Dysregulation of NIPBL leads to impaired RUNX1 expression and haematopoietic defects
Funding information
The work described in this paper was supported by Individual My first AIRC grant (MFAG #18714) and Piano di Sostegno alla Ricerca 2018 Linea 2, Dept. Biotecnologie Mediche e Medicina Traslazionale, Università degli Studi di Milano (PSR Linea2 2018) to AP, and AIRC grant IG-21999 to GC.
Abstract
The transcription factor RUNX1, a pivotal regulator of HSCs and haematopoiesis, is a frequent target of chromosomal translocations, point mutations or altered gene/protein dosage. These modifications lead or contribute to the development of myelodysplasia, leukaemia or platelet disorders. A better understanding of how regulatory elements contribute to fine-tune the RUNX1 expression in haematopoietic tissues could improve our knowledge of the mechanisms responsible for normal haematopoiesis and malignancy insurgence. The cohesin RAD21 was reported to be a regulator of RUNX1 expression in the human myeloid HL60 cell line and during primitive haematopoiesis in zebrafish. In our study, we demonstrate that another cohesin, NIPBL, exerts positive regulation of RUNX1 in three different contexts in which RUNX1 displays important functions: in megakaryocytes derived from healthy donors, in bone marrow samples obtained from adult patients with acute myeloid leukaemia and during zebrafish haematopoiesis. In this model, we demonstrate that alterations in the zebrafish orthologue nipblb reduce runx1 expression with consequent defects in its erythroid and myeloid targets such as gata1a and spi1b in an opposite way to rad21. Thus, also in the absence of RUNX1 translocation or mutations, additional factors such as defects in the expression of NIPBL might induce haematological diseases.
1 INTRODUCTION
In all vertebrates, the RUNX family of transcriptional regulators containing the runt domain (RD) comprises three isoforms: RUNX1, RUNX2 and RUNX3 that, together with the non-DNA-binding CBFβ subunit, regulate many developmental processes.1, 2 The RUNX members specify their functions depending on their cellular and tissue expression: RUNX1 plays a key role in blood development, primarily in the haematopoietic stem cells (HSCs), RUNX2 is manly involved in bone morphogenesis, and RUNX3 in cell growth of neurons, epithelial cells and T cells. However, the three RUNX proteins could exert biological activities also in other organs3-5; for example, RUNX2 and RUNX3 are known to play a role during haematopoiesis together with RUNX1. In addition, all the RUNX genes are transcribed by a distal and a proximal promoter (P1 and P2, respectively) in two main isoforms that differ in the 5′UTR and in the coding sequence of the first exon.6, 7 The P1 and P2RUNX transcripts are differentially expressed in diverse cell types and during specific developmental stages. Indeed, P1 and P2RUNX1 promoters have been reported to have specific activity patterns in the different haematopoietic lineages during development.8
RUNX1 function during haematopoiesis is strictly regulated by post-transcriptional and post-translational modifications such as alternative splicing, acetylation, methylation, phosphorylation and ubiquitination.9 As transcription factor, RUNX1 targets multiple genes, many of which are also pivotal transcriptional regulators involved in the formation of all haematopoietic lineages including the haematopoietic-specific member of E-twenty-six (ETS) family, PU.1.10, 11 Furthermore, the activity of RUNX1 is carried out by its interaction with different proteins fundamental during haematopoiesis such as GATA1, PU.1, CEBPA, PAX5 and ETS1.10, 12-14
