Plasmodium and Host Lactate Dehydrogenase Molecular Function and Biological Pathways: Implication for Antimalarial Drug Discovery
Abstract
Lactate dehydrogenase is an enzyme that catalyses the interconversion of pyruvate and lactate with concomitant interconversion of NADH and NAD+. Lactate dehydrogenase is present at high levels in humans and Plasmodium spp. However, the function of lactate dehydrogenase in malarial infection is not well characterized. In this investigation, a new gene ontology technology is used to predict molecular function and biological pathways of lactate dehydrogenase. In comparison with human lactate dehydrogenase, the P. falciparum lactate dehydrogenase has similar molecular functions such as l-lactate dehydrogenase activity. Furthermore, P. falciparum lactate dehydrogenase has l-malate dehydrogenase activity. Although the amino acid sequences for human and P. falciparum lactate dehydrogenase are very different, the molecular functions are similar. This suggests that any non-selective therapeutic treatment aimed at blocking P. falciparum lactate dehydrogenase function may affect human lactate dehydrogenase. In contrast, a selective lactate dehydrogenase inhibitor targeting the l-malate dehydrogenase function of P. falciparum and its corresponding tricarboxylic acid cycle provides an attractive therapeutic opportunity.
Lactate dehydrogenase (LDH) is an enzyme that catalyses the interconversion of pyruvate and lactate with concomitant interconversion of NADH and NAD+. LDH is present at high levels in both humans and Plasmodium spp. The existence of LDH has been demonstrated in P. falciparum (1) and P. vivax (2). Generally, P. falciparum LDH is an essential enzyme for energy generation within the parasite (3–5). A Plasmodium LDH assay, based on detection of the Plasmodium intracellular metabolic enzyme, has provided a new advance for the diagnosis of malarial infection (5). As LDH is an essential Plasmodium protein, it has become a potential antimalarial drug target (1–3). Gossypol, a disesquiterpene natural product isolated from cotton plants, exemplifies a lead compound that targets P. falciparum LDH (4). Furthermore, simple naphthalene-based compounds, representing the core of the gossypol structure, exhibit weak inhibition of the parasite LDH (4).
Vivas et al. have proposed that trophozoites possess relatively higher P. falciparum LDH enzyme activity and P. falciparum LDH-RNA expression levels than rings (6,7). This is possibly linked to their increased energy requirements and is consistent with glycolysis being an essential metabolic pathway for parasite survival within the erythrocyte (6,7). Vivas et al. have proposed that the knowledge on the parasite LDH could lead to the identification of compounds that target P. falciparum through such a novel mechanism and with more potent and selective antimalarial activity (6,7). However, the function of the LDH in malarial infection is not well characterized. It is noted that a full understanding host and parasite LDH may provide unique opportunities to advance malarial therapeutic treatment. In this investigation, a new gene ontology technology is used to predict the molecular function and biological pathways of this enzyme.
Materials and Methods
Mining LDH amino acid sequences and alignments
The database Unitprot (8) was used for data mining of the amino acid sequence for human host and P. falciparum LDH. Further, alignment of human and P. falciparum LDH sequences was performed using sim software on EXPASY Tools (http://www.expasy.org/tool).
Prediction of molecular function and biological pathways
The author performed prediction of molecular function and biological pathways of human and P. falciparum LDH using a novel gene ontology prediction tool, GoFigure (9). GoFigure is a computational algorithm tool which was recently developed in gene ontology (9). The tool accepts an input DNA or protein sequence, and uses BLAST to identify homologous sequences in gene ontology annotated databases (9). The approach uses a BLAST search to identify homologues in public databases that have been annotated with gene ontology terms (9), including SwissProt, Flybase (Drosophila), the Saccharomyces Genome Database (SGD), Mouse Genome Informatics (MGI) and Wormbase (nematode). Overall, this process will provided prediction of molecular function as well as biological pathways for the studied protein (9).
Results and Discussion
The sequences of human and P. falciparum LDH were determined from searching the database Uniprot. An alignment of human and P. falciparum LDH sequences showed a 30.7% sequence homology (Figure 1). Using GoFigure, the molecular function and biological pathways in human and P. falciparum LDH were predicted. The molecular function and biological process of human and P. falciparum LDH are illustrated in 2, 3, respectively. The summary on the molecular function and biological process comparing between human and P. falciparum LDH is presented in Table 1.

An alignment of human and Plasmodium falciparum lactate dehydrogenase sequences.
