Complete Mitochondrial Genomes Provide Evidence of a Killer Whale Refugium Off the Coast of Japan During the Last Glacial Maximum
Funding: This work was supported by the Human Frontier Science Program and National Geographic Society.
ABSTRACT
Aim
During glacial periods, highly mobile species often shifted to warmer, ice-free regions known as refugia, which frequently maintained higher genetic diversity than newly colonised areas after glacial retreat. We analyse complete mitogenome sequences from 11 killer whale samples in Nemuro Strait to test the hypothesis that waters around Japan may have preserved a refugial population of killer whales that retained historical genetic diversity.
Location
Western North Pacific.
Taxon
Orcinus orca ater, Cetacea.
Methods
Sequencing of complete mitochondrial genomes (16,387 bp) from 11 killer whale samples collected in the Nemuro Strait, southern Okhotsk Sea. Distribution of haplotypes in the social network was visualised using the data on social associations. The probability of individuals belonging to one or more possible reproductive groups was estimated based on 17 microsatellite loci.
Results
Seven samples shared a haplotype common in the western North Pacific, one had a haplotype previously found only in the eastern North Pacific, and three exhibited novel haplotypes. Killer whales with different haplotypes were connected into a single social network, but some degree of social segregation is evident within the network. No significant genetic clustering based on microsatellite markers was detected between Nemuro Strait and areas near the Kamchatka Peninsula.
Main Conclusions
With five distinct mitogenomes, Nemuro Strait now ranks second in mitogenomic diversity after the central Aleutian Islands. These findings support the hypothesis of a glacial refugium off Japan preserving a portion of pre-glacial genetic diversity. The lack of genetic clustering between Nemuro Strait and areas near the Kamchatka Peninsula indicates that all R-type killer whales in the western North Pacific belong to a single population. The low mitogenomic diversity north of Nemuro Strait likely reflects a founder effect, where a few groups colonised the region after the LGM, while most of the population with higher genetic diversity remained near Japan.
1 Introduction
Understanding the history of species' vicariance and range expansion offers critical insights into climate change, temporal variations in environmental and oceanographic conditions, and species' population dynamics. Mapping geographic patterns of genetic diversity is essential for reconstructing species' evolutionary histories. This is particularly relevant for cetaceans, whose ability to traverse vast distances and the absence of clear physical barriers in the open ocean make their evolutionary trajectories more complex and less predictable than those of terrestrial mammals.
The killer whale (Orcinus orca) is globally distributed, with distinct local forms or ecotypes in many regions (Ford et al. 1998; Saulitis et al. 2000; Pitman and Ensor 2003). In the North Pacific, three reproductively isolated ecotypes have been identified: the fish-eating “residents” (R-type) and mammal-eating “transients” or Bigg's killer whales (T-type) (Bigg 1987; Ford et al. 1998), and the “offshore” killer whales, which likely specialise in hunting sharks (Ford et al. 2011). Recently, the fish-eating and mammal-eating ecotypes were recognised as distinct subspecies: O. orca ater and O. orca rectipinnus, respectively (Morin et al. 2024).
The origins of these ecotypes remain debated, with some suggesting sympatric speciation (Moura et al. 2015) and others proposing secondary contact between distinct lineages (Foote et al. 2011; Foote and Morin 2015, 2016). The latter hypothesis posits that fish-eating residents may have colonised the North Pacific later than the mammal-eating transients, though their colonisation patterns have not been extensively studied.
Globally, killer whales exhibit low mtDNA variation (Hoelzel et al. 2007), with resident killer whales showing especially low diversity. In the eastern North Pacific, only three complete mitogenome haplotypes have been identified from Washington State, British Columbia, and Alaska (Morin et al. 2015), while just two haplotypes have been found in the western North Pacific from northeastern Kamchatka to the northern Kuril Islands (Filatova et al. 2018). The only notable hotspot of genetic diversity was found in the Aleutian Islands, where 11 mitogenome haplotypes have been documented (Morin et al. 2015). This pattern, with higher diversity in the central North Pacific and lower diversity at the periphery, led Filatova et al. (2018) to suggest a founder effect, proposing that resident killer whales persisted in the central Aleutian Islands during the last glacial maximum (LGM) while surrounding areas were covered by ice. As the ice receded, small groups of whales colonised peripheral regions, resulting in reduced genetic diversity. Consistent with this, coalescent-based estimations of effective population size through time, based on a single Alaskan resident genome, infer an order of magnitude decline during the last glacial period (Moura et al. 2014; Foote et al. 2021).