Given the high complexity in RUNX1 expression and function, its deregulation is commonly associated with haematopoietic diseases. Depletion of Runx1 in mice and zebrafish models leads to severe defects or complete absence of definitive haematopoiesis.15-18 RUNX1 is frequently involved in chromosomal translocations observed in acute leukaemias, such as ETV6-RUNX1 in t(12;21) and RUNX1-EVI1 in t(3;21),19 while the formation of the chimeric protein RUNX1-CBF2T1 (AML1-ETO) is associated with the M2 subtype of acute myeloid leukaemia (AML).20, 21 RUNX1 mutations determine the familial platelet disorder with a propensity for AML (AML/FPD) and the minimally differentiated acute myeloid leukaemia (AML/M0).22 Importantly, regulation of RUNX1 dosage is essential for the maintenance of normal haematopoiesis23 and several haematopoietic transcription factors are deputed to regulate RUNX1 expression such as Gata2, Ets factors (Fli-1, Elf-1 and Pu.1) and the SCL/Lmo2/Ldb1 complex.24 In zebrafish, the subunit Rad21 of the cohesin complex has been identified as a regulator of runx1 through a forward genetic screen,25 and multiple predicted and in vivo validated binding sites of Rad21 have been shown to be involved in the regulation of the zebrafish runx1.26
In this work, we demonstrate that NIPBL, another member of the cohesin complex, positively regulates RUNX1 expression in two different contexts in which it exerts important functions: normal cord blood megakaryocytes derived from healthy donors and bone marrow samples derived from adult AML patients. In addition, we generate a zebrafish model in which the nipblb-mediated dysregulation of runx1 expression leads to haematopoietic defects resulting in decreased expression of the erythroid marker gata1a and reduction of mature circulating erythrocytes, and increased expression of myeloid precursors positive for the spi1b marker. Our data confirm the regulatory loop between RUNX1-GATA1 and PU.1 during haematopoiesis and highlight a new role of NIPBL on top of this route.
2 MATERIALS AND METHODS
2.1 Patients
Diagnostic bone marrow samples from 34 adult patients affected by AML were collected and characterized for specific molecular aberrancies, including translocations t(9;22), t(8;21) and inv(16), in accordance with specific clinical protocol requirements. The analysed patients belong to different French-American-British (FAB) classification systems (FABs), excluding M3; therefore, all patients were negative for translocation t(15;17) (Table 1). Bone marrow of healthy individuals was collected as controls for gene expression assays, upon appropriate informed consent ASG-A-052A approved on 8 May 2012 by Azienda Socio-Sanitaria of Monza (ASST-Monza). Human material and derived data were used in accordance with the Declaration of Helsinki.
| Age at onset | Karyotype | FAB classification | NPM | FLT3-ITD | t(9;22) | t(8;21) | inv(16) | |
|---|---|---|---|---|---|---|---|---|
| 1 | 47 | 46,XX,t(10;11)(p11;p15)[20] | M0 | NEG | NEG | NEG | NEG | NEG |
| 2 | 49 | 46,XY[20] | M0/M1 | NEG | NEG | NEG | NEG | NEG |
| 3 | 48 | 46,XX[20] | M1 | NEG | NEG | NEG | NEG | NEG |
| 4 | 72 | 47,XY,+mar[10]/46,XY[10] | M2 | NEG | NEG | NEG | NEG | NEG |
| 5 | 58 | 46,XX,t(3;5)(q25;q34)[20] | M2 | NEG | NEG | NEG | NEG | NEG |
| 6 | 59 | 46,XY[20] | NEG | POS | NEG | NEG | NEG | |
| 7 | 33 | 46,XY[15] | M1 | NEG | POS | NEG | NEG | NEG |
| 8 | 30 | 46,XY[20] | M5 | NEG | POS | nk | NEG | NEG |
| 9 | 58 | 46,XY,inv(16)(p13q22)[20] | M4 | NEG | POS | nk | NEG | POS |
| 10 | 76 | nk | M5 | NEG | POS | nk | NEG | NEG |
| 11 | 78 | 46,XX[27] | M4 | NEG | POS | nk | NEG | NEG |
| 12 | 53 | 46,XY[22] | M4 | NEG | POS | nk | NEG | NEG |
| 13 | 64 | 46,XX[20] | M5 | NEG | POS | nk | NEG | NEG |
| 14 | 75 | 46,XY[26] | M4 | NEG | POS | nk | NEG | NEG |
| 15 | 39 | 46,XY[20] | M1 | POS (A) | NEG | NEG | NEG | NEG |
| 16 | 47 | 46,XX[20] | M5 | POS (A) | NEG | NEG | NEG | NEG |