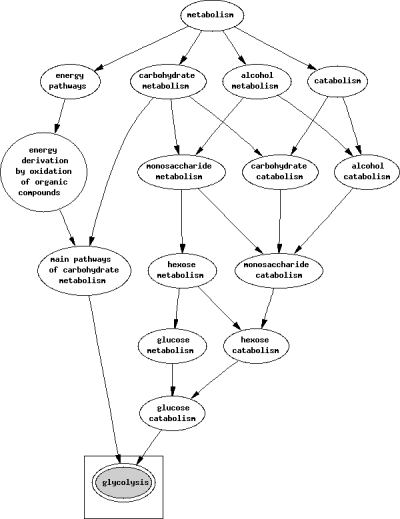
Predicted biological pathways of human lactate dehydrogenase.
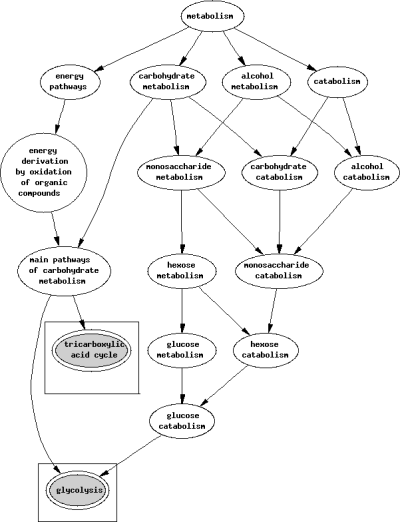
Expected biological process of Plasmodium falciparum lactate dehydrogenase.
| Summary | Molecular function | Biological process |
|---|---|---|
| Human LDH | l-LDH activity | Glycolysis |
| P. falciparum LDH | l-LDH activity, l-malate dehydrogenase activity | Glycolysis, tricarboxylic acid cycle |
Lactate dehydrogenase is the terminal enzyme of anaerobic Embden-Meyerhoff glycolysis and it plays an important role in the carbohydrate metabolism of human malaria parasites. LDH is believed to have a significant role in malarial infection and it has become a possible drug target for malarial treatment. Roles for both host and parasite LDH in cellular level metabolism during a malarial infection have been proposed (9). However, to date, the function of P. falciparum LDH correlating with human LDH is not well understood.
Based on recent advances in the genomics technology, microarray strategies permit examination of gene expression patterns for ten thousands of genes (8). A challenge facing the biologist interpreting such data are recognizing the functions of such a plethora of hits identified in a single experiment (8). In addition to literature searching, a rapid approach to gather potential function(s) of a gene product is through exploiting gene ontology technology (8), and many genes ontology tools have been constructed (10). In this investigation, the gene ontology tool GoFigure is used to probe the predicted functions of human and P. falciparum LDH.
Relative to human LDH, the P. falciparum LDH has similar molecular functions in terms of l-LDH activity. However, P. falciparum LDH also has l-malate dehydrogenase (MDH) activity. Although the amino acid sequences for human and P. falciparum LDH are very different, their molecular functions are similar. This implies that any non-selective treatment aimed at blocking P. falciparum LDH activity may affect human LDH. It is further noted that two of five LDH isozymes detected in mammalians have been identified in P. falciparum (11). However, parasite LDH exhibits high specificity for pyruvic acid, even more restricted than the specificity of human LDH M4 and H4 (12). In addition, the structure of active sites and cofactor-binding pockets of parasite LDH is different from human LDH (1). Inhibition studies confirm that Chloroquine acts as a weak inhibitor of LDH, with mild selectivity for the parasite LDH (13). Again, target selectivity between host and parasite LDHs would be desirable for such drug development.
In contrast to LDH targeting, a selective inhibitor of the l-MDH activity and its corresponding tricarboxylic acid cycle of P. falciparum is predicted to be a promising opportunity for drug discovery. As related to this concept, Chan and Sim have recently successfully cloned a gene belonging to the NADH-dependent l-MDH type from the P. falciparum genome (14). Furthermore, the cDNA corresponding to the l-MDH gene, and identified on chromosome 6 of the P. falciparum genome, was amplified by RT-PCR, cloned and overexpressed in Escherichia coli (15). Moreover, Tripati et al. found that P. falciparuml-MDH transcripts were detectable in trophozoites while P. falciparum MDH protein levels remained high in trophozoites as well as schizonts (15). In addition, they found that treatment of culture of P. falciparum trophozoites with gossypol caused induction in expression of both l-MDH and LDH activities (15). Hence, these studies implicated that dual inhibitors of both MDH and LDH activities might be required to target this pathway and to develop potential new antimalarial drugs (15).
In conclusion, it is predicted that antimalarial drug discovery might be successfully targeted on blocking P. falciparuml-MDH activity.




