Differentiation and Classification of Phytoplasmas Associated with Almond and GF-677 Witches'-Broom Diseases using rRNA and rp Genes
Abstract
Stone fruits are affected by several diseases associated with plant pathogenic phytoplasmas. Previous studies have been shown that phytoplasma agents of almond and GF-677 witches'-broom (AlmWB and GWB, respectively) diseases belong to pigeon pea witches'-broom (16SrIX) phytoplasma group. In this study, partial biological and molecular characterization was used to compare and classify phytoplasma agents of Khafr AlmWB (KAlmWB) and Estahban GWB (EGWB) diseases. Production of different symptoms in periwinkle indicated that agents of KAlmWB and EGWB are differentiable. Expected fragments were amplified from diseased almond and GF-677 trees in direct PCR using phytoplasma universal primer pairs P1/P7 and rpF1/rpR1 and nested PCR using P1/P7 followed by R16F2n/ R16R2 primer pair. 16S-rDNA Restriction fragment length polymorphism (RFLP) as well as phylogenetic analysis of rplV-rpsC and 16S–23S rRNA spacer region sequences classified KAlmWB and EGWB phytoplasmas within 16SrIX-C (rpIX-C) and 16SrIX-B (rpIX-B) subgroups, respectively.
Introduction
Phytoplasmas, previously referred to as mycoplasma-like organisms (MLOs) are plant-pathogenic mollicutes associated with diseases in several hundred plant species. They are non-culturable, phloem-limited and insect-transmissible bacterial pathogens causing diseases characterized by flower malformation, growth aberrations, yellowing and/or decline (Doi et al. 1967; McCoy et al. 1989; Weisburg et al. 1989; Seemüller et al. 1998; Lee et al. 2000).
Several molecular markers have been employed for differentiation and classification of phytoplasmas. The 16S rRNA gene is the most widely used marker in the phytoplasma research and proves to be very useful in preliminary classification of these pathogens. For finer differentiation and classification of phytoplasmas, additional phylogenetic markers such as ribosomal protein (rp), secY and tuf genes and the 16S–23S rRNA, intergenic spacer region (SR) sequences have been used as supplementary tools (Martini et al. 2007). As there are fewer evolutionary constraints on SR portion of the rRNA operon, there is generally greater variation in the SR sequence than in that of the 16S rRNA gene (Barry et al. 1991). The 16S rRNA plus SR also improved the separation of closely related strains that could not be well resolved by use of the 16S rRNA gene alone (Langer and Maixner 2004). Ribosomal protein (rp) genes are more variable than 16S rRNA gene and have more phylogenetically informative characters, which substantially enhance the resolving power in delineating distinct phytoplasma strains (Martini et al. 2002).
Stone fruits are affected by several diseases associated with plant pathogenic phytoplasmas (Abou-Jawdah et al. 2002). Witches'-broom disease of almond (AlmWB) caused by a pigeon pea witches'-broom (16SrIX)-related phytoplasma (‘Candidatus Phytoplasma phoenicium’) is an economically important disease in Iran (Salehi et al. 2006) and Lebanon (Verdin et al. 2003). Grafting experiments have been revealed that AlmWB may also affect peach, nectarine, apricot, plum and sour cherry (Abbasian and Salehi 2010).
Witches'-broom of GF-677 (GWB) trees is also reported as an important disease from Beedzard and Estahban areas in Fars province of Iran (Salehi et al. 2011). Previous studies have been shown that phytoplasmas associated with AlmWB and GWB are members of 16SrIX phytoplasma group (Salehi et al. 2006, 2011). For quarantine purposes, it was necessary to know whether AlmWB and GWB phytoplasmas belong to the same subgroup or different subgroups in 16SrIX phytoplasma group.
In this study, phytoplasmal agents of almond witches'-broom from Khafr (KAlmWB) and GF-677 witches'-broom from Estahban (EGWB) were compared using partial biological and molecular characteristics.
Materials and Methods
Dodder and graft transmission
Healthy seed-grown periwinkle plants were separately dodder (Cuscuta campestris Yank.)-inoculated with KAlmWB and EGWB agents, as reported earlier (Salehi et al. 2006).
In another experiment, young periwinkle of same age were side-grafted with scions (small branches with two to three leaves) taken from periwinkle plants previously infected with KAlmWB and EGWB agents via dodder inoculation.
