Multiplex PCR to Assay the Effect of Nucleic Acid-Based Inhibitors on Prothrombin Transcript Level
Abstract
We compared an antisense-oligodeoxynucleotide and four DNAzymes directed to the prothrombin mRNA for their efficiency to reduce prothrombin transcript level in HepG2 cells. The DNAzymes have different binding arm symmetry and cleavage sites, but are directed to the identical target site of the antisense-oligodeoxynucleotide. The nucleic acid-based inhibitors were transfected into HepG2 cells and prothrombin transcript level was quantified and normalized to the β-actin transcript level by multiplex PCR. All nucleic acid-based inhibitors reduced prothrombin transcript level and the effect was in almost all cases, strongest 24 h after transfection, but still remarkable up to 68 h after transfection. The antisense-oligodeoxynucleotide was more effective than the DNAzymes tested.
DNAzymes and antisense-oligodeoxynucleotides (AS-ODNs) are both considered to be useful agents in the treatment of various diseases because of their specific mode of target recognition (1,2). They both depend on Watson Crick base pairing to the target mRNA, but on a different mode of action. While AS-ODNs may inhibit gene expression by activation of RNAse H that cleaves specifically RNA of RNA/DNA hybrids, several DNAzymes have been reported to cleave their substrate mRNA successfully in cell-free activity test systems (3–5). Their mode of action in living cells may depend on both, activation of RNAse H and DNAzyme nuclease activity. However, there are few reports on DNAzymes that are effective in cell culture (6,7). The reasons for this are well known. DNAzymes have to overcome the same hurdles like any other nucleic acid-based agent to be active in living cells, like delivery into the cells, stability versus nucleases and of course accessibility of the given target region (8). To overcome these problems, researchers stabilized, for example, AS-ODNs as well as DNAzymes by site-specific phosphorothioate modifications. Furthermore, DNAzymes can be stabilized by the incorporation of the 10–23 catalytic motif in a circular loop–stem-loop structure (9) and their activity at unfavourable cleavage sites can be increased by the covalent incorporation of an intercalator in the catalytic loop (10). However, for the therapeutic use of DNAzymes, there is an additional fact that have to be considered. While cell-free test systems for DNAzymes utilize optimal conditions for the cleavage reaction, mostly dependent on divalent metal ion concentration and pH (1), these optimal conditions may not be found in cell cytoplasm. This indicates that the selection of DNAzymes by cell-free test systems may not be the right way to find active DNAzymes for the use in living cells.
We wanted to know if the inhibition of prothrombin gene expression observed for an AS-ODN in cell culture can be excelled by DNAzymes directed to the identical target region. We directed DNAzymes to an accessible site within the prothrombin mRNA that was found by a combination of mRNA secondary structure prediction and experimental selection procedure (11). The substrate binding arms of the respective DNAzymes always bind to the exact target sequence of the original AS-ODN (Figure 1A). This target region comprises six possible DNAzyme cleavage sites characterized by a purine nucleotide followed by a pyrmidine nucleotide in 5′ to 3′ direction of the respective mRNA (Figure 1A). We designed four DNAzymes directed to the marked cleavage sites 2–5 (Figure 1A). This led to four DNAzymes with unsymmetrical substrate binding arms but with the identical catalytic loop motif of the 10–23 DNAzyme (Figure 1B).

(A) The AS-ODN and DNAzyme target region (prothrombin mRNA gene bank accession number: NM_000506, target site nucleotides 513–532) within the prothrombin mRNA comprises six 10–23 DNAzyme cleavage sites. (B) DNAzymes directed to cleavage sites 2–5 of the given target region hybridized with their target sequence. The substrate binding arms are unsymmetrical and all DNAzymes share the catalytic loop motif of the 10–23 DNAzymes.
We transfected the hepatoma-derived cell line HepG2 that is endogenously expressing prothrombin and was characterized for the expression of coagulation-related proteins (12) with these DNAzymes and determined the time-dependent reduction of prothrombin transcripts caused by DNAzymes in comparison to an AS-ODN by a multiplex-PCR procedure. This experimental setting allows to judge non-specific effects of the studied oligonucleotides as they would also affect β-actin expression.
