Comparative Proteomic Analysis of Arabidopsis Mature Pollen and Germinated Pollen
Supported by a competitive research grant (30421002) for Creative Research Groups sponsored by the National Natural Science Foundation of China.
Abstract
Proteomic analysis was applied to generating the map of Arabidopsis mature pollen proteins and analyzing the differentially expressed proteins that are potentially involved in the regulation of Arabidopsis pollen germination. By applying 2-D electrophoresis and silver staining, we resolved 499 and 494 protein spots from protein samples extracted from pollen grains and pollen tubes, respectively. Using the matrix-assisted laser desorption ionization time-of-flight mass spectrometry method, we identified 189 distinct proteins from 213 protein spots expressed in mature pollen or pollen tubes, and 75 new identified proteins that had not been reported before in research into the Arabidopsis pollen proteome. Comparative analysis revealed that 40 protein spots exhibit reproducible significant changes between mature pollen and pollen tubes. And 21 proteins from 17 downregulated and six upregulated protein spots were identified. Functional category analysis indicated that these differentially expressed proteins mainly involved in signaling, cellular structure, transport, defense/stress responses, transcription, metabolism, and energy production. The patterns of changes at protein level suggested the important roles for energy metabolism-related proteins in pollen tube growth, accompanied by the activation of the stress response pathway and modifications to the cell wall.
Sexual reproduction of flowering plants comprises several sequential steps from pollination to fertilization. Pollen landing on a stigma is the first step, followed by pollen hydration, germination, and pollen tube growth through intercellular spaces in the pistil. When the pollen tube reaches the embryo sac of the ovary, it delivers sperm cells for double fertilization (Franklin-Tong 1999a; McCormick 2004). The critical steps in this continuous process include pollen germination on the stigma and pollen tube growth. The investigation of the regulatory mechanisms for pollen germination and tube growth is important for fundamental studies of fertility and reproduction in flowering plants. In addition, pollen germination is an ideal model system for the investigation of important issues in cell biology, such as polarized tip growth, cell-cell interactions, and signal transduction (Franklin-Tong 1999b).
Increasing efforts have been made to investigate the genetic and molecular mechanisms of pollen germination and tube growth, and at least 150 genes involving in pollen development and pollen tube growth have been studied (Twell 2002). Functional composition analysis of the Arabidopsis pollen transcriptome has revealed that the mRNAs specifically or preferentially presented in pollen mainly encode proteins potentially involved in cell wall metabolism, vesicle transport, cytoskeleton, and signaling (Honys and Twell 2003; Honys and Twell 2004; Pina et al. 2005; Wang et al. 2008). These results may reflect the functional specialization of mature pollen in the commitment of germination and tube growth. However, gene expression at the mRNA expression level lacks a direct correlation with protein level and activity (Greenbaum et al. 2003). Although mature pollen grains may contain pre-synthesized mRNA for germination and other mRNAs may be synthesized during pollen germination (Wang et al. 2008), newly transcribed mRNAs are not necessarily translated into the corresponding proteins and post-translational modifications may be also crucial to pollen function (Mascarenhas 1993; Taylor and Hepler 1997). Thus, detailed analysis at the protein level is an essential step toward the further identification of regulatory components involving pollen germination and tube growth.
Proteomic analyses of pollen development and germination can provide new insights on the whole genome level into fascinating mechanisms of pollen development and tip-growth regulation in higher plants (Chen et al. 2007; Dai et al. 2007b). Proteomic analyses of rice anthers provided comprehensive understanding of the proteins expression changes during microspore and pollen development and in response to environment stresses (Imin et al. 2001, 2004; Kerim et al. 2003). By analyzing rice pollen proteins, Dai et al. (2006) identified several novel proteins that may be involved in signal transduction, protein synthesis, assembly and degradation, and wall remodeling and metabolism. Proteomic analysis of tomato pollen showed that many of the identified proteins have designated roles in defense mechanisms, energy conversion, pollen germination, and pollen tube growth, and some possibly in sperm cell formation (Sheoran et al. 2007). By conducting a proteomic analysis of Arabidopsis pollen coat proteins, Mayfield et al. (2001) reported that oleosins and lipases may play important roles in initiating pollination. More recently, three independent proteomic analyses of Arabidopsis mature pollen were conducted (Holmes-Davis et al. 2005; Noir et al. 2005; Sheoran et al. 2006), which provide a broad analysis of the Arabidopsis pollen proteome and complement and extend the analysis of the pollen transcriptome.