DNA extraction
Total DNA was extracted using 0.3 g fresh midribs tissue from witches'-broom-affected almond and GF-677 trees and periwinkle plants infected with KAlmWB and EGWB disease agents according to the method described by Zhang et al. (1998). Total DNA extracted from a healthy seed-grown periwinkle plant and a periwinkle plant infected with lime witches'-broom phytoplasma was used as negative and positive controls, respectively.
PCR amplification
The phytoplasma universal primer pair P1/P7 (Deng and Hiruki 1991; Schneider et al. 1995) was used to amplify 1800 bp of rDNA operon from the 16S rRNA gene to the beginning of the 23 rRNA gene including internal spacer region (SR). Nested PCR assay was performed on P1/P7 primed PCR products diluted at 1:30 with sterilized water using R16F2n/R16R2 primer pair (Gundersen and Lee 1996) to amplify a 1250-bp fragment of 16S rRNA gene.
The rpF1/rpR1 primer pair (Lim and Sears 1992) was used to amplify approximately 1250 bp of the rp operon including rpl22 and rps3 genes. PCR was performed in 50 μl reaction mixture containing 100 ng total DNA, 0.4 μm of each primer, 0.2 mm of dNTP mix, 1.25 U Taq DNA polymerase (Cinnagen, Tehran, Iran) and 1X PCR buffer.
For PCR amplification, 35 cycles were carried out in a Bio-Rad (USA) thermal cycler with heated lid using the following thermal profile: 60s (120s in the first round) at 94°C, 120s at 55°C (42°C for rp amplification) and 180s at 72°C (10 min for the final cycle).
Six microlitres of each PCR product was electrophoresed through 1% agarose gel in 1X TBE buffer (67 mm Tris–HCl, 22 mm boric acid, 10 mm EDTA, pH 0.8). PCR products in gel were stained with ethidium bromide and visualized with a UV transilluminator.
Restriction fragment length polymorphism analysis
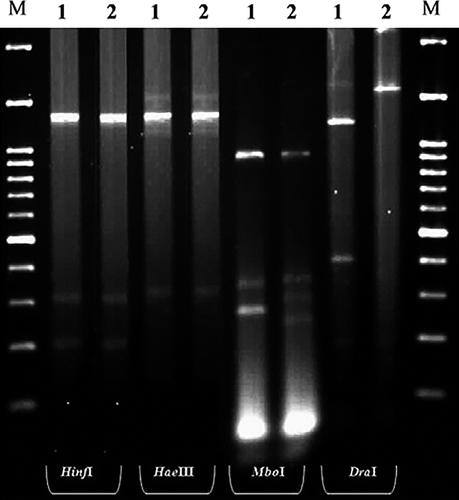
The amplicons obtained with the P1/P7 (1800 bp) primer pair were separately digested with restriction enzymes HinfI, DraI, MboI and HaeIII (Fermentas, Vilnius, Lithuania) following the manufacturer's instructions. Digested products were analysed using electrophoresis through 2.5% agarose gel, stained and visualized as described above.
Sequencing and phylogenetic analysis
The P1/P7 and rpF1/rpR1 primed PCR products were separately ligated into PTZ57R/T vector and cloned into Echerichia coli DH5α cells using InsT/A clone™ PCR Products Cloning Kit (Fermentas) according to manufacturer's instructions. Plasmid DNA from cultures of recombinant colonies was purified using High Pure Isolation Kit (Roche, Mannheim, Germany). Sequencing of both strands was performed by Macrogene (Seoul, South Korea). BLAST searches were performed at the NCBI website (http:// www.ncbi.nlm.nih.gove/) to determine the closest phytoplasma relatives of each isolate.
Virtual RFLP analysis
Virtual Restriction fragment length polymorphism (RFLP) was conducted using AluI, BamHI, BfaI, BstUI (ThaI) DraI, EcoRI, HaeIII, HhaI, HinfI, HpaI, HpaII, KpnI, Sau3AI (MboI), MseI, RsaI, SspI and TaqI restriction enzymes on the sequences obtained with R16F2n/R16R2 primer pair in iphyclasifier program (Zhao et al. 2009).
