Efficacy of a whole slide image-based prediction model for lymph node metastasis in T1 colorectal cancer: A systematic review
Declaration of conflict of interest: None.
Author contribution: K. I., Y. K., S. K., and Y. T. contributed to the study concept and design. K. I. and Y. K. drafted the manuscript. K. I. obtained funding. K. I., Y. K., S. K., Y. T., T. N., J. W., M. T., Y. M., K. G. Y., H. M., and M. M. contributed to the interpretation of data. S. K., K. G. Y., and M. M. contributed to study supervision. All authors of this article contributed to the data collection and critical revision of the manuscript and have read and approved the final version submitted.
Financial support: This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant number 22K16500).
Abstract
Background and Aim
Accurate stratification of the risk of lymph node metastasis (LNM) following endoscopic resection of submucosal invasive (T1) colorectal cancer (CRC) is imperative for determining the necessity for additional surgery. In this systematic review, we evaluated the efficacy of prediction of LNM by artificial intelligence (AI) models utilizing whole slide image (WSI) in patients with T1 CRC.
Methods
In accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines, a systematic review was conducted through searches in PubMed (MEDLINE), Embase, and the Cochrane Library for relevant studies published up to December 2023. The inclusion criteria were studies assessing the accuracy of hematoxylin and eosin-stained WSI-based AI models for predicting LNM in patients with T1 CRC.
Results
Four studies met the criteria for inclusion in this systematic review. The area under the receiver operating characteristic curve for these AI models ranged from 0.57 to 0.76. In the three studies in which AI performance was compared directly with current treatment guidelines, AI consistently exhibited a higher area under the receiver operating characteristic curve. At a fixed sensitivity of 100%, specificities ranged from 18.4% to 45.0%.
Conclusions
Artificial intelligence models based on WSI can potentially address the issue of diagnostic variability between pathologists and exceed the predictive accuracy of current guidelines. However, these findings require confirmation by larger studies that incorporate external validation.
Introduction
The absence of lymph node metastasis (LNM) makes colorectal intramucosal cancer a candidate for endoscopic resection, whereas colectomy with lymph node dissection is the standard approach to invasive cancer extending beyond the muscularis propria layer (T2).1 Submucosal invasive cancer (T1), which is between these two stages, presents a clinical dilemma because approximately 10% of these patients have extraintestinal LNM, necessitating choosing between endoscopic treatment and surgery.1, 2
Current European, US, and Japanese guidelines advocate secondary surgical resection with lymph node dissection after endoscopic resection of T1 colorectal cancer (CRC), depending on the risk of LNM.1, 3-6 Risk factors include deep submucosal invasion (depth of submucosal invasion ≥ 1000 μm; T1b), high-grade histological type (poorly differentiated adenocarcinoma, mucinous carcinoma, or signet-ring cell carcinoma), lymphovascular invasion, and high-grade tumor budding. Lesions with these characteristics typically require radical resection. Prior research has validated the efficacy of these guidelines for patients at low risk of LNM, that is, without these risk factors, including from a prognostic perspective.7, 8 However, there are two persistent primary challenges.9 Firstly, the accuracy of prediction of LNM by current guidelines is suboptimal. The rate of LNM when determined according to the guidelines is only 10%; the remaining 90% do not have LNM, resulting in overtreatment. Secondly, pathologists identify pathological risk factors inconsistently, particularly lymphovascular invasion, a crucial predictor of LNM in T1 CRC.10 These data suggest that stratification of LNM risk is heavily dependent on pathologists' subjective.11
In response to these challenges, researchers have recently focused on attempting to create whole slide image (WSI)-based models for predicting LNM in patients with T1 CRC.2, 12 These models aim to provide a more objective and potentially accurate means of predicting LNM from hematoxylin and eosin (HE)-stained virtual slides, independent of pathologists' findings.11 In this systematic review, we aimed to assess the effectiveness of WSI-based models in predicting LNM risk in patients with T1 CRC.
Methods
This systematic review was meticulously conducted and reported in alignment with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses guidelines (Data S1). Additionally, the study protocol was registered with the International Prospective Register of Systematic Reviews (registration number CRD42022356097) on September 6, 2022 (Data S2).
Search strategy
To ensure a comprehensive and methodical literature search, we collaborated with a medical sciences librarian in designing our search strategy. We conducted electronic searches across several databases, including MEDLINE (PubMed), Embase (ProQuest), and the Cochrane Library (Cochrane Central Register of Controlled Trials), spanning from their inception to December 2023. The detailed search strategy, including specific terms and combinations used, is thoroughly outlined in Data S3.
