, but missing one marker, were OH-MRL1, SC-S, and RI-P3 (ITS data missing), and PL-24-2008 (COI data missing). Only one specimen from S. arcuata (TW-10) and S. exigua (BG-1) had all three genes and was included in the data set. Batrachospermum gelatinosum was used as the outgroup to root the concatenated tree without the ITS data because this region is difficult/impossible to align across distantly related taxa. The concatenated data set produced a tree with similar topology to the rbcL tree using BI and ML analyses and only the BI tree is shown (Fig. 5). Sheathia exigua and S. arcuata did not form a clade in the BI analysis; however, they were weakly supported (62% bootstrap) as sister species in the ML analysis. As in the rbcL analysis, S. americana and S. confusa were supported as sister taxa (Clade II), and the addition of the COI and ITS data to the rbcL data resulted in strong support for S. americana in the concatenated analysis. Sheathia involuta, S. heterocortica, S. grandis, and S. boryana formed a well-supported clade as in the rbcL analyses (Clade III). With the addition of the COI and ITS data, the S. heterocortica lineage received 0.92 pp support. Overall posterior probabilities support values on the concatenated tree were similar or better than those on the rbcL tree; however, the ML bootstrap values were slightly lower than those for some clades in the rbcL analysis.
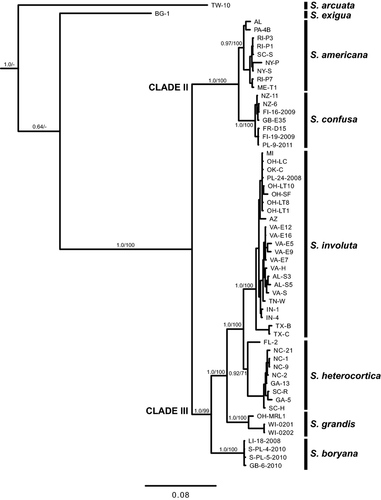
All specimens used in molecular analyses were examined for breeding system (monoecy/dioecy), heterocortication, and distinguishing qualitative characteristics. Breeding system was examined for trends corresponded to species. Although most species contained both monoecious and dioecious specimens, all species were predominately one or the other with a few exceptions (Table 1). Heterocortication was present and easily seen in all specimens in Clades II and III from the molecular analyses (Figs. 4 and 5). In addition to those specimens, one specimen from S. exigua, FR-R1, was also observed to have heterocortication. Therefore, all specimens from S. exigua (BG-1, BG-2, PL-5-2010, and LI-19-2008) were examined, and these also had heterocortication, but only on the thickest axis of the thallus near the attachment point to the substratum. There was only a single specimen from S. exigua without heterocortication (EST), but it was very small (~1 cm in length) and without a large axis. In addition, B. arcuatum specimens from Vis et al. (2010) and this study, with vouchers present at BHO, were reexamined and found to be lacking heterocortication (BG-3, FR-V3, GB-1-2010). The character of spermatangia on the involucral filaments of the carpogonial branch has been previously used to distinguish S. confusa (as B. confusum). All specimens of S. confusa included in the molecular analyses were examined and confirmed to have this trait, including a specimen identified as B. confusum f. anatinum (BHO A-0004). Although specimens were examined in detail, no other qualitative characteristics were observed that might distinguish the species.