Phylogenetic analysis
Sequences of SR (~250 bp) and rp (~1200 bp containing rplV and rpsC genes) of KAlmWB and EGWB isolates were trimmed from respective amplicons obtained in this study or from the GenBank databases (accession numbers in phylogenetic trees) and used for phylogenetic analysis. Acholeplasma laidlawii was used as out-group to root the tree. Sequences were aligned with CLUSTALW from the Molecular Evolutionary Genetics Analysis program – MEGA5 (Tamura et al. 2011). Phylogenetic trees were constructed using the neighbour-joining plot option of MEGA5 software. DNAMAN software, version 4.02 (Lynnon Biosoft, Quebec, Canada) was used to generate sequence homologies.
Results
Dodder and graft transmission
Agents of KAlmWB and EGWB diseases each were successfully transmitted from affected bitter almond and GF-677 seedlings to at least three healthy plants of periwinkle via dodder inoculation and from periwinkle to 10 young periwinkle of same age by grafting. Typical symptoms of KAlmWB agent in periwinkle were severe yellowing, little leaf, internode shortening and stunting and those of EGWB agent were virescence, phyllody, mild yellowing, little leaf and internode shortening (Fig. 1).

PCR amplification
In direct PCR using P1/P7 and rpF1/rpR1 primer pairs, expected fragments (1800, and 1254-1386, respectively) were amplified from affected almond (2/10) and GF-677 (5/10) trees, all periwinkle plants graft and dodder inoculated with KAlmWB and EGWB phytoplasmas and positive control (not shown). In nested PCR using P1/P7 followed by R16F2n/R2 primer pair, approximately 1250 bp of 16Sr RNA gene from all naturally witches'-broom-affected KAlmWB and EGWB trees and all dodder- and graft-inoculated periwinkle plants were amplified (not shown). No amplification occurred with collections of asymptomatic almond, GF-677 and periwinkle plants.
Actual RFLP analysis
The RFLP profiles were generated by restriction enzymes for the P1/P7 primed products (1800 bp) of KAlmWB and EGWB isolates (Fig. 2). In RFLP profiles, KAlmWB and EGWB isolates were differentiable on the basis of DraI and MboI enzymes. Collective RFLP patterns of KAlmWB and EGWB phytoplasmas were similar to those of pigeon pea witches'-broom, 16SrIX phytoplasmas group (Lee et al. 1998).
Nucleotide sequence
Spacer region and rp nucleotide sequences of KAlmWB and EGWB were determined and submitted to GenBank databases. BLAST search showed that SR sequences of KAlmWB (KP297391) and EGWB (KP297392) phytoplasma isolates had higher than 97.5% homology with 16SrIX phytoplasma group. This analysis revealed that among 16SrIX group phytoplasmas, KAlmWB and EGWB had closest homology with Brassica rapa phyllody phytoplasma from India (16SrIX-C, GU111554) and ‘Ca. P. phoenicium’ (16SrIX-B, AF515637), respectively.
BLAST search also showed that rp sequences of KAlmWB (KP400599) and EGWB (KP400598) phytoplasma isolates had more than 97.5% homology with rp gene sequences of 16SrIX group phytoplasmas. This analysis revealed that KAlmWB and EGWB had closest homology with Knautia arvensis phyllody (rpIX-C, EF186801) and almond witches'-broom strain A112 (rpIX-B1, EF186803), respectively.
The percentage homology between SR and rp sequences was determined. Based on SR sequences (Table 1), KAlmWB and EGWB isolates had 86.8% homology. KAlmWB was most similar to Indian Brassica rapa phyllody phytoplasma (16SrIX-C) and EGWB to ‘Ca. P. phoenicium’ (16SrIX-B). Based on rp sequences (Table 2), KAlmWB and EGWB phytoplasmas had 77.6% homology. KAlmWB was most similar to Knautia arvensis phyllody phytoplasma (rpIX-C) and EGWB to almond witches'-broom phytoplasma strain A112 (rpIX-B1).