During the LGM, the only ice-free areas in the North Pacific, besides the central Aleutians, were in the south, around California in the east, and Japan in the west. The southernmost resident population in the eastern North Pacific, known as the Southern Resident Community, ranges from central California to southeast Alaska, but primarily inhabits the waters in and around the Salish Sea. Although their range overlaps with another resident population, the two populations do not appear to interbreed, despite belonging to the same ecotype (Parsons et al. 2013). Filatova et al. (2018) suggested that Southern Residents may be descendants of whales that survived the LGM off California and subsequently lost contact with the descendants of whales from the central Aleutians.
In the western North Pacific, until recently, killer whale samples were available only as far south as the northern Kuril Islands. Mitani et al. (2021) analysed Japanese killer whale samples but did not provide sequences of enough length to distinguish ecotypes. Recently, Filatova et al. (2023) reported two new mitochondrial control region (D-loop) haplotypes in the Nemuro Strait between Hokkaido and Kunashir Islands, indicating a potential glacial refugium in this area. In this study, we analyse complete mitogenome sequences from 11 killer whale samples collected in Nemuro Strait to test the hypothesis that waters around Japan may have preserved a refugial population of killer whales that retained historical genetic diversity. We discuss the implications of our findings for understanding the history of killer whale colonisation in the North Pacific.
2 Methods
2.1 Data Collection
The data for this study were collected from free-ranging killer whales in Nemuro Strait in May–June 2021 and 2022. We approached the whales in a 5 m inflatable boat with a 30 hp outboard engine. To identify the individual whales and prevent sampling the same animals twice, we photographed the whales with a Canon 5D Mark III camera with a 70–200 mm lens for further identification. We identified the whales individually from the photographs using their natural markings (shape of the dorsal fin and saddle patch, scars, and scratches). Skin samples for genetic analysis were obtained through remote biopsy using a crossbow and a floating arrow. The mitogenome sequences for the cross-Pacific comparison of complete mitogenomes were downloaded from Genbank (see Table S1 for accession numbers).
2.2 Genetic Analysis
The samples were placed in 96% ethanol for storage and transportation. DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen, Valencia, CA) according to the protocol provided by the manufacturers. The ecotype of the whales was confirmed with Sanger sequencing of the mitochondrial control region as described in Filatova et al. (2023). For the whole mitogenome sequencing, genomic DNA was fragmented with sonication (Covaris) to approximately 300 bp, and quantified using a Tape Station (Agilent). Libraries were built with the NebNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs Inc.) following the manufacturer's instructions, amplified with individually indexed primers and equimolar pools. The pooled libraries were then sequenced across a partial S4 run of the Illumina Novaseq 6000 platform using 150-bp paired-end Illumina sequencing.
Following the protocol of Jourdain et al. (2024), demultiplexed reads from the enriched libraries were processed using AdapterRemoval2 (Schubert et al. 2016) to trim residual adapter sequence contamination and to remove adapter dimer sequences as well as low-quality (Q < 30) stretches at the read ends. Filtered reads greater than 50 bp were then mapped using the BWA-MEM algorithm (Li 2013) to a fasta file for the mitochondrial genome sequence of a Norwegian killer whale with haplotype ENAHN1 (NCBI accession: NC_023889.1; Morin et al. 2010), requiring a mapping quality greater than 30. Clonal reads were collapsed using the rmdup function of SAMtools v. 1.13 (Li et al. 2009). All sites were covered by at least three independent sequencing reads.
Mitochondrial sequences were then converted from BAM to FASTA format using the -dofasta option in ANGSD (Korneliussen et al. 2014). We followed this same conservative approach in our mapping of mitogenomes and identification of sequence variation in this study, as Morin et al. (2010, 2015) and Jourdain et al. (2024). Polynucleotide repeat regions; one of between 9 and 14 Cs in a row (positions 1129–1137), and another region of seven to eight As in a row (positions 5211–5217) were shortened to a fixed set of nine Cs and seven As, respectively, to avoid introducing potentially erroneous variation into downstream analyses.
Complete mitogenome sequencing revealed a 4A sequence at position 15,470 in all 11 samples, whereas all other resident haplotypes have 5As at that position. To resolve this issue, we performed Sanger sequencing of the control region, which is less prone to misidentification of polynucleotide repeat regions than high-throughput sequencing (see Filatova et al. 2023 for details), which yielded a 5A sequence at that position. Therefore, the complete mitogenome sequences were edited accordingly.
We used the “haploNet” function in the “pegas” library (Paradis 2010) in R to build a haplotype network. By default, the haplotype network is built using an infinite site model (i.e., uncorrected or Hamming distance) of DNA sequences and pairwise deletion of missing data.