Statistical analysis using ANOVA was conducted on quantitative morphometric characters to determine traits that could be used for distinguishing between clades (Table 1). Since S. arcuata has the distinct characteristic of regular cortication and voucher specimens were not available, it was not considered in the morphometric analysis. Both S. grandis and S. americana had significantly larger whorl diameters (P < 0.05) than S. heterocortica. Likewise, the number of cells per fascicle in S. grandis was significantly greater (P < 0.05) than those of S. heterocortica. There were no other significant differences between species based on whorl diameter or fascicle cell number. Comparisons of carposporophyte diameter showed that carposporophytes in S. grandis were significantly larger (P < 0.01) than those of S. involuta, S. americana, and S. confusa, but did not differ from S. heterocortica or S. exigua. Additionally, S. heterocortica had a significantly larger (P < 0.05) carposporophyte than S. confusa. There were no significant differences among species based on the carpogonium diameter. However, the carpogonium length of S. grandis was significantly longer (P < 0.01) than all other species except S. exigua. In addition, S. confusa had a significantly shorter (P < 0.05) carpogonium length than S. exigua, but did not significantly differ from all other species. The number of cells in the carpogonial branch of S. heterocortica was significantly fewer (P < 0.01) than those of S. grandis. There were no other significant differences between species based on carpogonial branch length. The larger carposporangial diameters of S. grandis and S. exigua were significantly different (P < 0.001) from other species of Sheathia, but they did not significantly differ from each other. Additionally, the carposporangium diameter of S. americana was significantly smaller (P < 0.05) than all other species of Sheathia except S. confusa. The carposporangium length of S. grandis was significantly longer (P < 0.05) than those of the other Sheathia species examined. The carposporangium length of S. exigua was significantly longer (P < 0.05) than those of S. boryana, S. involuta, S. americana, and S. confusa. The carposporangium length of S. confusa was significantly shorter (P < 0.05) than those of S. heterocortica, S. involuta, S. grandis, and S. exigua.
Efforts to obtain sequence data from type and historically important specimens were unsuccessful (Table S4 in the Supporting Information). Numerous attempts were made to amplify fragments of the three molecular markers used in this study. For most specimens, the results were no PCR products. In a few cases, PCR product was obtained but sequencing revealed a contaminant. Lastly, sequence data from three specimens were acquired, but these data could not be verified, as the results were not repeatable.
Since molecular data were unavailable for most type specimens, morphological data were utilized to link species epithets with the molecular clades. Type specimens of species with heterocortication were compared to the morphometric data from this study (Table 1). Most morphometric measurements for the type specimen of B. involutum were comparable to specimens from this study in the S. involuta clade. However, the upper values for carpogonium length and carposporangium length were slightly greater in the type specimen (Table 1). Measurements of the type specimen of B. heterocorticum were similar to S. heterocortica specimens from this study, except that the maximum carposporangium diameter was slightly greater in the type specimen. Measurements from the type specimens of B. anatinum and B. boryanum were mostly within the range of measurements of S. boryana specimens from this study. These types did differ from the specimens in this study in the following: greater maximum carpogonium diameter for B. anatinum and B. boryanum types, and greater maximum carposporangium length and diameter for the B. anatinum and B. boryanum types. All morphological measurements of the type specimen of B. confusum were within the range of measurements for specimens identified as S. confusa from this study.
Taxonomic proposals Sheathia Salomaki et M.L. Vis gen. nov.
Type species: Sheathia boryana (Sirodot) Salomaki et M.L. Vis. Heterotypic synonym: Batrachospermum section Helminthoidea Sirodot ex De Toni, Syll. Alg. 4(1): 55 (1897).
Diagnosis: Gametophytes similar in gross morphology to Batrachospermum, differing by cortication of main thallus axis composed of both bulbous and cylindrical cells (heterocortication) in all species, except S. arcuata. Segregated from Batrachospermum by rbcL, COI, and ITS sequences.
Comments: Carpogonium production per whorl is abundant, and carpogonial branches are composed of cells undifferentiated from fascicle cells and typically arise from fascicle cells, but carpogonia may be sessile on periaxial cells. Carposporophytes are spherical within or exerted from whorls and typically are much smaller than whorls. However, there is significant overlap in these morphological traits with Batrachospermum section Batrachospermum, and Sheathia cannot be distinguished from this section based on these characteristics. To date, only heterocortication has been found to be unique to this genus.
Etymology: This genus is named in honor of Dr. Robert G. Sheath, who has conducted extensive research on many of the species in this genus as well as other freshwater red algae.