| 17 | 63 | 46,XY,t(8;14)(q24;q32),add(13q34)[18]/46,XY[9] | nk | POS (D) | NEG | nk | NEG | NEG |
| 18 | 58 | 46,XY/47,XY,+8[7/10] | nk | POS (QM) | NEG | nk | NEG | NEG |
| 19 | 50 | 46,XX[20] | M4 | POS (A) | NEG | nk | NEG | NEG |
| 20 | 77 | 46,XY[20] | nk | POS (A) | NEG | nk | NEG | NEG |
| 21 | 54 | 46,XX,t(9;22)(q34;q11)[14]/46,XX[6] | M4 | POS (A) | NEG | POS | NEG | NEG |
| 22 | 60 | 46,XX[6] | nk | POS | NEG | nk | NEG | NEG |
| 23 | 62 | 46,XX[25] | M5 | POS (A) | NEG ITD/POS D835/D836 | nk | NEG | NEG |
| 24 | 58 | 46,XX[20] | nk | POS (A) | NEG | nk | NEG | NEG |
| 25 | 48 | 46,XX[20] | M4 | POS (A) | POS | NEG | NEG | NEG |
| 26 | 51 | 46,XX[20] | M5 | POS (A) | POS | NEG | NEG | NEG |
| 27 | 68 | 46,XX[20] | M4 | POS (A) | POS ITD/POS D835/D836 | NEG | NEG | NEG |
| 28 | 46 | 46,XY[20] | M2 | POS | POS | NEG | NEG | NEG |
| 29 | 39 | 46,XX[22] | M1 | POS (A) | POS | nk | NEG | NEG |
| 30 | 58 | 46,XY | M5 | POS (A) | POS | nk | NEG | NEG |
| 31 | 35 | 46,XY,?r(18)(?)[16]/47,idem,+8[3]/46,XY[1] | nk | POS (B) | POS | nk | NEG | NEG |
| 32 | 58 | 46,XY[24] | M1 | POS (A) | POS | nk | NEG | NEG |
| 33 | 70 | 46,XY[20] | M5 | POS (A) | POS | nk | NEG | NEG |
| 34 | 12 | 46,XY[24] | nk | POS (A) | POS | NEG | NEG | NEG |
2.2 Animals
Zebrafish embryos were raised and maintained according to international (European Union Directive 2010/63/EU) and national (Italian decree no. 26 of 4 March 2014) guidelines on the protection of animals used for scientific purposes. The fish were maintained under standard conditions in the fish facilities of Bioscience Dept, University of Milan, Via Celoria 26-20133 Milan, Italy (Aut. Prot, n. 295/2012-A—20 December 2012). We express the embryonic ages in hours post-fertilization (hpf) and days post-fertilization (dpf). Zebrafish AB strains obtained from the Wilson laboratory (University College London, London, UK) and Tg(fli1a:EGFP)y1 27 were maintained at 28°C on a 14-h light/10-h dark cycle. Embryos were collected by natural spawning, staged according to Ref. 28 and raised at 28°C in fish water (Instant Ocean, 0,1% Methylene Blue) in Petri dishes, according to established techniques. To prevent pigmentation, 0,003% 1-phenyl-2-thiourea (PTU, Sigma-Aldrich) was added to the fish water prior to 24 hpf. Before observations and picture acquisitions, embryos were washed, dechorionated and anaesthetized, with 0.016% tricaine (ethyl 3-aminobenzoate methanesulfonate salt; Sigma-Aldrich).
2.3 Reverse transcription and real-time quantitative polymerase chain reaction assays (RT-qPCR)
RNA was extracted from human and zebrafish embryos using TRIzol reagents (Life Technologies), following the manufacturer's protocol. For human samples and RT-qPCR experiments, Superscript II enzyme (Life Technologies) was used for cDNA synthesis. For this set of experiments, a LightCycler 480II (Roche Diagnostics, Basel, Swiss) was used. Probes were selected according to the Software Probe Finder (Roche Diagnostics) and are reported in Table 2. hGUS gene was used as reference gene in human patients and cells derived from healthy donors as standard control. For zebrafish samples, DNase I RNase-free (Roche Diagnostics) treatment was performed to avoid possible genomic contamination and 1 μg of RNA was reverse-transcribed using the “ImProm-II™ Reverse Transcription System” (Promega). RT-qPCRs were carried out in a total volume of 20 μl containing 1X iQ SYBR Green Supermix (Promega), using proper amount of the RT reaction and a mixture of oligo(dT) and random primers according to manufacturer's instructions. RT-qPCRs were performed using the Bio-Rad iCycler iQ Real-Time Detection System (Bio-Rad). For normalization purposes, rpl8 expression levels were tested in parallel with the gene of interest. Primers are reported in Table 3. Expression levels in the Y-axis were relative to the control.