For the experiments, the hepatoblastoma cell line HepG2 (German Collection of Microorganism and Cell Cultures, Braunschweig, Germany) was cultured in RPMI 1640 Medium with Glutamax-I (Invitrogen, Karlsruhe, Germany) containing 10% FCS (Biochrom KG, Berlin, Germany) and 80 mg/l gentamicin (Biochrom) and was passaged once a week after trypsination.
Cells were seeded at a density of 6 × 105 cells/well in 6-well testplates and were transfected 24 h after seeding. Cells were transfected for 4 h with 3-ml cell culture media without FCS containing 14 μg/ml lipofectamin reagent (Invitrogen) complexed with 2 μg/ml of the respective oligonucleotide [see (12) for details].
Total RNA was isolated at the respective time after transfection (Figure 2) using Trizol Reagent (Life Technologies, Karlsruhe, Germany) following the manufacturer's instructions and was unspecifically reverse transcribed with oligo dT primer (5′-(T)17-3′) using a standard protocol. The cDNA was amplified in a PCR-reaction with specific prothrombin primers (sense primer 5′-TAGCACCAGGGTCCCGTGGT-3′, antisense primer 5′-TCCTCAGCAAGCACGGTCGC-3′) at 65 °C annealing temperature and 25 cycles. The generated prothrombin PCR-product consists of 420 bases. The housekeeping gene β-actin was co-amplified with specific primers (sense primer 5′-CAATGAGCGGTTCCGCTG-3′ and antisense primer 5′-CGCCAGACAGCACTGTGTTG-3′). The generated β-actin PCR-product consists of 150 bases. All primers were custom primers (Invitrogen) and all enzymes and buffers for reverse transcription and PCR were supplied by Fermentas (St Leon Roth, Germany).
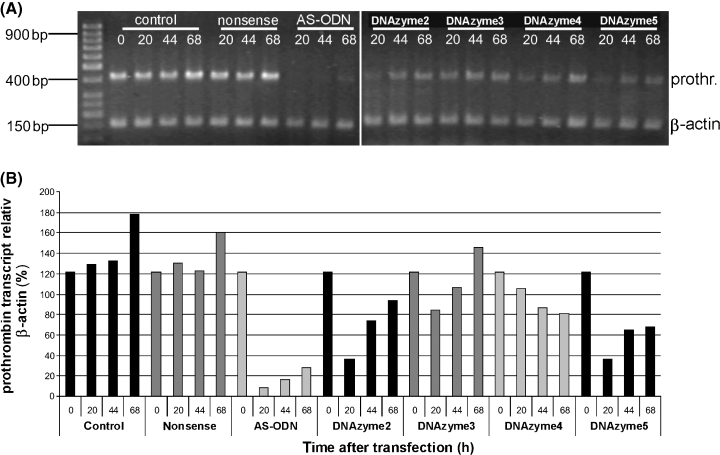
DNAzymes and an AS-ODN directed against the same target region of the prothrombin mRNA reduce prothrombin transcripts in transfected HepG2 cells. (A) Total RNA was isolated from untreated cells (control), and cells transfected with nonsense-ODN, AS-ODN, and DNAzymes 2–5 at distinct time points (0, 22, 44 and 68 h after transfection). Total RNA was unspecifically reverse transcribed and a multiplex PCR with specific prothrombin (420 bp) and β-actin (150 bp) primers was carried out. PCR products were separated on a 2% agarose-gel. (B) Densitometric analyses were performed using β-actin as internal standard.
By the described procedure, we were able to analyse the effect of prothrombin specific nucleic acid-based inhibitors on prothrombin transcript level in comparison to β-actin transcript level. We addressed the influence of transfection on prothrombin gene expression to exclude the possibility of non-specific reduction of prothrombin transcript level by transfection of a nonsense-oligonucleotide (nonsense-ODN; sequence 5′-ATGCATGCATGCATGCATGC-3′) that is not able to bind to the prothrombin mRNA.