Study selection
The inclusion criteria were as follows: (i) prospective and retrospective cohort studies and case–control studies and (ii) studies reporting associations between HE-stained virtual slides and LNM in patients with T1 CRC using artificial intelligence (AI). The exclusion criteria were as follows: (i) case reports, reviews, and meta-analysis; (ii) full text not accessible; (iii) published in languages other than English; and (iv) data not extractable. In cases of overlapping study cohorts reported by the same authors or institutions, only the most recent study was included.
This review focused on adults (age ≥ 18 years) diagnosed with T1 CRC who had undergone primary or secondary surgical resection with lymph node dissection. Patients who had received preoperative chemotherapy and/or radiotherapy and those who had not undergone lymph node dissection were excluded. The definitive standard for determining the presence or absence of LNM was operative specimens.
Outcomes
The primary outcome of this study was the accuracy of WSI-based AI prediction of LNM in patients with T1 CRC. Specifically, we evaluated the sensitivity, specificity, and area under the receiver operating characteristic curve (AUC) of the tools used to predict LNM in patients with T1 CRC compared with those of the Japanese guidelines. For the process of assessing diagnostic accuracy, in the guideline models, the presence of one or more following risk factors resulted in LNM being predicted as positive: (i) depth of submucosal invasion ≥ 1000 μm (T1b); (ii) positive lymphovascular invasion; (iii) poorly differentiated adenocarcinoma, mucinous carcinoma, or signet-ring cell carcinoma; and (iv) high-grade tumor budding (plus5 positive resection margin in Song et al.14). The presence of LNM was diagnosed by examining HE-stained sections of surgically dissected lymph nodes.
Data extraction
The references were initially screened independently by three reviewers (Y. K., Y. T., and J. W.) who evaluated the titles and abstracts of all retrieved studies. Any study identified as a potential candidate by at least one reviewer was listed for further evaluation. The full texts of these studies were then independently reviewed by two authors (Y. K. and Y. T.) to determine their eligibility according to the predefined review criteria. When these two reviewers disagreed, resolution was sought through consensus discussion, including input from a third reviewer (K. I.) when necessary. Additionally, we requested clarification or additional information from the original authors regarding missing relevant data. The extracted data from each study included the first author's name, publication year, country of origin, study design, number of patients, specific inclusion and exclusion criteria, reported outcome events, materials utilized for analysis, and details of the AI algorithm employed in predicting LNM.
Results
Study selection and characteristics
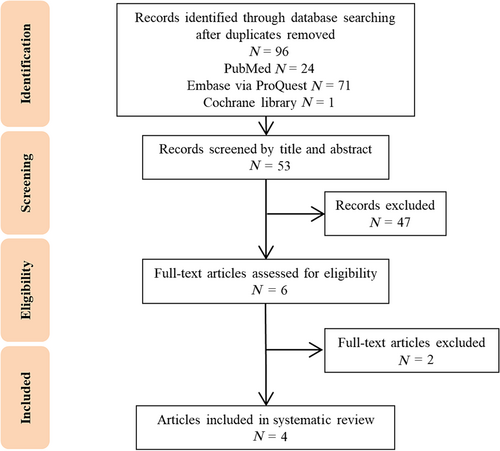
The initial search yielded 96 articles, as depicted in our flow diagram (Fig. 1). After removing 43 duplicates, thorough full-text evaluations resulted in the selection of four studies, encompassing 1703 patients with T1 CRC, that met our inclusion criteria. Detailed characteristics of these studies are presented in Table 1.
|
First author (year) |
Country | Study design | Algorithm | Type of scanner | Material selection for assessment | Total cohort, n (LNM-positive, %) | Training cohort, n (LNM-positive, %) | Validation cohort, n (LNM-positive, %) |
|---|---|---|---|---|---|---|---|---|
|
Brockmoeller13 (2022) |
Denmark |
Multicenter Retrospective |
DNN | Aperio XT Scanner | One HE slide with widest and deepest areas of invasion |
203 (16.3) |
Not divided | Not divided |
|
Takamatsu12 (2022) |
Japan |
Single-center Retrospective |
CNN, RF | NanoZoomer | Slides including all submucosal invasive areas |
783 (7.8) |
548 (7.8) |
235 (7.7) |
|
Song14 (2022) |
Korea |
Single-center Retrospective |
DCNN, attention-based learning | VENTANA iScan HT | N/A |
400 (17.8) |
320 (17.8) |
80 (17.5) |
|
Takashina15 (2023) |
Japan |
Single-center Retrospective |
CNN, RF | NanoZoomer | One HE slide with deepest area of invasion |
585† (35.2) |
485† (39.4) |
100 (15.0) |
- † Including some T2 colorectal cancers (n = 268).