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. KlmAWBa | 100 | |||||||||
| 2. EGWB | 86.0 | 100 | ||||||||
| 3. EchinWB | 90.0 | 93.3 | 100 | |||||||
| 4. LachPh | 89.0 | 92.9 | 97.6 | 100 | ||||||
| 5. JunWB-2A | 94.9 | 84.8 | 91.0 | 89.1 | 100 | |||||
| 6. BraR | 98.6 | 84.4 | 89.6 | 87.9 | 96.4 | 100 | ||||
| 7. PPWB | 88.5 | 92.0 | 96.4 | 96.0 | 89.6 | 88.1 | 100 | |||
| 8. ‘Ca. P. phoenicium’ | 83.5 | 99.2 | 94.2 | 93.8 | 82.4 | 82.0 | 92.9 | 100 | ||
| 9. NalmWB | 83.4 | 99.2 | 94.2 | 93.8 | 82.3 | 81.9 | 92.9 | 100 | 100 | |
| 11. Acholeplasma laidlawii | 42.9 | 41.0 | 42.1 | 42.6 | 41.7 | 42.8 | 42.1 | 41.7 | 41.9 | 42.5 |
- a KAlmWB, Khafr almond witches'-broom (16SrIX-C, KP297391); EGWB, Estahban GF-677 witches'-broom (16SrIX-B, KP297392); EchinWB, Echinops witches'-broom (16SrIX-D, GU902973); LachPh, Iranian Lactuca sativa phyllody (16SrIX-D, DQ195212); JunWB-2A, Juniperus occidentalis witches'-broom strain 2A (16SrIX-E, GQ925919); BraR, Brassica rapa phyllody phytoplasma (16SrIX-C, GU111554); PPWB, Pigeon pea witches'-broom (16SrIX-A, AF248957); Ca. P. phoenicium, Candidatus Phytoplasma phoenicium (16SrIX-B, AF515637); NalmWB, Neyriz almond witches'-broom (16SrIX-B, DQ195210); A. laidlawii, Acholeplasma laidlawii (D13260).
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| 1. KAlmWBa | 100 | |||||
| 2. EGWB | 77.6 | 100 | ||||
| 3. AlmWB-112 | 82.9 | 91.9 | 100 | |||
| 4. KAP | 93.0 | 82.3 | 88.6 | 100 | ||
| 5. PEY | 88.7 | 82.7 | 89.3 | 94.7 | 100 | |
| 6. PPWB | 88.4 | 84.1 | 88.8 | 94.3 | 94.8 | 100 |
| 7. Acholeplasma laidlawii | 57.9 | 56.8 | 60.8 | 60.2 | 60.2 | 66.6 |
- a KAlmWB, Khafr almond witches'-broom (16SrIX-C, KP400599); EGWB, Estahban GF-677 witches'-broom (16SrIX-B, KP400598); AlmWB-112, Almond witches'-broom strain A112 (16SrIX-B, EF186803); KAP, Knautia arvensis phyllody (16SrIX-C, EF186801); PEY, Picris echioides yellows phytoplasma (16SrIX-C, EF186802); PPWB, Pigeon pea witches'-broom (16SrIX-A, EF193383); A. laidlawii, Acholeplasma laidlawii (M74771).
Phylogenetic analysis
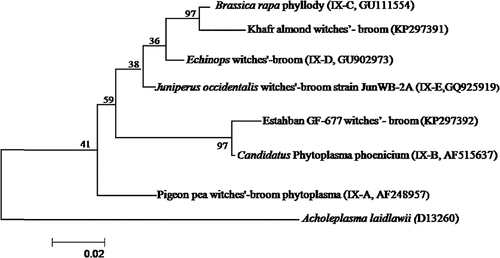
A phylogenetic tree constructed with SR sequences of seven phytoplasmas belonging to 16SrIX group (Fig. 3) revealed that KAlmWB clustered with Brassica rapa phyllody phytoplasma (16SrIX-C, GU111554) and EGWB with ‘Candidatus P. phoenicium’ (16SrIX-B, AF515637). Based on phylogenetic tree constructed with rp (covering rpl22 and rps3) sequences of 22 phytoplasmas belonging to various groups (Fig. 4), KAlmWB clustered with Knautia arvensis phyllody phytoplasma (rpIX-C2, EF186801) and EGWB with almond witches'-broom strain A112 (rpIX-B1, EF186802).