Species of Sheathia: The following species are here transferred from Batrachospermum to Sheathia. De Toni (1897) originally placed four species in Batrachospermum section Helminthoidea, but B. crouanianum is a heterotypic synonym of B. confusum, and B. helminthosum Sirodot is a nom. illeg. Batrachospermum boryanum is here transferred to Sheathia and B. anatinum is here considered a synonym of S. boryana. In addition, taxa that have previously been shown to belong to Batrachospermum section Helminthoidea in molecular analyses (Entwisle et al. 2009, present study), or have been reported to have the characteristic of heterocortication (Vis et al. 1995, Vis and Sheath 1996) are here transferred.
Sheathia arcuata (Kylin) Salomaki et M.L. Vis comb. nov.
Basionym: Batrachospermum arcuatum Kylin (1912: 22, fig. 7, a–e).
Type Locality: Hör, Skåne, Sweden (Kylin 1912).
Sheathia boryana (Sirodot) Salomaki et M.L. Vis comb. nov. Basionym: Batrachospermum boryanum Sirodot (1874: 1366).
Type Locality: Lectotype - Caniveau de la Trottinais, near Bourg-des-Comptes, south of Rennes, France (Vis et al. 1995).
Heterotypic synonyms: Batrachospermum anatinum Sirodot (1884: 249, pl. 32, figs. 1–7, pl. 33, figs. 1–5) emend. M.L. Vis, Sheath et Entwisle (1995: 54) (Type Locality: Lectotype – Ruisseau de Vau-deMeu, au patis Saint-Lazare, Montfort, France, 10 iv 1872); Batrachospermum confusum forma anatinum (Sirodot) S.A. Stewart et M.L. Vis (2007b: 743).
Representative DNA Barcode: GenBank JX669707
Sheathia carpoinvolucra (Sheath et M. L. Vis) Salomaki et M.L. Vis comb. nov.
Basionym: Batrachospermum carpoinvolucrum Sheath et M.L. Vis (Vis and Sheath 1996: 128, figs. 16–24).
Type Locality: Montezuma Well outflow canal, Arizona, USA (Vis and Sheath 1996).
Sheathia confusa (Bory) Salomaki et M.L. Vis comb. nov.
Basionym: Batrachospermum ludibundum var. confusum Bory de Saint-Vincent (1808: 320), pl. 29 fig. 3.
Homotypic synonym: Batrachospermum confusum (Bory) Hassall (1845: 105).
Type Locality: near Fougères, Brittany, France (Bory de Saint-Vincent 1808).
Representative DNA Barcode: GenBank JX669682
Sheathia fluitans (Kerner) Salomaki et M.L. Vis comb. nov.
Basionym: Batrachospermum fluitans Kerner (1882: 134), Flora Exsiccata Austro-Hungarica No. 397).
Type Locality: near Mühlau, Innsbruck, Austria (Kerner 1882).
Sheathia heterocortica (Sheath et K.M. Cole) Salomaki et M.L. Vis comb. nov.
Basionym: Batrachospermum heterocorticum Sheath et K.M. Cole (1990: 566, figs. 9–18).
Type Locality: Mormon Creek, Marion County, Florida, USA (Sheath and Cole 1990).
Representative DNA Barcode: GenBank JX669718
Sheathia involuta (M.L. Vis et Sheath). Salomaki et M.L. Vis comb. nov.
Basionym: Batrachospermum involutum M.L. Vis et Sheath (1996: 128, figs. 25–34).
Type Locality: San Marcos, Texas, USA (Vis and Sheath 1996).
