NaMYB8 regulates distinct, optimally distributed herbivore defense traits
Summary
When herbivores attack, plants specifically reconfigure their metabolism. Herbivory on the wild tobacco Nicotiana attenuata strongly induces the R2R3 MYB transcriptional activator MYB8, which was reported to specifically regulate the accumulation of phenolamides (PAs). We discovered that transcriptional regulation of trypsin protease inhibitors (TPIs) and a threonine deaminase (TD) also depend on MYB8 expression. Induced distributions of PAs, TD and TPIs all meet predictions of optimal defense theory: their leaf concentrations increase with the fitness value and the probability of attack of the tissue. Therefore, we suggest that these defensive compounds have evolved to be co-regulated by MYB8.
-
Edited by: Yonggen Lou, Zhejiang University, China
As primary producers, plants have developed intricate strategies to defend themselves against herbivores, including chemical defenses that act as anti-digestives, toxins, or repellents, or attract predators and parasitoids of herbivores. These responses are often tailored to particular herbivores, who reveal themselves by the elicitors they secrete, and other feeding-associated traits. The tailoring of defense in response to herbivore elicitation helps plants to respond appropriately to different attackers, and to avoid fitness costs incurred by unnecessary production of defensive compounds (Baldwin 1998).
In the wild tobacco, Nicotiana attenuata, the recognition of fatty acid-amino acid conjugates, elicitors present in the oral secretions (OS) and regurgitant of its specialist herbivore Manduca sexta (Halitschke et al. 2001), induce the rapid accumulation of jasmonate (JA) hormones, which can activate the transcription of secondary regulators like MYB8 via transcription factors such as MYC2, to induce specific defense responses (Woldemariam et al. 2013). MYB8 was discovered as a homolog of the R2R3-type MYB transcription factor NtMYBJS1 from cultivated tobacco (Nicotiana tabacum) and shown to regulate the accumulation of phenolamides (PAs) in BY-2 tobacco cell cultures in a JA-dependent manner (Galis et al. 2006). In N. attenuata, MYB8 transcripts accumulate after herbivory, resulting in the transcription of genes related to PA biosynthesis, in particular the three hydroxycinnamoyl-coenzyme A: polyamine transferases AT1, DH29 and CV86 (Kaur et al. 2010; Onkokesung et al. 2012). Plants rendered deficient in MYB8 by RNA interference (RNAi, irMYB8) have drastically lower levels of PAs including caffeoylputrescine, but are similar to wild-type (WT) plants in their accumulation of other phenolic compounds, including rutin and chlorogenic acid. Kaur et al. (2010) showed that M. sexta and Spodoptera littoralis caterpillars grow faster on irMYB8 than on WT plants and that spraying caffeoylputrescine on irMYB8 plants reduces M. sexta growth, suggesting a defensive function of the PAs regulated by MYB8 (Kaur et al. 2010).
The microarray data presented by Kaur et al. (2010) also shows a reduction in trypsin proteinase inhibitor (TPI) transcripts, indicating that MYB8 might additionally regulate other plant defense responses. To follow up on that observation, we present experimental data that implicate MYB8 as a regulator of the within-plant distribution of multiple plant defense compounds. For detailed information about the experimental conditions see Supplemental Materials and Methods.
To confirm the influence of MYB8 silencing on TPI transcript accumulation and activity, we conducted qPCR analysis (Figure 1A) and radial diffusion TPI activity assays (Figures 1B, S1). Similar to the microarray results from Kaur et al. (2010), which were obtained with another, independent transgenic line, the TPI transcript levels were attenuated to less than half of wild type (WT) levels, as was basal and induced TPI activity. However, hemizygous crosses with TPI-deficient plants indicate that the increased growth of M. sexta caterpillars on irMYB8 plants can be largely attributed to other MYB8-regulated factors, such as PAs, as proposed by Kaur et al. (2010) (Figure S2). The absence of a TPI-mediated effect on caterpillar growth (Figure S2) was also described in some other publications, for example, Schuman et al. (2012) and might be caused by compensatory feeding or a high protein content of plant tissue. The dependence of TPI expression on MYB8 can elegantly explain some previous findings; for example, the primed induction of MYB8, PAs and TPIs but no other JA-mediated traits in N. attenuata plants on which Spodoptera exigua oviposited (Bandoly et al. 2015).
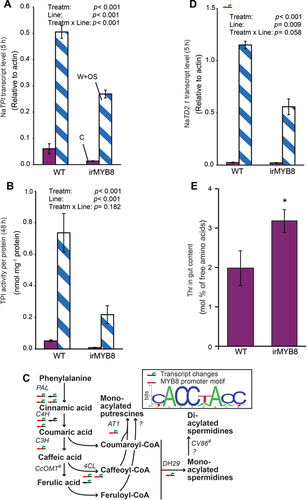
To identify other potential MYB8-regulated herbivory responses, we conducted a complete microarray analysis using a whole-transcriptome array. This comprehensive analysis confirmed the effect of MYB8 on genes related to PAs (Figures 1C, S3–S8). To identify further targets of MYB8, we conducted a promoter motif analysis for regions 2 kb upstream of genes. The identified motif (Figure 1C) is similar to the motif for the tobacco homolog NtMYBJS1 identified by gel mobility shift assays (Galis et al. 2006). Forty out of 61 genes (65.6%) that were downregulated in irMYB8 plants contained the promoter motif (Figure 1C, Table S2). We did not identify this motif in the 2kb region upstream of the TPI gene. The TPI gene contains repetitive regions (Wu et al. 2006) and thus the assembly of the gene sequence and upstream elements is less certain than for other target genes. Therefore, further investigation is required to determine whether the regulation of TPI by MYB8 is direct or indirect.