CLUSTALW alignment between SR sequences of 16SrIX subgroups showed several deletions, insertions and substitutions. Insertion of A and C bases at 17–18 positions, eight bases at positions 207–214 and 28 bases at positions 73–100 was characteristic of KAlmWB and Indian Brassica rapa phyllody phytoplasmas, members of 16SrIX-C subgroup. EGWB, Neyriz AlmWB and ‘Ca. P. phoenicium’, members of 16SrIX-B subgroup were differentiable from other 16SrIX subgroups based on deletion of eight bases at positions 207–214 and 28 bases at positions 73–100.
Virtual RFLP
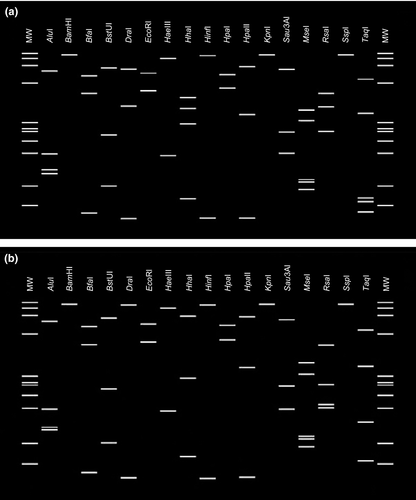
Virtual RFLP analysis was carried out on R16F2n/R16R2 sequences from KAlmWB and EGWB isolates. Visualization and comparison of plotted patterns of virtual gel showed that based on DraI, HhaI, RsaI and TaqI restriction enzymes, KAlmWB and EGWB phytoplasmas are differentiable. KAlmWB patterns were most similar to Picris echioides yellows phytoplasma (Y16389) representative of 16SrIX-C subgroup and EGWB to ‘Ca. P. phoenicium’ (AF515637) representative of 16SrIX-B subgroup (Fig. 5)
Discussion
Almond witches'-broom (AWB) caused by Candidatus Phytoplasma phoenicium (Verdin et al. 2003) is a lethal disease of almond in Fars and certain provinces of Iran (Salehi et al. 2006) and Lebanon (Choueiri et al. 2001; Abou-Jawdah et al. 2002). GF-677 (Prunus amygdalus × Prunus persica) with strong roots and a good potential for pests and diseases resistance (Fasolo et al. 1987) is one of the most suitable rootstocks for almond and peach used in calcareous soils to overcome lime-induced chlorosis (Kester 1970; Fasolo et al. 1987; Hartmann et al. 1990). GF-677 cuttings were firstly imported from France to Iran and planted in different regions of Fars province including Beedzard, Estahban and Neyriz areas for further propagation and distribution to other Iranian stone fruit growing areas. witches'-broom is an important disease of GF-677 in Fars province (Salehi et al. 2011). Previous studies have been shown that phytoplasmas associated with KAlmWB and EGWB are members of 16SrIX phytoplasma group (Salehi et al. 2006, 2011). Our results also confirmed that KAlmWB and EGWB phytoplasmas belong to 16SrIX phytoplasma group. For quarantine purposes, it was necessary to know whether KAlmWB and EGWB phytoplasmas belong to the same subgroup or different subgroups in 16SrIX phytoplasma group. Production of different symptoms in periwinkle indicated that agents of KAlmWB and EGWB are differentiable. This study showed that these phytoplasmas could be distinguished from each other on the basis of comparison of RFLP patterns of P1/P7 PCR products and sequence analysis using 16S–23S rRNA spacer region (SR) and rp genes. Based on these molecular analyses and virtual RFLP, KAlmWB and EGWB phytoplasmas belong to 16SrIX-C and 16SrIX-B subgroups, respectively. Phytoplasmas related to B subgroup of 16SrIX group previously were reported as causal agent of AlmWB in Iran and Lebanon (Molino Lova et al. 2011; Lee et al. 2012). Source of infection of imported GF-677 trees probably is witches'-broom-affected almond trees in Iran. Epidemiological studies are required to understand the role of ‘Ca. P. phoenicium’ in infection of GF-677 trees in Fars province. It is known that the leafhopper Asymmetrasca decedens is natural vector of ‘Ca. P. phoenicium’ in Lebanon (Abou-Jawdah et al. 2014), but it is not known whether it also transmits ‘Ca. P. phoenicium’ to GF-677 trees or whether another vector is responsible. It is yet to be determined whether KAlmWB phytoplasma is transmissible to GF-677 trees.