Representative DNA Barcode: GenBank JX669717
Emended description: Thalli monoecious or dioecious with spherical- to barrel-shaped whorls. Mature whorls ranged from 245 to 2,635 μm in diameter. Thallus axis of bulbous and cylindrical cells (heterocortication). Spermatangia terminal on fascicles. Gonimoblast filaments of 2–4 cylindrical cells with terminal obovoid carposporangia 7.8–17.5 μm diameter and 9–18 μm long. Carpogonia with clavate trichogynes 6.3–16.4 μm diameter and 17.4–54 μm long. Carpogonium-bearing branch undifferentiated from fascicle cells, 6–19 cells long. Female whorls contain one to many peripheral or exerted carposporophytes. Carposporophytes spherical, pedicellate, 58–254 μm in diameter. Representative unique molecular sequences have been deposited to GenBank: rbcL JX669786; COI JX669717; ITS JX669658. GenBank accession numbers for additional rbcL, COI, and ITS sequences attributed to S. involuta are available in Table S1.
Comments: Some of the characteristics in the original description of this taxon appear to be environmentally induced or at least not present in specimens subsequently collected from the type locality. The characteristics of paired fascicle tips turned inward toward each other (Vis and Sheath 1996: fig. 26), and rhizoidal outgrowths from mid-fascicle cells (Vis and Sheath 1996: figs. 27, 28) are not present on all specimens that have identical DNA sequences for the rbcL, COI barcode, and ITS regions. In addition, none of the specimens examined in this study showed spermatangia on the involucral filaments and it is unclear from the protologue (Vis and Sheath 1996: fig. 33) that this is a valid characteristic for this species.
The following are also assigned to the new genus Sheathia:
Sheathia exigua Salomaki et M.L. Vis sp. nov. (Fig. 6, a–f)
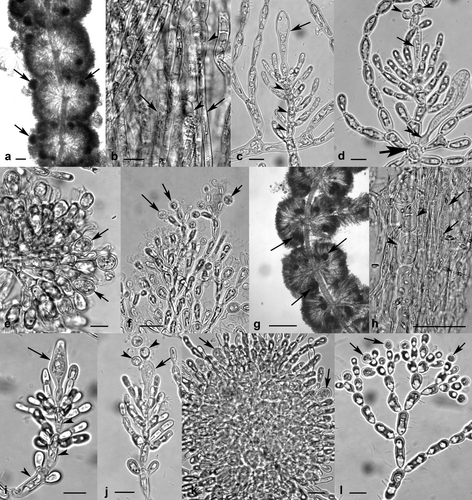
Thalli dioecious with spherical- to barrel-shaped whorls (Fig. 6a). Mature whorls 301–1,236 μm diameter. Thickest thallus axis of bulbous and cylindrical cells (heterocortication; Fig. 6b) and of cylindrical cells (regular cortication) on thinner axes. On thalli <1 cm long only regular cortication is present. Spermatangia terminal on fascicles of male thalli (Fig. 6f). Gonimoblast filaments 2–4 cylindrical cells long with terminal spherical to obovoid carposporangia 7–14 μm diameter and 11–18 μm long (Fig. 6e). Carpogonia with clavate trichogynes 5.5–12.2 μm diameter and 22.1–49.4 μm long (Fig. 6, c and d). Carpogonium-bearing branches undifferentiated from fascicle cells, 5–13 cells long (Fig. 6c), occasionally a carpogonium arising directly from pericentral cell (Fig. 6d). Female whorls contain one to many peripheral or exerted carposporophytes (Fig. 6a). Carposporophytes spherical, pedicellate, 66–187 μm diameter. Representative unique molecular sequences deposited to GenBank: rbcL GU457345; COI JX669616; ITS JX669623. GenBank accession numbers for additional rbcL, COI, and ITS sequences attributed to S. exigua available in Table S1.
Type designated herein: Holotype: France; near Corseul, stream flowing in meadow adjacent to Chateau de Montafilan ruins parking lot, 48.487444° N, 2.190694° W. Coll. W. B. Chiasson & E. D. Salomaki 6.x.2011 (Holotype NY – Voucher Barcode 01840460, DNA Aliquot Barcode 1295193; Isotypes BHO A-0905 MICH).