| PRIMER | length | sequence | PROBE |
|---|---|---|---|
| hGUS-L | 20 | CGCCCTGCCTATCTGTATTC | 57 |
| hGUS-R | 20 | TCCCCACAGGGAGTGTGTAG | |
| hNIPBL-L | 19 | CTATGCGAACAGCCCAAAA | 55 |
| hNIPBL-R | 24 | TTCACCTTGCTTACTACCACATTT | |
| hRAD21-L | 20 | ATTGACCCAGAGCCTGTGAT | 62 |
| hRAD21-R | 20 | GGGGAAGCTCTACAGGTGGT | |
| HRUNX1-L | 18 | ACAAACCCACCGCAAGTC | 21 |
| HRUNX1-R | 23 | CATCTAGTTTCTGCCGATGTCTT | |
| HSPI1-L | 20 | CTGGAGTTCCCCAATCACAT | 25 |
| HSPI1-R | 23 | TGATTTCAGACATGACAAAAGGA |
| PRIMER | Length | Sequence |
|---|---|---|
| zrpl8-L | 21 | CTCCGTCTTCAAAGACCATGT |
| zrpl8-R | 21 | TCCTTCACGATCCCCTTGATG |
| zP1-runx1-L | 20 | ATGGCCTCCAACAGCATCTT |
| zP2-runx1-L | 20 | GAGCCGAAACTCACGGAGAC |
| zrunx1 common-R | 20 | GCAAACCCTCGCTCATCTTC |
| zspi1b-L | 19 | GCCATTTCATGGACCCAGG |
| zspi1b-R | 19 | ACACCGATGTCCGGGGCAA |
| zgata1a-L | 26 | AACGACATCTTCAATACTACACTTGC |
| zgata1a-R | 18 | GGACACCCAACGAGAAGG |
2.4 In situ hybridization, o-dianisidine and immunofluorescence analyses
Whole-mount in situ hybridization (WISH) experiments were carried out as described by Thisse et al.29 For quantification of the observed phenotypes, WISH experiments were done at least in 3 independent batches of embryos (minimum 15-20 embryos for each category). Embryos were fixed overnight in 4% paraformaldehyde (PFA, Sigma-Aldrich) in phosphate-buffered saline (PBS) at 4°C, and then dehydrated stepwise to methanol and stored at −20°C. Antisense riboprobes were previously in vitro labelled with modified nucleotides (i.e. digoxigenin, Roche Diagnostics). runx1,30 spi1b31 and gata1a32 probes were synthesized according to literature. To detect haemoglobin activity, o-dianisidine (Sigma) staining was performed as described in Ref. 33. Controls and MO-injected embryos at the same developmental stage were scored from 1 to 3 according to the intensity of the staining by microscopy, and o-dianisidine-positive cells on the yolk surface and in the Caudal haematopoietic tissue (CHT) were compared.
2.5 Injections
Injections were carried out on one- to two-cell stage embryos. Details of concentration and sequence of nipblb morpholino (nipblb-MO, Gene Tools, Oregon, US) and rad21-MO (Gene Tools) are described in Ref. 34 and Ref. 25, respectively. In all experiments, MO-injected embryos were compared to embryos at the same developmental stage injected with the same amount of a ctrl-MO that has no target in zebrafish (Gene Tools LLC). The runx1/PCS2+ construct was kindly provided by C.E. Burns18 and injected at a concentration of 200 pg/embryo.