We defined the allelic composition of 17 microsatellite loci of nuclear DNA for 5 samples from Nemuro Strait: KWM12 (Hoelzel et al. 1998), EV1, EV5 (Valsecchi and Amos 1996), TtruGT39, TtruGT48 (Caldwell et al. 2002), Dde66, Dde72 (Coughlan et al. 2006), 464/465 (Fullard et al. 2000), DlrFCB12, DlrFCB13, DlrFCB17 (Buchanan et al. 1996), MK5, MK9 (Krützen et al. 2002), Ttr04, Ttr34, Ttr11, Ttr48 (Rosel et al. 2005). One of the primers in each pair was labelled with a fluorescent dye (FAM, R6G, ROX, or TAMRA). Amplification of the selected regions was performed using Mag Mix 2025 PCR master mix (Dialat Ltd., Russia). All primers were synthesised by JSC Syntol (Russia). We performed capillary electrophoresis on a 3500 Genetic Analyser (Applied Biosystems, Thermo Fisher Scientific, USA) in the presence of the GeneScan 500 LIZ dye Size Standard (Applied Biosystems, Thermo Fisher Scientific, USA). Fragment lengths were determined using the Gene Mapper v.4.1 software (Applied Biosystems, Thermo Fisher Scientific, USA).
For the comparison of microsatellite markers with other areas of the western North Pacific, the data were extracted from the database that summarised the results of our previous studies in the area. There were a total of 72 samples: 47 from Avacha Gulf, southeastern Kamchatka, 2 from Karaginsky Gulf, northeastern Kamchatka, 15 from the Commander Islands, and 8 from the northern Kuril Islands.
We checked the possible presence of null alleles using MICRO-CHECKER 2.2.1 (Van Oosterhout et al. 2004). We estimated the expected and observed heterozygosity values in the total sample and in the four geographic groups (Commander Islands, Avacha Gulf, northern Kuril Islands, and Nemuro Strait) separately, as well as the level of differences between groups (Fst), using Arlequin 3.5.1 software (Excoffier and Lischer 2010). We estimated the average number of alleles per locus, allelic richness values, Fis, and the probability of deviation from Hardy–Weinberg equilibrium (heterozygote deficit) using the FSTAT v.2.9.3 program (Goudet 1995).
In these calculations, we excluded two individuals (one from the Commander Islands and one from the northern Kuril Islands sample) for which values were obtained for fewer than 12 loci. The level of missing data in the remaining sample was 2.0%.
We used Arlequin 3.5.1 to estimate haplotype and nucleotide diversity, conduct neutrality tests, and estimate distances based on the number of pairwise differences (Φst) and haplotype frequencies for the complete mitochondrial genome sequences of killer whales of the central Aleutian Islands, Nemuro Strait, and the Northwest Pacific (see below).
We performed pairwise relatedness estimation between all individuals in the total sample and separately in the Nemuro Strait sample using ML-Relate software (Kalinowski et al. 2006).
The probability of individuals belonging to one or more possible reproductive groups based on 17 microsatellite loci was estimated using the clustering algorithms implemented in STRUCTURE v. 2.3.3 (Pritchard et al. 2000). We assumed the “LOCPRIOR” model that uses the sampling location as default information to assist clustering.
To reconstruct the demographic history of the killer whale population in the waters off the western Pacific coast based on the allelic composition of microsatellite loci, we used the R package VarEff (Nikolic and Chevalet 2014). Given the lack of genetic differences between samples from different geographic areas, we performed the analysis for the generalised sample. We used 5 × 10−4 as the mutation rate (a value accepted for many mammal species) and 150 for the actual effective size prior (based on preliminary estimates using NeEstimator v.2 software, results not shown). We limited our analysis to the last 1000 generations, which, based on the estimated generation time for killer whales (see Taylor et al. 2007; Foote et al. 2019), corresponds to approximately 25,000 years. This period includes the LGM, through the Holocene, and on to the present. All parameters and priors used for the analysis are given in Table S2.
In addition, we tested the probability of postglacial founding of the R-type killer whale population in the Northwest Pacific by migrants (founders) from two potential refugial areas (central Aleutian Islands and waters around Japan) by evaluating the pattern of mitochondrial lineage distribution. For this, we applied the approximate Bayesian computation (ABC) method implemented in the DIYABC 2.1.0 program (Cornuet et al. 2014).