Representative DNA Barcode: GenBank JX669616
Etymology: The species epithet meaning “slight” was chosen because the distinguishing characteristic of the genus, heterocortication can be difficult to detect and present only slightly.
Sheathia grandis Salomaki et M.L. Vis sp. nov. (Fig. 7, a–g)
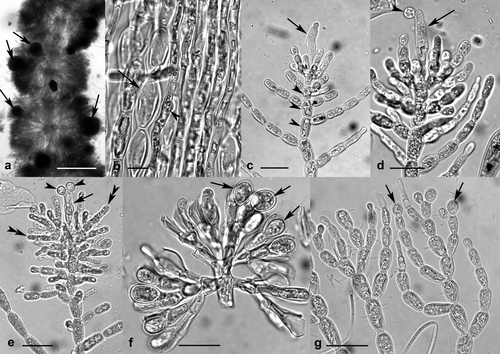
Thalli monoecious or dioecious with spherical- to barrel-shaped whorls (Fig. 7a). Mature whorls 291–2,105 μm in diameter. Thallus axis of bulbous and cylindrical cells (heterocortication; Fig. 7b). Spermatangia terminal on fascicles (Fig. 7g). Gonimoblast filaments 2–4 cylindrical cells long with terminal obovoid carposporangia 7.8–17.5 μm diameter and 11–24 μm long (Fig. 7f). Carpogonia with clavate trichogynes 6.1–12.8 μm diameter and 30.6–80.9 μm long (Fig. 7, c–e). Carpogonium-bearing branch undifferentiated from fascicle cells, 5–17 cells long (Fig. 7c). Female whorls of one to many peripheral or exerted carposporophytes (Fig. 7a). Carposporophytes spherical, pedicellate, 73–240 μm diameter. Representative unique molecular sequences deposited to GenBank: rbcL JX669762; COI JX669700; ITS JX669676. GenBank accession numbers for additional rbcL, COI, and ITS sequences attributed to S. grandis available in Table S1.
Type designated herein: Holotype: USA; Wisconsin, Racine County, 8 km south of Big Bend, Tichigan Creek at Ranke Rd crossing – 100 m east of parking lot, 42.792222° N, 88. 230278° W (estimated) 6.x.2008. Coll. P.A. Schwartz (Holotype NY – Voucher Barcode 01840459, DNA Aliquot Barcode 1295192; Isotypes BHO A-0092, MICH).
Representative DNA Barcode: GenBank JX669700
Etymology: The species epithet meaning “large” was chosen because the specimens have a larger whorl diameter, carpogonium length, and carposporangium length than other species examined in this study.
Sheathia americana Salomaki et M.L. Vis sp. nov. (Fig. 6, g–l)
Thalli dioecious with spherical- to barrel-shaped whorls (Fig. 6g). Mature whorls 178–2,172 μm diameter. Thallus axis of bulbous and cylindrical cells (heterocortication; Fig. 6h). Spermatangia terminal on fascicles of male plants (Fig. 6l). Female whorls with one to many peripheral or exerted carposporophytes (Fig. 6g). Carposporophytes spherical, pedicellate, 40–218 μm diameter. Gonimoblast filaments 2–4 cylindrical cells long with terminal spherical to obovoid carposporangia 5–10 μm diameter and 7–15 μm long (Fig. 6k). Carpogonia with clavate trichogynes 5.4–9.8 μm diameter and 16.8–43.9 μm long (Fig. 6, i and j). Carpogonium-bearing branch undifferentiated from fascicle cells, 4–15 cells long. Representative unique molecular sequences deposited to GenBank: rbcL JX669759; COI JX669697; ITS JX669637. GenBank accession numbers for additional rbcL, COI, and ITS sequences attributed to S. americana available in Table S1.