2.6 Statistical analyses
For RT-qPCR experiments, data were statistically analysed applying one-way analysis of variance (ANOVA), defining P ≤ .05 (*), P ≤ .01 (**) and P ≤ .001 (***) as statistically significant values.35 Data were analysed using the comparative ΔΔCt method. Both ANOVA and standard deviation (SD) values refer to data from triplicate samples. In zebrafish, at least three different experiments were done for each analysis.
The degree of linear relationship between RAD21, NIPBL, RUNX1, MPL and SPI1 expression levels was calculated using Spearman's correlation coefficient (r value).
2.7 TRAM analysis
TRAM (Transcriptome Mapper) software36 allows the import, decoding of probe set identifiers to gene symbols via UniGene data parsing,37 integration and normalization of gene expression data in tab-delimited text format for the generation and analysis of transcriptome maps. We analysed the transcriptome map previously obtained from a gene expression profile datasets for normal human megakaryocytes (MK) cells derived from healthy donors.38 The dataset is composed of 19 samples previously described (Pool D in Ref. 38). In particular, we used the function "Export" of TRAM software in order to obtain normalized expression values assigned to NIPBL, RAD21, RUNX1 and MPL genes for each sample. The degree of linear relationship between RAD21, NIPBL, RUNX1, MPL and SPI1 expression levels was calculated using Spearman's correlation coefficient (r value).
3 RESULTS
3.1 Positive correlation between NIPBL and RUNX1 expression in normal megakaryocytes derived from healthy donors and bone marrow cells derived from adult AML patients
RUNX1 expression has been reported to be regulated by the cohesin subunit RAD21 and the CTCF insulator in human myelocytic leukaemia cells HL-60.26 As RUNX1 is pivotal in the differentiation of megakaryocytes and myeloid lineages, we investigated the relative expression of RAD21 and RUNX1 in two different contexts in which RUNX1 exerts important functions: the differentiation of the megakaryocytes and myeloid compartments under physiological and pathological conditions. For the megakaryocytes compartment in physiological condition, we performed in silico analyses of quantitative transcriptome maps, using TRAM (Transcriptome Mapper) software, which allows import and effective integration of data obtained by different experimenters, experimental platforms and data sources.36 In megakaryocytes (MK) derived from healthy donors, RAD21 expression did not correlate with the expression levels of RUNX1 (Figure 1A). Conversely, we found a positive correlation between the expression of RUNX1 and that of NIPBL, another member of the cohesin complex (Figure 1B). To explore the myeloid compartment under pathological condition, we used bone marrow (BM) cells derived from adult AML patients. Similar to TRAM analyses, when RAD21 and RUNX1 expressions were investigated in a cohort of 34 AML adult patients without anomalies in chromosome 21 that contains the RUNX1 locus, no significant correlation was reported (Figure 1C). Conversely, we observed the positive NIPBL/RUNX1 correlation already detected in megakaryocytes (Figure 1D).
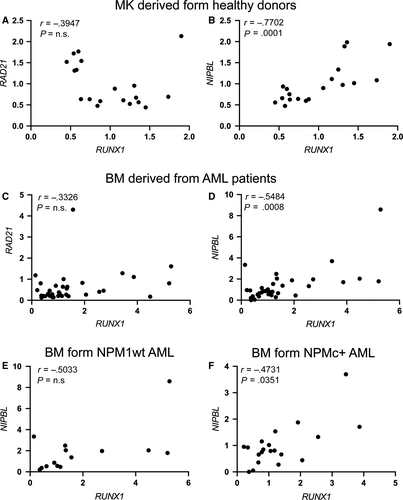
We previously showed that NIPBL transcript abundance is decreased in AML patients carrying the mutated NUCLEOPHOSMIN1 (NPM1), which transfers NPM1 in the cytoplasm (NPMc+), compared to the NPM1 wild-type (NPM1wt).34 Therefore, we analysed the correlation between the expression of NIPBL and RUNX1 in BM cells derived from 20 patients NPMc+, selected among the 34 AML patients, compared to 14 patients NPM1wt and found a significant positive correlation in NPMc+ but not in NPM1wt AML patients (Figure 1E-F). Taken together, these findings suggest a new role for NIPBL, different from that of RAD21, in the regulation of RUNX1 expression and that aberrant expression of NIPBL, such as in AML patients with NPM+ mutation, might lead to alteration in RUNX1 transcript levels.