We used complete mitochondrial genome sequences (16,388 bp, 17 haplotypes) of 19 individuals from the central Aleutian Islands, 11 individuals from Nemuro Strait, and 24 individuals from the Northwest Pacific north of Nemuro Strait. The mitogenome sequences from the central Aleutian Islands were downloaded from Genbank (see Table S1 for accession numbers), and sequences from the Northwest Pacific north of Nemuro Strait were obtained from our previous study (Filatova et al. 2018). In order to account for the deletion at position 5217, we designated it as a substitution.
- Scenario 1 assumed the formation of the modern pool of mitotypes of killer whales of the Northwestern Pacific as a result of their separation from the group from Nemuro Strait.
- Scenario 2 assumed the formation of the Northwest Pacific mitotypes pool from the group of the central Aleutian Islands, and
- Scenario 3 assumed formation of the Northwest Pacific mitotypes pool from a mixed group originating from both refugia.
We allowed high variation in the actual effective population size in three geographic groups (10–10,000), in the time of the beginning of the population decline and its separation into two lineages (4000–400 generations, approximately 100–10 kya), and in the bottleneck duration (up to 4000 generations, i.e., up to the present). We assumed the time of separation of the group of lineages that formed the Northwest Pacific pool in the range of 40–1000 generations (1–25 ky).
We used the HKY model for sequence analysis (Morin et al. 2010) and left the default values defining the mutation process. Specifically, a range of 1 × 10−9 to 1 × 10−7 per site per generation was set for the mean mutation rate parameter. This range includes values calculated previously for the total mitochondrial genome of killer whales (1.50 × 10−9 to 3.83 × 10−9 per site per year or 3.75 × 10−8 to 9.56 × 10−8 per site per generation; Morin et al. 2010).
Scenarios and all settings, and priors used are summarised in Table S3.
Subsequent analyses were performed based on a reference table built by simulating 6,000,000 data sets.
2.3 Social Network Analysis
For the social network analysis, we defined association as the presence in the same spatial group. A spatial group was defined as animals within four body lengths of each other. All animals photographed in the same spatial group at least once in the sampling period (1 day) were considered associated in that sampling period. To assess the social associations between the whales, we calculated the simple ratio association index for each pair of whales using SocProg 2.9 (Whitehead 2009). Based on these indices, a social network was visualised using the Fruchterman–Reingold layout algorithm in Gephi 0.10 (Cherven 2013).
3 Results
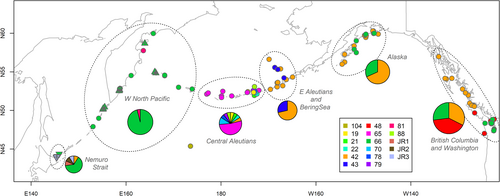
We successfully amplified and sequenced complete mitogenomes (16,387 bp) of 11 samples of resident (R-type) killer whales from Nemuro Strait (Table 1). Among the analysed samples, seven had haplotype 66, one had haplotype 42 previously found only in the eastern North Pacific, and three samples had new previously undescribed haplotypes. These sequence data have been submitted to the GenBank databases under accession numbers PQ878649, PQ878650, and PQ878651. New haplotypes were not clustered together and differed from haplotype 66 by one or two substitutions (Figure 1). With five different mitogenomes, Nemuro Strait is now the area with the second highest mitogenome diversity after the central Aleutian Islands (Figure 2).
| Date | Time | Lat | Lon | Sex | ID | Haplotype | Microsatellites |
|---|---|---|---|---|---|---|---|
| 08.05.2021 | 12:40 | 44.02197 | 145.47803 | Male | KU201 | 66 | N |
| 08.05.2021 | 13:13 | 44.05888 | 145.48677 | Female | KU203a | 66 | N |