Type designated herein: Holotype: USA; Maine, Hancock County, East Hancock, Tunk Mountain, 44.64472° N, 68.10472° W (estimated) Coll. M.L. Vis 16.vi.1997 (Holotype NY – Voucher Barcode 01840458, DNA Aliquot Barcode 1295191; Isotypes BHO A-1110, MICH).
Representative DNA Barcode: GenBank JX669697
Etymology: The species epithet was chosen because this species appears to be present only in North America.
Discussion
Morphological separation of Batrachospermum section Helminthoidea from Batrachospermum section Batrachospermum, which includes B. gelatinosum, the type specimen for the genus, has historically proven difficult and remains difficult based on morphology alone (Necchi and Entwisle 1990, Entwisle et al. 2009). However, the application of DNA sequences has clearly demonstrated that the two sections are distantly related and that Batrachospermum is polyphyletic and in need of taxonomic revision (Vis et al. 1998, Entwisle et al. 2009). The rbcL phylogeny for the Batrachospermales provides strong support for the new genus Sheathia, as a distinct clade from B. gelatinosum, regardless of the morphological similarities. It is unfortunate that a definitive synapomorphy has yet to be found across all species of Sheathia. However, the presence of heterocortication is unique to the genus and appears to have been lost in S. arcuata. It is clear that in the Batrachospermales there is much morphological plasticity and great potential for cryptic diversity, such that combining molecular data with morphological characters, when possible, has enabled a revision that accurately reflects the evolutionary history within the order.
Morphological identification of species in Batrachospermum section Helminthoidea (genus Sheathia), which has been based primarily on presence/absence of heterocortication, breeding system, and subjective qualitative characteristics, resulted in the recognition of a few broadly, defined taxa. The rbcL gene has been the standard marker for species delimitation in the Batrachospermales (Vis et al. 1998, Vis and Entwisle 2000, Entwisle et al. 2009), and over the past several years, COI has been increasingly verified as a reliable DNA barcode for Rhodophytes (Le Gall and Saunders 2010, Clarkston and Saunders 2012). Previous work has shown the highly variable ITS region to be informative in some studies, though difficult and occasionally impossible to align in Rhodophyta (Hu et al. 2009, Vis et al. 2012). In Sheathia, the effort to sequence the ITS region proved to be quite laborious, though its inclusion, along with the COI, in the concatenated data set provided greater nodal support for many of the proposed species. The application of three species-level molecular markers in combination with detailed morphological analysis on a large number of specimens has provided fine-scale resolution of the existing diversity. These results indicate that there are seven well-defined heterocorticate species based on molecular data and at least one non-heterocorticate species within Sheathia.
In addition to elucidating species diversity in Sheathia, the molecular analyses provided the insight that heterocortication is an underlying synapomorphy for the genus. Morphological investigation of a clade of specimens previously identified as S. arcuata (Vis et al. 2010) revealed that, although not present throughout the specimen, heterocortication was found on the thickest axis. The rbcL phylogenetic analysis provided strong support for this clade (S. exigua) being basal to S. arcuata. From this result, it is inferred that heterocortication was shared by a common ancestor of taxa in Sheathia and has since been lost by S. arcuata.
It is preferable to use both morphological and molecular data to assign species epithets to molecular clades. However, molecular data from type specimens or locations were only available for a few recently described species. Although type localities for several species were visited, no specimens could be found. Likewise, several attempts to sequence material from type specimens were unsuccessful. As a result, species epithets have been assigned to molecular clades by a combination of morphological analysis of type and contemporary specimens, use of historical notes, and assessment of geographic distributions. The inability to get sequence data from type specimens was a source of frustration in this study, but also has been problematic in general for rhodophytes (Saunders and McDevit 2012). Nevertheless, other researchers have been successful in obtaining reliable sequence data (Hughey and Gabrielson 2012). Therefore, it may be possible in the future to obtain trustworthy data for older specimens of Sheathia and other batrachospermalean taxa.