The products of the multiplex-PCR reaction were separated on a 2% agarose gel and densitometry of these bands, with the use of β-actin as internal standard, showed that the prothrombin-specific PCR-product, and therefore the amount of prothrombin transcripts in HepG2 cells was reduced after transfection with AS-ODN and DNAzymes, but was not reduced after transfection with nonsense-ODN in comparison to untreated cells (Figure 2).
The AS-ODN reduced prothrombin transcripts much stronger than any DNAzyme at the respective time. The reduction was most prominent 20 h after transfection for the AS-ODN and DNAzymes 2, 3 and 5. The amount of prothrombin transcripts increased with time after transfection, but was still remarkably reduced up to 68 h after transfection by the AS-ODN and DNAzymes 2, 4 and 5. Prothrombin transcripts of cells transfected with DNAzyme 4 were stronger reduced with the ongoing experiment and reduction was most prominent 68 h after transfection (Figure 2).
Our results show that the inherent sequence-specific endonuclease activity of DNAzymes did not lead to an increased inhibition of prothrombin gene expression compared with an AS-ODN directed to the same target region. Instead all DNAzymes tested were less active inhibitors than the AS-ODN.
It is not possible to determine by our assay, if the reduction of prothrombin transcripts in DNAzyme-transfected cells is dependent on the catalytic activity of the DNAzymes or at least in parts on the action of RNAse H. The reduction of prothrombin transcripts by DNAzymes in cell culture may not be because of the nuclease activity of the tested DNAzymes. It is possible to interpret our data in a way that AS-ODN/target-mRNA hybrids are far better substrates for RNAse H than DNAzyme/target-mRNA hybrids, because of the bulky catalytic loop of DNAzymes that may avoid the association of RNAse H with the hybrid. Therefore, one may conclude that in the presented case, DNAzymes are less affectively recruiting RNAse H when compared with the AS-ODN.
The influence of DNAzyme substrate binding arm length respectively its symmetry have been in focus of other studies, but mainly the effects of symmetric DNAzymes have been analysed (1,4,13). These studies tried to find an optimal DNAzyme design with respect to the need for the specific recognition of a substrate by a DNAzyme that is reflected by long substrate binding arms resulting in a stable hybrid. However, the longer the binding arms are, the lower will be the activity of the DNAzyme under multiple turnover conditions, because the DNAzyme will not dissociate from the cleaved substrate (13). Perhaps, this discussion is of interest in case of kinetic studies using cell-free activity assays, but not if the degradation of a target mRNA hybridized with a DNAzyme in living cells is mainly because of RNAse H.
DNAzymes 2 and 5 reduced prothrombin transcripts more effectively than DNAzymes 3 and 4 at 20 h after transfection. The substrate binding arms of DNAzyme 2 were most unsymmetric (5′-three nucleotides, 3′-sixteen nucleotides), whereas the substrate binding arms of DNAzyme 5 differed only in three bases (5′-eight nucleotides, 3′-eleven nucleotides). Interestingly, there are two DNAzymes in our set (4 and 5) were the binding arm symmetry is just switched and these two showed an opposite time-dependent reduction of prothrombin transcripts when transfected into HepG2 cells. DNAzyme 4 was most active 68 h after transfection, whereas DNAzyme 5 was most active 20 h after transfection. These data are as confusing as the data gained by other authors, who tried to find systematic rules for arm length design of DNAzymes (1,13). It seems that the optimal design of DNAzymes mainly depends on the target region to which they are directed and has to be evaluated empirically.
Our results show that the incorporation of the 10–23 DNAzyme catalytic loop in the sequence of an effective AS-ODN directed to the prothrombin mRNA reduces the inhibitory activity. This effect can be assayed by multiplex PCR.
Acknowledgments
This work was supported by the Sächsisches Landesamt für Umwelt und Geologie (Az: 13-8811.61/123).