- CNN, convolutional neural network; DCNN, deep convolution neural network; DNN, deep neural network; HE, hematoxylin and eosin-stained; LNM, lymph node metastasis; N/A, not available; RF, random forest.
The identified studies did not include any randomized trials or prospective cohort studies. Three of the included studies were conducted in Asia (total of 1500 cases of T1 CRC) and one in Europe (203 cases of T1 CRC). Brockmoeller et al. focused on patients with pT1 CRC (n = 203) who had undergone resection with known lymph node status.13 Takamatsu et al. gathered data on patients with T1 CRC treated by endoscopic resection followed by surgery (n = 271) or surgery alone (n = 512).12 Song et al. studied patients with T1 CRC who had undergone endoscopic resection followed by surgery (n = 400).14 Takashina et al. analyzed a training cohort comprising patients with T1 (n = 217) and T2 CRC (n = 268) and a validation group (n = 100).15 All studies utilized HE-stained slides, with WSIs obtained using a digital slide scanner.
Algorithm of artificial intelligence
Brockmoeller et al
The process of AI development began with selecting the most representative HE-stained slide for each tumor, focusing on the widest and deepest areas of invasion. High-resolution whole slide scanning (Aperio XT Scanner; Aperio Technologies, San Diego, CA, USA) was performed to digitize the slides. In cases where tumor areas varied between slides, the one with the larger area was chosen. Digital annotations of invasive tumor areas were conducted to ensure accurate analysis. The experimental design involved training deep neural networks on all available WSIs, without restrictions on tissue types or areas, to ensure unbiased detection of predictive features. Models were trained using a threefold cross-validation approach to ensure robustness. Image preprocessing included the extraction of tiles (512 × 512 pixels in size) with background and artifacts removed. The tiles underwent Macenko normalization to ensure color consistency across samples. The deep learning network, based on ShuffleNet, employs transfer learning for predicting LNM status. Training datasets were balanced to address class imbalance.
Takamatsu et al
This model was developed using two steps. The first step utilized convolutional neural networks (CNNs) to classify images into cancerous and non-cancerous tiles from WSIs (NanoZoomer, Hamamatsu Photonics, Hamamatsu, Japan). The CNN was trained and validated using a dataset of 783 cases to achieve high accuracy in identifying relevant histological features. The second step employed a random forest (RF) algorithm that used CNN output as input features. The model aggregated these features to calculate a predictive score for LNM, focusing on the probability and distribution of classified tiles. The main variables in the RF model included tumor location, total number of cancer-class tiles, number of tiles classified as metastatic or non-metastatic, percentages of tiles classified as metastatic or non-metastatic, average probabilities, standard deviations of cancer-class probabilities, and metastatic or non-metastatic probabilities, and a probability score summary for each tile.
Song et al
This study included patients with at least one conventional risk factor, such as positive resection margin, deep submucosal invasion (≥ 1 mm), poorly differentiated histology, presence of lymphovascular invasion, and tumor budding. The model operates in two primary steps. First, a deep CNN was trained to extract features from individual patches of the WSIs (Roche Diagnostics, Basel, Switzerland), learning to recognize histopathological patterns associated with LNM. Then, these features were aggregated using an attention mechanism that weights the importance of each patch based on its contribution to the final prediction. This method ensures that the model pays more attention to the most informative areas of the slide, enhancing the accuracy of LNM prediction without the integration of clinical data. Additionally, the study utilized an attention mechanism to highlight regions of interest on WSIs, indicating that the AI focused on areas such as immature stroma and tumor budding for its predictions.
Takashina et al
Hematoxylin and eosin-stained slides from T1 and T2 CRC cases were observed, and the slide with the deepest invasion was selected for analysis. The WSIs (NanoZoomer, Hamamatsu Photonics) were then cropped into small patches, which were analyzed using unsupervised machine learning techniques. Specifically, the patches were clustered using the k-means algorithm, allowing the AI to learn and identify patterns associated with LNM without explicit labeling of the training data. This method aimed to leverage the heterogeneity within and across the slides, acknowledging that cancerous regions within a slide could vary significantly in their appearance and histological features. Using the extracted features, a predictive model for LNM was built employing the RF algorithm. This model used the proportion of patches from each cluster in a WSI as features, combined with additional patient information such as patient sex and tumor location, to perform the analysis.