3.2 Knock-down of nipblb specifically reduces runx1 expression in zebrafish
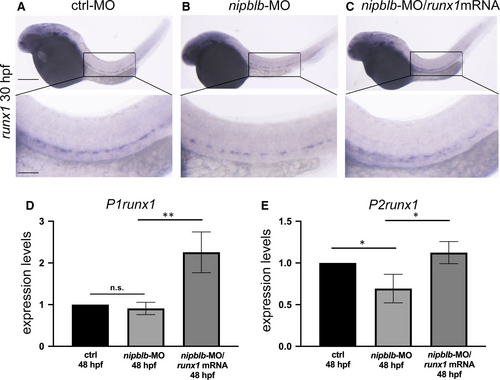
To confirm the positive correlation between NIPBL and RUNX1 observed in human, we took advantage of a zebrafish model with down-regulation of nipblb, the orthologue of the human NIPBL, previously generated in our laboratory.34 The expression of runx1 was analysed in embryos at 30 and 48 hpf as definitive HSCs arise from the vascular endothelium from these developmental stages. Moreover, we verified that both P1-P2runx1 isoforms were highly expressed from 24 hpf (Figure S1). WISH analyses showed a reduction of the runx1 transcript in the aorta-gonad mesonephric (AGM) tissue in nipblb-MO-injected embryos compared to controls at the same developmental stage. The injection of the full-length runx1/mRNA rescued this phenotype as expected (Figure 2A-C). As the full-length runx1 riboprobe does not distinguish between the P1- and P2runx1 isoforms present in zebrafish,8 we performed RT-qPCR analysis of both isoforms, revealing a significant reduction exclusively in P2runx1 transcript levels following nipblb down-regulation. The expression of both isoforms was increased in embryos injected with nipblb-MO and runx1mRNA, confirming the efficacy of the runx1 overexpression (Figure 2D-E). These results provide evidence that nipblb knock-down causes the reduction of runx1 in zebrafish, confirming the positive correlation between NIPBL and RUNX1 expression observed in normal megakaryocytes and in BM of AML patients.
3.3 NIPBL-mediated RUNX1 down-regulation impairs the expression of RUNX1 target genes
We further verified whether the NIPBL-mediated RUNX1 reduction affects the expression of RUNX1 haematopoietic downstream targets. In MK cells derived from healthy donors, we observed a positive correlation between the expression of RUNX1 and that of MPL gene, the marker of megakaryocyte/platelet differentiation (Figure 3A).39 In BM cells derived from AML human patients, we showed a positive correlation between the expression of RUNX1 and its targets SPI1, the marker of myeloid precursors (Figure 3B).40 The expression of runx1 targets gata1a and spi1b was investigated also in zebrafish in nipblb-MO-injected embryos and controls at 48 hpf. The expression of gata1a, analysed by RT-qPCR, was significantly decreased following nipblb down-regulation as a result of runx1 reduction. Indeed, the injection of the runx1mRNA in the nipblb-MO-injected embryos rescued the gata1a expression (Figure 3C). Conversely, the expression of spi1b was significantly increased in both nipblb-MO- and nipblb-MO/runx1mRNA-injected embryos (Figure 3D). Consistent with the role of runx1 in the positive regulation of the erythroid lineage, mature circulating erythrocytes, visualized by o-dianisidine staining at 48 hpf, were drastically reduced in nipblb-MO-injected embryos (70%; N = 140), compared to controls (Figure 3E-F). This phenotype is not caused by alterations in vascular tree development as shown in the Tg(fli1a:EGFP)y1 embryos (Figure S2), or absence of blood flow (data not shown). The reduction of o-dianisidine-positive erythrocytes was rescued in the 75% of the nipblb-MO-runx1mRNA-injected embryos (N = 113) (Figure 3G), confirming that the phenotype is dependent on nipblb-mediated runx1 reduction.