Proteomic analysis of the Arabidopsis flower presented the importance of post-translational regulation of proteins in the flower and provided new understanding about flower development and physiology (Feng et al. 2009). Proteomic analyses of the differentially expressed proteins during pollen tube growth in Pinus strobus and Picea meyeri provide an important insight into the molecular basis of pollen tube growth and the effects of actin cytoskeleton disruption on global protein patterns in pollen tube (Fernando 2005; Chen et al. 2006). Proteomic identification of differentially expressed proteins associated with pollen germination and tube growth in rice revealed that these differentially expressed proteins involve different cellular and metabolic processes with obvious functional skew toward wall metabolism, protein synthesis and degradation, cytoskeleton dynamics, and carbohydrate/energy metabolism (Dai et al. 2007a). Obviously, a more comprehensive pollen proteomic analysis of Arabidopsis, the model plant of dicots of angiosperms, particularly the changes in protein expression profiles during the transitions from desiccated mature pollen to germinating pollen and to growing pollen tubes, is needed to reveal the complex molecular mechanisms of pollen germination and tube growth. Along with the development of methods for collecting large quantities of Arabidopsis pollen (Johnson-Brousseau and McCormick 2004) and high rates of pollen germination in vitro (Fan et al. 2001; Wang et al. 2008), it now becomes possible to experimentally investigate changes in proteomic profiles during the process of pollen germination and pollen tube growth. This study reports proteomic analysis of changes in protein expression profiles between the mature pollen and growing pollen tubes. In addition, potential roles of the differentially expressed proteins involved in the regulation of pollen germination and tube growth are discussed.
Results
Proteomic maps of Arabidopsis pollen and pollen tubes
After the 2-D electrophoresis (2-DE) gels were aligned and matched, the protein spots shown on the gels of mature pollen and pollen tubes were analyzed. There were in total 499 and 494 reproducible protein spots detected in the gels loaded with protein samples extracted from pollen grains and pollen tubes, respectively. These proteins cover the pI (isoelectric point) range from 5 to 8, and their MW (molecular weight) ranged from 10 to 110 kDa (1-3). There were in total 303 protein spots excised from two sets of gels for mass spectrometry (MS) analysis to generate peptide mass fingerprints (PMFs). As shown in Table 1, 213 protein spots, representing 189 different proteins, were identified after searching with the National Center for Biotechnology Information (NCBI) database. The identified proteins corresponding to the spot numbers are shown in 1-3. The calculated MW of the identified proteins ranged from 11.9 kDa to 108.9 kDa, and the calculated pI range was from 4.36 to 9.25, which is close to the experimental data as judged from the location of the spots on the 2-DE gels (Table 1; 1-3).
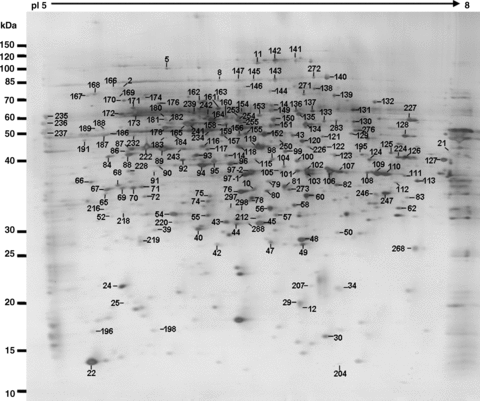
A representative proteome map of mature pollen. All labeled spots are proteins without changes in intensity between pollen and the pollen tube 2-D gels (see Table 1 for details).