Diagnostic performance of artificial intelligence compared with guidelines
Table 2 presents the sensitivity, specificity, and AUC of each of the AI models alongside the Japanese guidelines. Notably, three of these studies used different datasets for internal validation than for training. At a fixed sensitivity of 100%, reported specificities ranged from 18.4% to 45.0%, whereas accuracy ranged between 24.7% and 63.8%. In terms of AUC, the AI models performed better than the guidelines across three studies.
| WSI models | Guidelines | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
First author (year) |
Subject N |
Sensitivity | Specificity | Accuracy | AUC | Sensitivity | Specificity | Accuracy | AUC |
|
Brockmoeller13 (2022) |
Total 203 |
N/A | N/A | N/A | 0.57 | N/A | N/A | N/A | N/A |
|
Takamatsu12 (2022) |
Test 235 |
100 | 18.4 | 24.7 | 0.76 | 100 | 20.1 | 25.8 | 0.60 |
|
Song14 (2022) |
Test 80 |
100 | 45.0 | 63.8 | 0.76 | 100 | 0 | 17.5 | 0.50 |
|
Takashina15 (2023) |
Test 100 |
100 | 24.7 | 36.0 | 0.75 | 100 | 4.7 | 9.4 | 0.52 |
- AUC, area under the receiver operating characteristic curve; N/A, not available; WSI, whole slide image.
Discussion
In this systematic review, we aimed to evaluate the ability of WSI-based AI models to predict LNM in patients with T1 CRC. These AI models were found to predict LNM with greater accuracy and reproducibility than current treatment guidelines; this finding has significant implications for clinical practice and future research.
Novel predictive models in which AI is merged with digital pathology are currently under development. These pathologist-independent models aim to score LNM risk objectively from data extracted from HE-stained images, bypassing the need for human assessment. Assessment by WSI-based AI models involves several steps: (i) creation of digital slides: digital versions of pathology slides acquired; (ii) patch creation: segmentation of the digital slides into smaller patches for detailed analysis; (iii) application of AI: algorithms employed to assess the risk of LNM shown by each patch; and (iv) prediction of risk of LNM: an AI model utilized to aggregate these assessments and predict the overall risk of LNM. In all three studies that compared diagnostic accuracy with the current guidelines, WSI-based AI models showed better discrimination for the presence of LNM than did the guidelines.12, 14, 15 This innovative approach promises to minimize diagnostic variability and thus enhance the precision of assessment of the risk of LNM in patients with T1 CRC.
Why is a WSI-based model needed? The current guidelines for treatment of T1 CRC have two primary challenges to address: low diagnostic accuracy and poor reproducibility of pathological variables.9 Two prediction models that address the first of these issues by leveraging extensive T1 CRC data have recently emerged. The first is an AI model developed by the authors.16 This AI uses an artificial neural network and incorporates eight factors: patient sex and age; tumor size, location, and morphology; lymphatic and vascular invasion; and histological differentiation. This model was developed using data from 5131 cases of T1 CRC from seven Japanese centers (1997–2017), six being involved in training and one in external validation. The artificial neural network demonstrated significantly greater accuracy than did the Japanese guidelines (AUC 0.83 vs 0.57; P < 0.001). The second model is a nomogram developed by Kajiwara et al.17 This nomogram visualizes predictive probabilities, offering clear insight into each variable's weight. It was developed using data from 4673 cases of T1 CRC across 27 Japanese centers (2009–2016), 18 centers (3080 cases) for development and 9 (1593 cases) for testing. The nomogram, which includes six variables, achieved a concordance statistic (C-statistic) of 0.790 for LNM prediction in the validation cases, surpassing the 0.777 of the guidelines.
The large-scale studies under discussion have highlighted a critical issue that needs to be addressed by future research, namely, the reproducibility of pathological diagnoses, which form the basis of LNM prediction models. Variations in pathological assessments among different pathologists examining the same lesion are concerning. This variability impacts the accuracy and reproducibility of prediction models built upon these assessments. Despite each study validating its model using different test and development datasets, thereby ensuring a degree of accuracy, consistent results have not always been achieved because of variations in validation data. It is reasonable to infer that discrepancies in pathological findings directly impact variability in the prediction model's accuracy. Two primary factors contribute to this challenge.