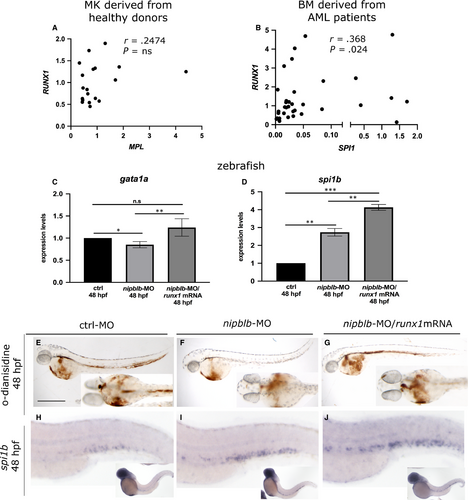
WISH analyses of spi1b expression showed the increase of the transcript in the CHT of nipblb-MO-injected embryos (Figure 3H-I) confirming the RT-qPCR data and our previous findings.34 In agreement with the positive regulation exerted by runx1 on spi1b, the injection of the runx1/mRNA further enhanced this phenotype (Figure 3J).40
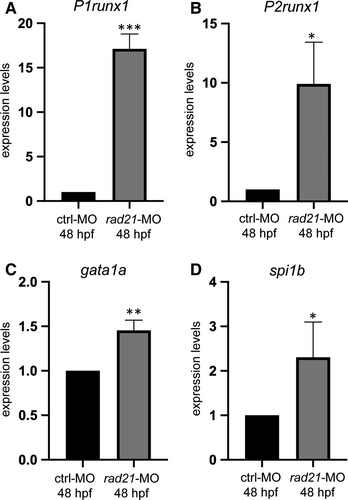
As it has been previously demonstrated that rad21, another member of the cohesin complex, regulates runx1 in zebrafish embryos during primitive haematopoiesis,25, 26 we further verified the expression of runx1 during definitive haematopoiesis following rad21 down-regulation by means of morpholino injection.25 rad21-MO-injected embryos at 48 hpf showed an increased expression of both P1 and P2runx1 isoforms and a consequent increase in the expression of the runx1 downstream targets gata1a and spi1b (Figure 4A-D). These data are in agreement with the negative regulation exerted by RAD21 on RUNX1 expression reported in the myeloid HL60 cell line.26
4 DISCUSSION
The transcription factor RUNX1 is a pivotal gene in the development and differentiation of HSCs: as transcription factor, it controls the expression of master genes involved in megakaryocytes and myeloid lineages differentiation, and it interacts with different proteins fundamental during haematopoiesis. Somatic translocations and mutations of RUNX1 are causative of haematological diseases such as myelodysplastic syndrome, acute myeloid leukaemia, acute lymphoblastic leukaemia, chronic myelomonocytic leukaemia and acute megakaryoblastic leukaemia with familial platelet disorder. In addition, dysregulation of RUNX1 expression might lead to impaired haematopoiesis and the insurgence of a pathological condition. Among the genes discovered to regulate RUNX1, there is RAD21, a member of the cohesin complex, and the CTCF insulator.26 In human K562 cells and murine and zebrafish models, RAD21 and CTCF bind to a cis-regulatory element (CRE) enhancer located in an intron between the P1 and P2RUNX1 promoters, associated with RNApolII.41 As cohesins preferentially bind to transcriptionally active genes and recruit RNAPolII and chromatin modifiers to activate gene transcription,42 it would have been expected that RAD21 positively regulates RUNX1 transcription by binding to the CRE between P1 and P2 promoters. This finding is reported by Supernat and colleagues 43 in patients with endometrial cancers. However, in a zebrafish mutant for Rad21 the expression of runx1 was reduced 25 and the P1 and P2runx1 isoforms were differently expressed: P1 isoform was increased, while P2 was not varied or even decreased following Rad21 depletion.26 Moreover, the silencing of RAD21 in the human HL60 leukaemic cell line leads to an enhanced expression of RUNX1 indicating that RAD21 might also repress RUNX1 expression.26 In our study, we did not observe a significant correlation between the expression of RAD21 and RUNX1 neither in megakaryocytes derived from healthy donors, nor in bone marrow cells derived from a selected cohort of adult AML patients. However, we showed that during definitive haematopoiesis, the down-regulation of rad21 in zebrafish enhances the expression of both P1 and P2runx1 isoforms leading to impaired expression of the runx1 downstream targets gata1a and spi1b.