Sheathia confusa is distinguished by the presence of heterocortication and spermatangia on the involucral filaments of the carpogonial branch. In this study, we reexamined specimens from New Zealand that were originally identified as B. anatinum and that led to the placement of B. anatinum in synonymy with B. confusum (Stewart and Vis 2007a). Here, we morphologically identified those specimens as S. confusa. Although sparse, spermatangia were observed on the involucral filaments of some carpogonial branches of all specimens examined. Therefore, we do not consider B. anatinum to be conspecific with S. confusa.
Batrachospermum boryanum was first described from France for specimens that were dioecious, with relatively large carposporophytes, and with ovoid and often irregular or bumpy trichogynes and was distinguished from B. anatinum (Type locality: Saint-Lazare, Montfort, France) by breeding system (Sirodot 1884). Additionally, Sirodot noted that the dioecious B. boryanum was much less common from his collections in France than the monoecious B. anatinum, which he described in his 1884 monograph. However, contemporary collections throughout Europe have yielded more dioecious than monoecious specimens. In this study, most molecular clades of Sheathia contain specimens that are both monoecious and dioecious and therefore, breeding system cannot be considered as a taxonomically informative characteristic for species in this genus. Sequence data provided strong support for a clade containing both monoecious and dioecious specimens distributed throughout Europe. Since there were no other consistent distinguishing morphological characters separating the type specimens and the modern collections, we propose that B. anatinum be placed in synonymy with S. boryana.
Sheathia involuta is a widespread North American species that also includes specimens found at a single watershed in Poland. Molecular data suggest that this species probably has a more variable morphology than provided in the protologue. For example, the distinguishing characters of dichotomous involute fascicle tips and rhizoidal growths from mid-fascicle cells (Vis and Sheath 1996) were not present on any of the specimens in this study including a topotype specimen. We conclude that these morphological characters are most likely environmentally induced.
The S. heterocortica clade included sequence data from the type locality of B. heterocorticum as well as other locations in southeastern United States. The type locality specimen was on a separate branch in the ITS phylogeny (data not shown), but in all other analyses it was within the clade (high pp support in concatenated analysis). This result raises some questions about attributing all the specimens to a single species. Nevertheless, when considering all lines of evidence, the molecular data and morphological similarities, it is recommended that the morphological circumscription and geographic range of S. heterocortica (previously known only from the type locality) be expanded to include specimens from Georgia, North Carolina, and South Carolina. Future studies with greater sampling, especially from isolated springs, like the type locality of this species, may provide additional support for further species delineation of specimens from the southeastern United States.
There were more clades of heterocorticate specimens from the molecular analyses than could be accommodated by previously recognized species epithets. Therefore, three new species are proposed, S. grandis, S. americana, and S. exigua. Sheathia grandis is distinguished by large whorl diameter in combination with longer and wider carpogonia and carposporangia than other heterocorticate taxa. Unlike most of the other species examined, this taxon had equal numbers of monoecious and dioecious specimens supporting the assertion of other researchers that breeding system alone cannot be reliably used for species identification (Entwisle et al. 2004, Ji et al. 2011). Sheathia americana is morphologically very similar to S. boryana, S. involuta, and S. heterocortica and unfortunately does not appear to have a discrete morphological characteristic that enables this species to be distinguished from others except by DNA sequences. In addition, the geographic ranges overlap such that collection location may not be used to provide identification.