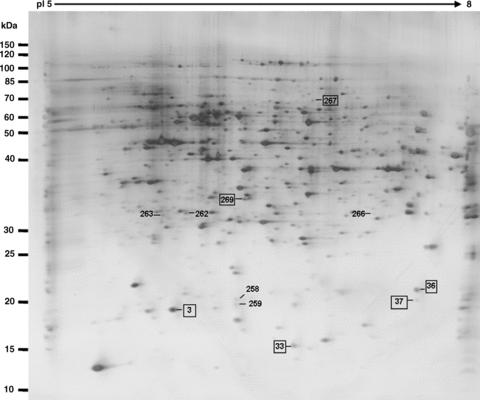
Present and upregulated protein spots during the transition from mature pollen to pollen germination and pollen tube growth. The spots marked by rectangles are the identified proteins (listed in Tables 1, 2).
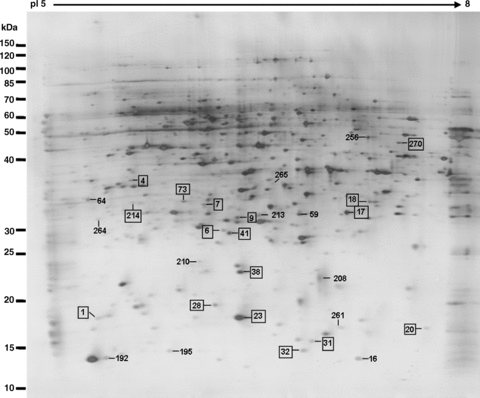
Disappeared and downregulated protein spots during the transition from mature pollen to pollen germination and pollen tube growth. The spots marked by rectangles are the identified proteins (listed in Tables 1, 2).
| Spot no.a | AGI name | Protein identity | pI/MW (kDa)d | Sequence coverage | Mascot score | |
|---|---|---|---|---|---|---|
| Experimental | Theoretical | |||||
| Cell fate (5)b | ||||||
| 28c | At1g26630 | Eukaryotic translation initiation factor 5A, putative | 5.55/17.4 | 5.81/17.1 | 42% | 79 |
| 56c | At1g63000 | Expressed protein | 5.73/33.9 | 5.78/33.6 | 31% | 114 |
| 137 | At3g17240 | 2-oxoglutarate dehydrogenase, E3 subunit | 6.0/50.3 | 7.03/53.9 | 18% | 107 |
| 236c | At1g56340 | Calreticulin 1 (CRT1) | 4.50/47.7 | 4.20/48.5 | 26% | 99 |
| 237c | At1g09210 | Calreticulin 2 (CRT2) | 4.36/48.4 | 4.12/48.1 | 16% | 71 |
| 297c | At1g63000 | Expressed protein | 5.73/33.9 | 5.78/33.6 | 13% | 67 |
| 298c | At1g63000 | Expressed protein | 5.73/33.9 | 5.78/33.6 | 13% | 72 |
| Cell type differentiation (2) | ||||||
| 22Ac | At2g19770 | Profilin 4 | 5.02/14.6 | 4.84/14.5 | 50% | 80 |
| 22Bc | At4g29340 | Profilin 3 | 5.02/14.6 | 4.84/14.4 | 57% | 82 |
| Cell structure (12) | ||||||
| 20 | At1g48020 | Invertase /pectin methylesterase inhibitor family protein | 7.57/19.2 | 7.82/18.9 | 32% | 100 |
| 23c | At4g24640 | Invertase /pectin methylesterase inhibitor family protein | 5.54/20.1 | 5.65/19.9 | 40% | 153 |
| 87A | At3g12110 | Actin 11 (ACT11) | 5.23/41.9 | 5.05/41.6 | 31% | 91 |
| 89c | At5g59370 | Actin 4 (ACT4) | 5.37/42.0 | 5.29/41.7 | 55% | 213 |
| 94c | At3g02230 | Reversibly glycosylated polypeptide-1 (RGP1) | 5.61/41.1 | 5.70/40.6 | 55% | 224 |
| 96c | At5g15650 | Reversibly glycosylated polypeptide-2 (RGP2) | 5.76/41.4 | 5.98/40.9 | 25% | 111 |
| 126 | At4g37990 | Mannitol dehydrogenase, putative (ELI3-2) | 6.79/39.4 | 7.24/38.9 | 16% | 109 |
| 187c | At1g04820 | Tubulin alpha-2/alpha-4 chain (TUA4) | 4.93/50.2 | 4.68/49.5 | 36% | 128 |
| 196c | At4g25590 | Actin-depolymerizing factor, putative | 5.08/15.4 | 4.82/15.8 | 50% | 70 |