The first of these factors is inter-pathologist discrepancies. The concordance rate between pathologists in assessing T1 CRC varies considerably. A previous study examining this reported relatively low kappa values, indicative of moderate to fair agreement: 0.33 for lymphovascular invasion, 0.48 for histological grade, 0.29–0.44 for tumor budding, and only 0.21 for depth of submucosal invasion.9, 18 Differences in pathologists' findings lead to inconsistency in diagnoses, prejudicing the reliability of the prediction models that rely on those diagnoses. The second of these factors is differences in pathology procedures. For example, when evaluating lymphovascular invasion in patients with T1 CRC, the application of immunostaining varies significantly, lacks uniform guidelines, and is often subject to individual institutional practices or pathologists' discretion. Assessment of markers like D2-40 for lymphatic invasion and Victoria Blue/Elastica van Gieson for vascular invasion has been shown to increase accuracy and consequently the odds ratio for LNM.19 Similarly, there is no standard methodology for assessing histological grade, particularly when a lesion exhibits multiple levels of differentiation. Whether the predominant histology (component covering the largest area) or the least differentiation (highest grade component) is prioritized is inconsistent. In Japan, practices vary between institutions.20-22 Notably, the AI model described earlier utilizes the least differentiation approach, whereas the nomogram employs the predominant histological differentiation.16, 17 The lack of standardization in both immunostaining practices and assessment of histological grade introduces significant variability in diagnostic evaluations, further complicating the process of making treatment decisions for patients with T1 CRC. These variations can lead to differences in weighting of variables within a predictive model. Clearly, a standardized approach to pathological diagnosis would enhance the reproducibility and effectiveness of AI-based LNM prediction models.
Although WSI-based AI is reproducible and potentially improves diagnostic accuracy in patients with T1 CRC, several critical issues still require resolution. First is the need for external validation. The four AI studies included in this systematic review were all validated internally. Their accuracy has yet to be validated on an independent external dataset. Variations in staining methods, conditions, and types of scanners used in different institutions can affect results. Furthermore, acquisition magnification, selection of the deepest section for analysis, and procedures for specimen preparation need to be standardized. Because of the variation in methodologies and subject characteristics across the studies, we determined that integrating them into a single analysis might not yield appropriate results. Therefore, we opted for a systematic review, focusing on describing the algorithms and other details of each individual study. The number of studies included is too small, and results can be limited. Also, there may be a difference in quality of HE-stained images between primary endoscopic resection and primary surgical resection cases, leading to a potential difference in degree of submucosal invasion depth and width. The real target of WSI-based AI is primary endoscopic resection cases. When conducting external validation, it is also necessary to consider that the number of retrieved lymph nodes is important to ensure the quality of the surgical specimens. The second issue is the interpretation of AI heat maps. Understanding the basis upon which AI models, visualized through heat maps of HE-stained images, determine whether the risk of LNM is high or low is crucial. Do the areas identified by AI as high-risk correlate with conventional findings such as lymphovascular invasion, tumor budding, or poorly differentiated adenocarcinoma? Or is the AI identifying other factors, such as desmoplastic reactions? Clarifying the explanatory variables linking virtual slide images to the output variable (LNM risk) is imperative for enhancing a model's reproducibility and accuracy. The third issue is the integration of pathology AI into clinical practice. Once the accuracy of pathology AI is established, its practical application will raise several questions. How will it be integrated with existing guidelines for risk assessment? What will its relationship with pathologists be? Studies on AI in colonoscopy have compared the diagnostic accuracy of AI with that of experts/trainees. A similar approach may be necessary for pathology AI. Addressing these challenges is pivotal for advancing AI for assessment of pathology and ensuring its efficacy, reproducibility, and practical utility in clinical practice.
To the best of our knowledge, this systematic review is the first comprehensive attempt to elucidate the utility of WSI-based AI models in predicting LNM in patients with T1 CRC. Our findings highlight the potential of AI models to address current challenges in diagnostic accuracy and mitigate discrepancies in pathological diagnoses and their implications, particularly concerning the necessity of additional surgical resection following endoscopic treatment. The deployment of these AI models in clinical settings necessitates further validation. Future studies, particularly large-scale prospective studies, are crucial for affirming the efficacy of these AI tools in guiding treatment decisions. The promise shown by WSI-based AI models in enhancing diagnostic precision marks a significant step forward in the field of oncological pathology and optimization of treatment strategy.
Acknowledgment
We thank Dr Trish Reynolds, MBBS, FRACP, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.