Instead, we found that one of four threonine deaminase (TD) homologs (TD2.1) had the motif located within the 2kb upstream region and its transcripts were reduced in irMYB8 plants (Figures 1D, S9). TDs catalyze the deamination of Thr to α-ketobutyrate, which is a precursor of Ile. TDs are thus required for the accumulation of JA-Ile (Kang et al. 2006). TD activity is usually limited by substrate-level feedback, but Chen et al. (2005) described a JA-inducible TD isoform in Solanum lycopersicum that lacks its regulatory domain after proteolytic digestion and deaminates Thr in M. sexta guts, depriving larvae of this essential amino acid (for a comprehensive characterization of TD in plant defense see Gonzales-Vigil et al. 2011). Since irMYB8 plants were shown not to be impaired in JA-Ile formation (Kaur et al. 2010; Figure S10), and the Thr and Ile levels were hardly affected in MYB8-silenced plants (Figure S10), we propose an anti-nutritive function of the MYB8-regulated TD similar to that reported by Chen et al. (2005). This hypothesis is supported by an analysis of amino acids extracted from the gut content of M. sexta larvae, showing an increase in Thr as a molar percent of amino acids for larvae feeding on irMYB8 versus WT plants (Figure 1E).
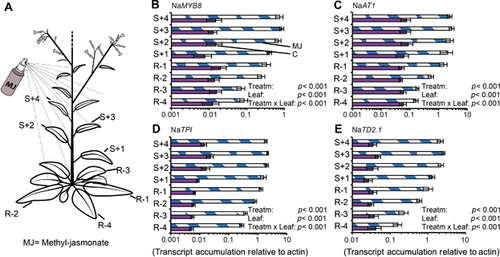
The data presented by Kaur et al. (2010) and Onkokesung et al. (2012) showed that the induced accumulation of PAs is specifically localized within the plant. The distribution follows patterns similar to that described by the optimal defense hypothesis (ODH), which states that tissues that face a high attack risk and contribute the most to fitness should be most defended (McKey 1974). Recently, it was shown that cytokinins can regulate these PA distribution patterns, likely via MYB8 (Schäfer et al. 2015; Brütting et al. 2017; Figure 2B, C). The TPI transcript levels were also associated with cytokinin content in leaves and had a distribution pattern similar to that of PAs (Schäfer et al. 2015; Brütting et al. 2017, Figure 2D). Here, we show that TD2.1 transcripts also accumulate to higher levels in younger leaves than older leaves after JA induction (Figure 2E; for a negative control see Figure S11, NaEF).
Steppuhn and Baldwin (2007) demonstrated that TPI activity induced compensatory feeding in S. exigua, increasing the susceptibility of larvae to nicotine-producing plants. MYB8 regulates not only the accumulation of compounds with a potential toxic effect (PAs, Kaur et al. 2010), but also one known and one potential anti-nutritive defense (TPI, Zavala et al. 2004; TD, Chen et al. 2005; Kang et al. 2006). This raises the question whether similar synergisms occur for MYB8-regulated defense responses. We hypothesize that TPI and TD activity increase the susceptibility of herbivores to PAs, or, alternatively, that the combination of TPI, TD and PAs acts to reduce protein availability for herbivores more than the summed effect of each individually.
ACKNOWLEDGEMENTS
We thank Matthias Schöttner, Thomas Hahn, Antje Wissgott and Wibke Kröber for technical assistance. We thank Tamara Krügel, Andreas Weber, Andreas Schünzel and the entire glasshouse team for plant cultivation. We thank Thomas Brockmöller, Klaus Gase, Shree Pandey and Sarah Pottinger, and Jorge A. Zavala for helpful discussions. The authors acknowledge the following agencies for funding: the Max Planck Society (all), the Collaborative Research Centre “Chemical Mediators in Complex Biosystems - ChemBioSys” (SFB 1127) (M.S.) and Advanced Grant No. 293926 of the European Research Council to I.T.B. (C.B., M.C.S.), Swiss National Science Foundation (No. PEBZP3-142886) and the Marie Curie Intra-European Fellowship (IEF) (No. 328935) to S.X.
AUTHOR CONTRIBUTIONS
Conceptualization: M.S., C.B., S.X., A.S., I.T.B. and M.C.S.; Methodology: M.S., C.B., S.X., Z.L., I.T.B. and M.C.S.; Software: S.X. and Z.L.; Validation: M.S., C.B. and S.X.; Formal analysis: M.S., C.B., S.X. and Z.L.; Investigation: M.S., C.B. and M.C.S.; Resources: S.X., Z.L. and I.T.B.; Data curation: M.S., C.B., S.X., I.T.B. and M.C.S.; Writing – original draft: M.S., C.B., S.X., I.T.B. and M.C.S.; Writing – review & editing, M.S., C.B., S.X., Z.L., A.S., I.T.B. and M.C.S.; Visualization: M.S.; Supervision: I.T.B. and M.C.S.; Project administration: I.T.B. and M.C.S.; Funding acquisition: I.T.B., S.X. and M.C.S.