| 09.05.2021 | 10:38 | 44.36120 | 145.91419 | Female | KU213 | 66 | N |
| 15.05.2021 | 14:27 | 44.06199 | 145.49636 | Male | KU222 | 66 | N |
| 27.05.2021 | 11:18 | 43.97490 | 145.43383 | Male | KU225 | JR1 | N |
| 02.06.2021 | 14:08 | 43.93758 | 145.32731 | Female | KU245 | JR2 | N |
| 03.06.2022 | 14:15 | 43.97092 | 145.37049 | Male | KU307 | 66 | Y |
| 07.06.2022 | 15:19 | 43.96029 | 145.38185 | Female | KU340 | JR3 | Y |
| 08.06.2022 | 11:31 | 43.99672 | 145.45195 | Female | KU325 | 66 | Y |
| 09.06.2022 | 12:47 | 43.96192 | 145.37698 | Male | KU333 | 42 | Y |
| 09.06.2022 | 17:03 | 43.97950 | 145.47265 | Male | KU350 | 66 | Y |
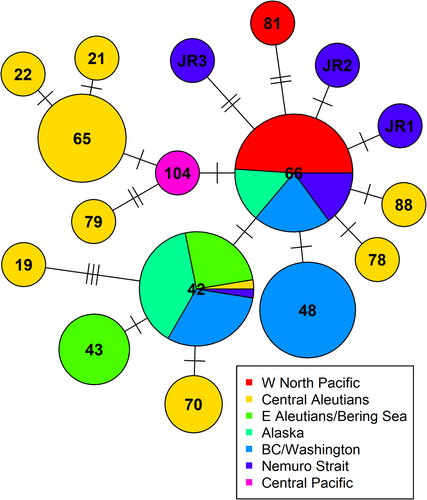
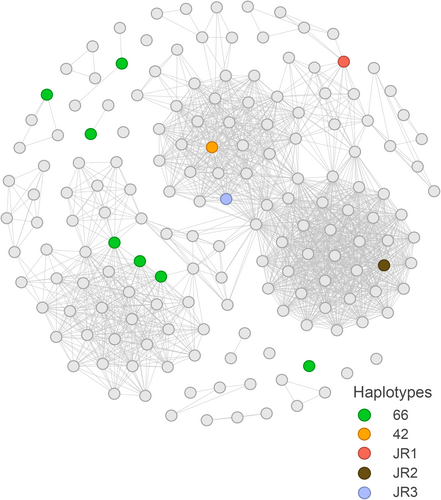
Haplotypes were distributed unevenly in the social network (Figure 3). Haplotype 66 was found in three associated whales in one of the major social clusters, as well as in four whales that were not assigned to any major social clusters. Haplotype JR2 was found in the largest social cluster; haplotypes 42 and JR3 were found in the third major cluster, closely associated with the cluster that included JR2, and haplotype JR1 was found in a loose cluster associated with the previous two. The major social cluster that included three animals with haplotype 66 and two other major clusters that included new haplotypes and haplotype 42 were connected through only two whales, while the social clusters that contained new haplotypes and haplotype 42 were substantially more interconnected (Figure 3).
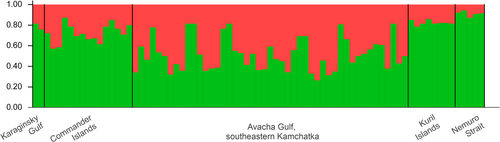
To test whether Nemuro Strait killer whales are a part of the larger population of the western North Pacific, we analysed 17 microsatellite loci of 75 R-type individuals from Kamchatka and the Commander and Kuril Islands, including five whales from Nemuro Strait (Figure 4). The number of alleles per locus ranged from 2 to 5; the mean was 3.5. No evidence for scoring error due to stuttering, for large allele dropout, and for null alleles was found for any of the 17 loci.
Allelic diversity indices and Fst values estimated for microsatellite loci are shown in Tables 2 and 3, respectively. No significant deviation from Hardy–Weinberg equilibrium was observed for any of the loci in any of the geographic samples, nor in the total sample. All groups had similarly low levels of heterozygosity. No statistically significant differences in allele frequencies were found for any pair of geographic groups.
| Com | Av | Kur | NWP | Nem | Total | |
|---|---|---|---|---|---|---|
| n | 14 | 47 | 7 | 70* | 5 | 75 |
| A | 2.941 | 3.235 | 2.765 | 3.412 | 2.471 | 3.529 |
| AR | 2.210 | 2.290 | 2.287 | 2.278 | 2.219 | n/a |
| He | 0.474 | 0.499 | 0.511 | 0.498 | 0.506 | 0.499 |
| Ho | 0.485 | 0.493 | 0.535 | 0.497 | 0.524 | 0.498 |
| Fis | −0.024 | 0.011 | −0.051 | 0.001 | −0.038 | 0.002 |
| p-value | 0.666 | 0.347 | 0.759 | 0.491 | 0.657 | 0.463 |
| Com | Av | Kur | NWP | |
|---|---|---|---|---|
| Com | ||||
| Av | −0.00118, p = 0.497 | |||
| Kur | 0.0085, p = 0.200 | 0.00087, p = 0.423 | ||
| NWP | n/a | n/a | n/a | |
| Nem | 0.01518, p = 0.123 | 0.01932, p = 0.115 | 0.0086, p = 0.263 | 0.01704, p = 0.150 |
Diversity indices and neutrality test results, as well as the estimated distances between groups of mitochondrial genomes from the three geographic regions (central Aleutian Islands, Nemuro Strait, and Northwest Pacific north of Nemuro Strait) are shown in Tables 4 and 5, respectively. The samples from the central Aleutians had the highest diversity, differing significantly from the two more western regions by both criteria. In contrast, the differences between the samples from the Nemuro Strait and the Northwestern Pacific north of Nemuro Strait were weak and not statistically significant (taking into account the Bonferroni correction).