The different members of the cohesin complex can exert similar or individual functions in the regulation of gene expression. For example, Zuin et al44 demonstrated that NIPBL binds to chromatin independently in time and space than other cohesins, revealing a new role for NIPBL as transcriptional regulator not linked to the cohesin complex. In this work, we demonstrate that NIPBL exerts a different regulation on RUNX1 expression than RAD21. Indeed, in three different contexts: normal megakaryocytes derived from healthy donors, bone marrow cells derived from adult AML patients and zebrafish embryos with nipblb down-regulation, we demonstrate a positive correlation between NIPBL and RUNX1 expression.
The NIPBL-mediated RUNX1 dysregulation affects the RUNX1 downstream targets responsible for the differentiation of the erythroid and myeloid lineages. RUNX1 augmented GATA1-mediated promoter activation; in this regard, the decrease in RUNX1 transcription/activity leads to down-regulation of the erythroid GATA1 transcription factor.45 Interestingly, cohesins-haploinsufficient cells presented enriched or depleted GATA1 consensus binding sites indicating that they can modulate GATA1 activity directly or through other molecules.12, 46
Also the SPI expression is positively regulated by RUNX1, facilitating the interaction between the SPI enhancer and its proximal promoter.47 Indeed, we observed a positive correlation between RUNX1 and SPI1 in human samples and in zebrafish when we forced runx1 expression. However, following nipblb down-regulation, we also observed an increase in spi1b expression according to our previous data.34 This result does not correlate with the runx1 reduction and its positive activity on spi1b expression and raises three possibilities: first that the increased number of myeloid precursors, previously reported in zebrafish following nipblb-MO injection,34 leads to an augmented number of cells expressing spi1b with a consequent total increase of spi1b transcript. Second, it has been reported that the chromatin structure at the spi1b/PU.1 locus could be differentially regulated during the different stages of haematopoiesis,11 suggesting the possibility that other mechanisms than RUNX1 might control spi1b expression. For example, we demonstrated that the canonical Wnt pathway, modulated by nipblb, has a pivotal role in regulating spi1b myeloid expression during definitive haematopoiesis in zebrafish.34 Moreover, in vitro and in vivo studies demonstrated that forced expression of gata1 down-regulates spi1b, while forced expression of spi1b down-regulates gata1.48-50 In this scenario, the nipblb-mediated runx1 down-regulation might lead to spi1b enforced expression that, in turn, reduces gata1a expression. Alternatively, the two P1 and P2runx1 isoforms might exert different functions on spi1b regulation. Indeed, as for the case of Rad21 zebrafish mutants,26 we demonstrated that the down-regulation of nipblb differently affects the two isoforms by significantly reducing only the P2runx1. Third, it has been demonstrated that NIPBL might regulate SPI1 by itself, encompassing the Runx1 regulation.44
Although in this work we did not address the mechanism through which NIPBL regulates RUNX1 expression, we demonstrated that NIPBL positively regulates RUNX1 transcription and that the link between NIPBL dysregulation and RUNX1-driven haematopoietic defects might explain haematological malignancy occurrence. Thus, also in the absence of RUNX1 translocation or mutations, additional factors such as defects in the expression of NIPBL observed in AML patients might contribute to haematological diseases.
ACKNOWLEDGMENTS
The authors thank Carol Burns for the runx1 mRNA. They also thank Dorela Meta and Giulia Salmoiraghi for technical help in data preparation.
CONFLICT OF INTEREST
The authors declare no competing financial interest.
AUTHOR CONTRIBUTION
MM, GF, CS, AR, AP, MCP, GG and LF contributed to the study. MP and MF provided patients samples. EB, AP, GC, AB, and AM designed and performed the experiments and analysed the data. AP designed and organized the experiments and analysed the data; and wrote the manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