| 198c | At5g52360 | Actin-depolymerizing factor, putative | 5.57/15.4 | 5.31/15.9 | 55% | 76 |
| 222Ac | At2g37620 | Actin 1 (ACT1) | 5.31/42.0 | 5.16/41.8 | 45% | 190 |
| 222Bc | At3g53750 | Actin 3 (ACT3) | 5.31/42.1 | 5.16/41.8 | 45% | 190 |
| Defense/stress responses (9) | ||||||
| 3 | At1g60740 | Peroxiredoxin type 2, putative | 5.33/17.6 | 5.23/17.4 | 65% | 135 |
| 3 | At1g65970 | Peroxiredoxin type 2, putative | 5.33/17.6 | 5.23/17.4 | 65% | 136 |
| 6 | At2g30870 | Glutathione S-transferase, putative | 5.49/24.2 | 5.39/24.2 | 25% | 68 |
| 7 | At4g08390 | L-ascorbate peroxidase, stromal (sAPX) | 8.31/40.5 | 8.57/40.4 | 33% | 133 |
| 12c | At4g11600 | Glutathione peroxidase, putative | 6.59/18.8 | 9.85/25.6 | 33% | 85 |
| 29c | At4g11600 | Glutathione peroxidase, putative | 6.59/18.8 | 9.85/25.6 | 43% | 82 |
| 34c | At3g06050 | Alkyl hydroperoxide reductase/thiol specific antioxidant (AhpC/TSA) | 8.99/21.3 | 9.39/21.4 | 52% | 158 |
| 44c | At1g07890 | L-ascorbate peroxidase 1, cytosolic (APX1) | 5.85/27.8 | 6.03/27.5 | 38% | 103 |
| 45c | At1g07890 | L-ascorbate peroxidase 1, cytosolic (APX1) | 5.85/27.8 | 6.03/27.5 | 60% | 178 |
| 48c | At2g47730 | Glutathione S-transferase 6 (GST6) | 6.09/24.1 | 8.93/29.2 | 48% | 92 |
| 49c | At2g47730 | Glutathione S-transferase 6 (GST6) | 6.09/24.1 | 8.93/29.2 | 75% | 191 |
| 122 | At3g52880 | Monodehydroascorbate reductase, putative | 6.86/46.6 | 6.80/46.5 | 47% | 205 |
| Development (1) | ||||||
| 216 | At5g66530 | Aldose 1-epimerase family protein | 5.64/34.0 | 5.73/33.7 | 21% | 77 |
| Energy (59) | ||||||
| 1c | At3g52300 | ATP synthase D chain-related | 4.97/17.7 | 4.81/19.6 | 43% | 87 |
| 5c | At4g15530 | Pyruvate phosphate dikinase family protein | 5.25/93.9 | 6.06/104.2 | 36% | 352 |
| 8c | At5g65690 | Phosphoenolpyruvate carboxykinase [ATP], putative | 5.98/69.3 | 6.37/72.9 | 14% | 78 |
| 11c | At4g26970 | Aconitate hydratase, cytoplasmic, putative | 6.71/108.9 | 7.15/108.5 | 21% | 175 |
| 13c | At2g44350 | Citrate synthase, mitochondrial, putative | 6.41/53.0 | 6.88/52.6 | 14% | 79 |
| 24c | At5g47030 | ATP synthase delta chain, mitochondrial | 6.20/21.5 | 6.68/21.5 | 39% | 70 |
| 33 | At1g08480 | Expressed protein/unknown protein | 6.28/15.8 | 6.80/15.8 | 19% | 61 |
| 39c | At3g55440 | Triosephosphate isomerase, cytosolic, putative | 5.24/27.4 | 5.16/27.1 | 44% | 137 |
| 40c | At3g55440 | Triosephosphate isomerase, cytosolic, putative | 5.24/27.4 | 5.16/27.1 | 82% | 234 |