| n | h | H | pi, % | Tajima's D | Tajima's D p-value | FS | FS p-value | |
|---|---|---|---|---|---|---|---|---|
| CAI | 19 | 9 | 0.731 | 0.0174 | −1.067 | 0.138 | −2.096 | 0.116 |
| Nem | 11 | 5 | 0.618 | 0.0055 | −1.791 | 0.013 | −2.310 | 0.009 |
| NWP | 24 | 2 | 0.083 | 0.0015 | −1.733 | 0.012 | 0.359 | 0.332 |
| CAI | Nem | NWP | |
|---|---|---|---|
| CAI | 0.2860, p = 0.0000 | 0.4014, p = 0.0000 | |
| Nem | 0.3155, p = 0.0000 | 0.0358, p = 0.1299 | |
| NWP | 0.6187, p = 0.0000 | 0.1775, p = 0.0293 |
The mean relatedness between individuals in the pooled sample was estimated to be 0.088, ranging from 0 to 0.633 (including pairs of individuals from different regions). For the five individuals from Nemuro Strait, the mean relatedness was 0.133, with a range from 0 to 0.364.
The best estimate of the number of clusters with the “Admixture-LOCPRIOR” model was 1, as the maximum value of the estimated Ln probability of data was reached at K = 1 and decreased with larger K values (tested up to K = 4). Therefore, all analysed individuals, including the whales from the Nemuro Strait, belong to the same population. Measures of Fst comparing five sampling locations are shown in Table 3.
Analysis of the population's demographic history based on microsatellite locus allele data showed that the population may have experienced a significant decline since approximately 300–500 generations (~7500–12,500 years) ago (Figure S1).
Among the three scenarios of postglacial founding of the R-type killer whale population in the Northwest Pacific by migrants from two potential refugia (central Aleutian Islands and waters around Japan) tested using the approximate Bayesian computation, the resulting posterior probability was determined as 0.6571 for scenario 1 (migration from Japan), 0.060 for scenario 2 (migration from the central Aleutian Islands), and 0.282 for scenario 3 (migration from both refugia) (Table S4, Figure S2). No values lower than 5% or greater than 95% were found neither by computing the proportion of simulated data that has a value below the value of the observed dataset for each summary statistic (pre-evaluate scenario-prior combination results, Table S5) nor for the proportion of datasets simulated from the posterior that have a value lower than the observed dataset (model checking results, Table S6). The posterior distribution of parameters estimated for scenario 1 is shown in Table S7.
4 Discussion
From 11 samples of resident (R-type) killer whales from Nemuro Strait, we identified three new and two previously known mitogenome haplotypes. This discovery establishes Nemuro Strait as the region with the second-highest mitogenome diversity of resident killer whales after the central Aleutian Islands. It supports the hypothesis by Filatova et al. (2018) that a glacial refugium existed off Japan (or in waters to the south of Japan) during the LGM, preserving a portion of the pre-glacial genetic diversity.
Haplotype 66 was the most common among the Nemuro Strait killer whales, appearing in seven out of the eleven samples. This is expected, as haplotype 66 is the most prevalent in the western North Pacific, with only one R-type whale with a different haplotype (81) previously reported in this area (Morin et al. 2015). More surprising was the presence of haplotype 42 in Nemuro Strait, which until now had only been found in the eastern North Pacific, ranging from northern Vancouver Island (Northern Resident population) to the eastern Aleutians.
Of note, haplotype 66 is also found in the eastern North Pacific, from the Gulf of Alaska to Washington State (Southern Resident population), but it is absent in the central and eastern Aleutians (Figure 2), despite being ancestral to two Aleutian haplotypes (Figure 1). This pattern suggests that most resident killer whales in the western North Pacific likely descended from the Japanese refugium rather than from the Central Aleutian refugium.
We observed no significant genetic clustering based on microsatellite markers in our sample that included R-type whales from most of their range in the western North Pacific (Filatova et al. 2019): Karaginsky Gulf (northeastern Kamchatka), Avacha Gulf (southeastern Kamchatka), Commander Islands, northern Kuril Islands, and Nemuro Strait. This likely indicates that all R-type killer whales in the western North Pacific belong to a single population. Similarly, microsatellite analysis by Parsons et al. (2013) found no population structure among the Russian samples but did reveal significant genetic boundaries at Buldir Pass between the western and central Aleutians.