In the literature, there are taxa that may be attributed to Sheathia based on the presence of heterocortication. One such species is B. carpoinvolucrum, known only from the outflow canal at Montezuma Well, Arizona (USA). In addition to having heterocortication, the important distinguishing character of B. carpoinvolucrum is carpogonia terminal on involucral filaments of the carpogonial branch. This “diagnostic” trait has been observed in specimens from multiple species of Sheathia and is potentially environmentally induced rather than a taxonomically informative. In this study, specimens were collected from the type locality and the sequence data were within the S. involuta clade. Unfortunately, the important diagnostic character of carpogonia on the involucral filaments was not observed such that this specimen could not unequivocally be assigned to B. carpoinvolucrum. Therefore, for the time being, this taxon is transferred to Sheathia as a separate species pending further molecular investigation of specimens from the type location matching the morphological description. A second, currently recognized, heterocorticate species that is attributed to Sheathia is B. fluitans. This species, known only from Europe, is monoecious, but distinguished by long carpogonia (52–65 μm) (Vis et al. 1995). No collections could be obtained from near the type location in Austria, nor were any European specimens in this study observed to have such long carpogonia. Therefore, this taxon is transferred to Sheathia, but contemporary collections are needed to determine if B. fluitans is a distinct species, is conspecific with S. boryana specimens with longer carpogonia, or is possibly a disjunct population of S. grandis.
Biogeographic ranges for the heterocorticate taxa in Sheathia show some species to be widespread and others to have more narrow geographic ranges. Sheathia confusa has a disjunct distribution, having been collected in numerous locations in Europe and New Zealand. Undoubtedly, a long-range dispersal event has occurred, but the vector is unknown. This species is not the only taxon within Sheathia with such a disjunct distribution; S. arcuata has closely related specimens (identical cox2-3 spacer and rbcL sequence) from the west coast of the United States and New Zealand (Vis et al. 2010). Likewise, S. involuta appears to be widespread in North America from Virginia to Texas and with a single watershed in Poland. It is interesting that this taxon is so prevalent in North America, but appears to be rare in European rivers. Perhaps, with a greater sampling effort, a more widespread distribution will become evident on that continent. Both S. exigua and S. boryana were collected only from Europe. These taxa and S. confusa overlap in distribution on that continent. Three taxa, S. americana, S. heterocortica, and S. grandis were only from locations in North America. The distribution of S. americana was centered in New England, S. heterocortica in the southeastern United States, and S. grandis from locations in the Midwest United States, making them somewhat distinct from each other in their ranges. However, the widespread S. involuta overlapped in range with all these species.
Molecular sequence data from this study suggest greater diversity of non-heterocorticate Sheathia than is recognized by the single species name S. arcuata. By examining the intergenic spacer region between the mitochondrial cytochrome oxidase 2 and cytochrome oxidase 3 genes, Vis et al. (2010) found 11 unique haplotypes of B. arcuatum specimens with >14% variable bases between haplotypes. Additionally, Vis et al. (2010) revealed >6% variation in rbcL sequences between specimens; however, little morphological variation was evident. Resolving this diversity will be complicated, as many of the specimens that have published molecular data do not have corresponding morphological vouchers or have been collected only in the taxonomically uninformative Chantransia stage. Future studies with increased sampling efforts targeted at these non-heterocorticate specimens are likely to result in further expansion of the species diversity in Sheathia.
Matt White and Harvey Ballard are thanked for critical reading of an earlier version of this manuscript and comments by Mike Wynne and Daryl Lam greatly improved the manuscript. Mike Wynne and Craig Schneider provided valuable assistance with Latin names and the International Code of Nomenclature. We thank Line LeGall and Bruno de Riviers for providing access to materials from the herbarium at the Muséum National d'Histoire Naturelle in Paris, France (PC) and Ken Karol for access to materials from the herbarium at the New York Botanical Garden (NY) along with advice on molecular methods. Sam Drerup helped with statistical analyses of morphometric data in R. We gratefully acknowledge the individuals listed in Table S1 for collection of specimens. We thank the staff of the Ohio University Genomics Facility for help in primer design and DNA sequencing. Emily Keil is gratefully acknowledged for extraction and PCR of samples from 2012 in Table S5. Funding for portions of the research was provided by the grant no. N N304 285937 from the Polish Ministry of Sciences and Higher Education, NSF DEB 0936855, the Ohio University Graduate Student Senate, and the Phycological Society of America.