The distribution of mitogenomes aligns with this finding, as haplotype 66 predominates in all areas west of Buldir Pass but is absent in the central and eastern Aleutians east of the pass. Together, these observations suggest that R-type killer whales likely colonised the western North Pacific from the glacial refugium off Japan, extending northwest up to the western Aleutians.
Microsatellite locus analysis does not appear to be suitable for testing this hypothesis through reconstruction of the population's demographic history. We currently lack allele frequency data for the central Aleutian Islands region, which is known to differ from populations further west (Parsons et al. 2013). Meanwhile, our data indicate that killer whales from various regions of the Northwest Pacific and the Nemuro Strait can presently be regarded as a single panmictic population. Therefore, comparing a sample representing one part of this population against the rest is unlikely to yield meaningful results.
At the same time, the social structure typical of R-type killer whales—where females remain in their natal groups for life and gene flow is primarily mediated by males (Bigg et al. 1990)—may contribute to the regional structuring of maternal (mitochondrial) lineages in the areas where these groups are distributed. Our approximate Bayesian computation analysis of the possible formation of the current geographic distribution of mitochondrial lineages, as well as the analysis of Fst and Φst distances between mitotype sets from different regions, supports a post-glacial colonisation of the Northwest Pacific originating from Japan rather than from the central Aleutian Islands.
The low mitogenome diversity in the western North Pacific north of Nemuro Strait is consistent with founder effects: i.e., if when ice retreated after the LGM, a few groups of killer whales moved north, colonising these vast areas and giving rise to all contemporary R-type killer whales in the Russian Pacific and western Aleutians. This aligns with the decline in effective population size estimated to have occurred approximately 7500 to 12,500 years ago, based on microsatellite allele data. A similar pattern of significant population decline during the latter half of the Late Pleistocene was also observed in R-type killer whales from the northeastern Pacific, as shown by genomic analysis (Foote et al. 2021). The authors proposed that this reduction in genetic diversity may have been driven by serial founder effects and/or ongoing range expansion during dispersal.
Lower genetic diversity in the western North Pacific has been shown for other marine species. Taguchi et al. (2010) reported decreased diversity of maternal lineages in the western North.
Pacific for the harbour porpoise Phocoena phocoena. Kim et al. (2007) found low haplotype diversity of chum salmon, Oncorhynchus keta, in Russian waters.
Filatova et al. (2018) identified 11 haplotypes in the Aleutian Islands based on the data from Morin et al. (2015). However, upon closer examination, the majority of this diversity is concentrated in a small hotspot near Seguam Island, where 9 of the 11 haplotypes are found (Figure 2). In contrast, only a single haplotype is present west of this hotspot, extending to Buldir Pass. To the east, a pronounced genetic boundary is observable, supported by both microsatellite data (Parsons et al. 2013) and mitochondrial genome analyses (Figure 2), with only two haplotypes identified in the eastern Aleutians.
The Seguam Island region is notable for its high abundance of Atka mackerel Pleurogrammus monopterygius (Lowe et al. 2017). Despite its small size, Atka mackerel is an important killer whale prey in some regions due to its high fat content and predictable occurrence during the breeding season (Filatova et al. 2023). Additionally, a 20-nautical-mile Trawl Exclusion Zone was established around the Steller sea lion rookery at Seguam Pass in 1992 (Fritz et al. 1995). The high abundance of prey, coupled with reduced fisheries activity, may render this region particularly attractive to fish-eating killer whales from a broad area of the Bering Sea, potentially contributing to the elevated haplotype diversity observed. Furthermore, the significant haplotype diversity in the central Aleutian Islands supports the hypothesis of a glacial refugium in this region.
Genetic boundaries between killer whale populations often align with biogeographic divisions, such as those observed between the oceanic (central Aleutian) and neritic (eastern Aleutian) ecosystems at Samalga Pass (Ladd et al. 2005; Parsons et al. 2013). However, in some instances, population boundaries occur in regions without clear ecological distinctions, such as the division between the western North Pacific and central Aleutian populations at Buldir Pass. This suggests that these boundaries may reflect historical demographic events rather than contemporary ecological specialisation. A comparable phenomenon has been documented in other marine species. For example, a sharp genetic division exists between Pacific herring (Clupea pallasii) populations in the Bering Sea and the Gulf of Alaska, likely marking a post-glacial contact zone between populations that were isolated in southern refugia during late Pleistocene coastal glaciation (Liu et al. 2012).
The mechanisms driving the isolation between killer whale populations of the same ecotype are not immediately apparent, especially given the documented movements of individually identified animals between the western and central Aleutian Islands (Fearnbach et al. 2014). It is probable that this isolation is maintained through behavioural factors, akin to the separation between Northern and Southern Resident killer whale populations in British Columbia and Washington, which do not interact or interbreed despite overlapping parts of their range (Ford et al. 2000).
In the Nemuro Strait, killer whales with different haplotypes were connected into a single social network (Figure 3), indicating their membership in the same community. However, some degree of social segregation is evident within the network, which is divided into three main clusters. One cluster included three whales with the most common haplotype 66, another contained whales with haplotypes 42 and JR3, and a third main cluster included a whale with haplotype JR2. Additionally, haplotype JR1 was found in an individual at the periphery of the social network. The cluster associated with haplotype 66 was connected to the other clusters via only two individuals, while the clusters containing haplotype 42 and the new haplotypes exhibited a higher level of interconnection. These findings suggest variability in the cohesiveness of social bonds across different maternal lines.
The social structure of killer whales is predominantly matrilineal, with maternal kinship playing a central role (Ford et al. 2000). This pattern is particularly pronounced among resident-type killer whales, which remain with their natal groups throughout their lives, irrespective of sex. Familial bonds between individuals descending from the same matriline may weaken over time, but they tend to remain stronger than associations between individuals from more distantly related matrilines for several generations (Ford et al. 2000). Furthermore, killer whales from different maternal lineages often exhibit highly distinct dialects (Yurk et al. 2002) and ranging patterns (Olsen et al. 2018; Yurk et al. 2010), which likely reinforces social cohesion within groups sharing the same haplotype.
It is surprising, however, that individuals with haplotypes 42 and JR3 were observed within the same social cluster, despite each being more closely related to haplotype 66 than to one another (Figure 1). The data for these observations are based on only 6 days of fieldwork in 2021 and 9 days in 2022, which cannot fully capture the long-term dynamics of killer whale social structure. The unique resident killer whale community in Nemuro Strait offers a valuable opportunity to study the interactions among descendants from several distinct maternal lines, examining their ecological and behavioural differences and providing insights into cultural transmission and gene-culture coevolution. Further research is needed to better understand these complex social relationships.
In conclusion, our results suggest the existence of a killer whale glacial refugium off Japan during the LGM. This aligns with historical demographic data, which, through coalescent-based estimations of effective population size, inferred a population decline by an order of magnitude during the last glacial period (Moura et al. 2014; Foote et al. 2021). Later, as the ice sheets retreated, some groups of killer whales gradually migrated north, colonising regions of the western North Pacific, including areas north of Japan such as the Kuril Islands, Kamchatka, the Commander Islands, and the western Aleutian Islands. This migration resulted in a reduction of genetic diversity due to the founder effect, leaving these groups with only the most common mitogenome haplotype from the ancestral population. In contrast, the groups that remained around Japan retained the higher diversity of the ancestral population, providing an excellent opportunity to study the pre-glacial genetic diversity of resident killer whales in the North Pacific.
Author Contributions
Olga A. Filatova: conceptualization, data curation, formal analysis, investigation, methodology, funding acquisition, project administration, visualization, writing – original draft, writing – review and editing. Ivan D. Fedutin: investigation, project administration, resources. Ekaterina A. Borisova: formal analysis, methodology. Ilya G. Meschersky: methodology, data curation, formal analysis, writing – review and editing. Marina V. Shitova: methodology, resources. Erich Hoyt: funding acquisition, project administration, writing – original draft, resources. Andrew D. Foote: methodology, formal analysis, writing – original draft, resources.
Acknowledgements
The field research was conducted in frames of a collaboration agreement with the Kurilsky Nature Reserve on the topic “Inventory of animals and plants of the reserve (zapovednik), zakaznik and adjacent territories”, which included permission for the fieldwork with whales and biopsy sampling. Data collection was funded by the National Geographic Explorer Grant NGS-61285R-19. Mitogenome sequencing was funded by a Laura Corrigan grant for female scientists awarded to the FEROP team. O.A.F. was supported by the Human Frontiers Science Program (grant no. RGP0045/2022). We are grateful to the staff of Kurilsky Nature Reserve, especially E. Linnik, A. Yakovlev, and M. Ragimov, as well as citizens of Yuzhno-Kurilsk, V. Leschenko and V. Zuev, for the equipment storage and logistical support. We are grateful to I. Bobyr, T. Ivkovich, and T. Pridorozhnaya, who participated in the fieldwork.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference number PQ878649, PQ878650, PQ878651.