| 47 | At5g54500 | Quinone reductase, putative | 5.96/21.8 | 6.35/21.8 | 50% | 105 |
| 57 | At4g02580 | NADH-ubiquinone oxidoreductase 24 kDa subunit, putative | 7.55/27.6 | 8.03/28.4 | 30% | 115 |
| 60c | At1g53240 | Malate dehydrogenase [NAD], mitochondrial | 8.54/36.0 | 8.58/35.8 | 15% | 72 |
| 69c | At3g59480 | Pfkb-type carbohydrate kinase family protein | 5.12/35.2 | 4.99/35.0 | 65% | 291 |
| 70 | At5g50850 | Pyruvate dehydrogenase E1 component beta subunit, mitochondrial / PDHE1-B (PDH2) | 5.67/39.4 | 5.55/39.2 | 38% | 133 |
| 71c | At2g31390 | Pfkb-type carbohydrate kinase family protein | 5.31/35.4 | 5.13/35.3 | 32% | 99 |
| 72c | At2g31390 | Pfkb-type carbohydrate kinase family protein | 5.31/35.4 | 5.13/35.3 | 53% | 230 |
| 76c | At3g15020 | Malate dehydrogenase [NAD], mitochondrial, putative | 8.3/36.0 | 8.43/35.8 | 26% | 87 |
| 80c | At3g47520 | Malate dehydrogenase [NAD], chloroplast (MDH) | 8.66/42.6 | 8.81/42.4 | 23% | 107 |
| 82 | At2g01140 | Fructose-bisphosphate aldolase, putative | 8.19/42.5 | 8.27/42.3 | 17% | 64 |
| 91 | At1g43670 | Fructose-1,6-bisphosphatase, putative | 5.28/37.7 | 5.12/37.3 | 26% | 94 |
| 92c | At1g79550 | Phosphoglycerate kinase, putative | 5.49/42.2 | 5.33/42.1 | 28% | 106 |
| 93Ac | At2g20420 | Succinyl-coa ligase [GDP-forming] beta-chain, mitochondrial, putative | 6.30/45.6 | 6.68/45.3 | 48% | 188 |
| 101c | At1g04410 | Malate dehydrogenase, cytosolic, putative | 6.11/35.9 | 6.51/35.6 | 46% | 171 |
| 102c | At3g52930 | Fructose-bisphosphate aldolase, putative | 6.05/38.9 | 6.40/38.5 | 52% | 203 |
| 106c | At5g43330 | Cytosolic malate dehydrogenase | 6.33/36.0 | 6.77/35.6 | 40% | 84 |
| 107c | At5g43330 | Cytosolic malate dehydrogenase | 6.33/36.0 | 6.77/35.6 | 31% | 78 |
| 108c | At5g43330 | Cytosolic malate dehydrogenase | 6.33/36.0 | 6.77/35.6 | 31% | 74 |
| 109c | At3g04120 | Glyceraldehyde-3-phosphate dehydrogenase,cytosolic (GAPC) | 6.62/37.0 | 7.14/36.9 | 33% | 120 |
| 110c | At1g13440 | Glyceraldehyde 3-phosphate dehydrogenase, cytosolic, putative | 6.67/37.0 | 7.21/36.9 | 49% | 161 |
| 111 | At1g79530 | Glyceraldehyde 3-phosphate dehydrogenase, cytosolic, putative | 8.75/45.0 | 8.97/44.8 | 18% | 81 |
| 113 | At5g08300 | Succinyl-coa ligase [GDP-forming] alpha-chain, mitochondrial, putative | 8.55/36.6 | 8.39/36.1 | 23% | 74 |
| 115 | At1g01090 | Pyruvate dehydrogenase E1 component alpha subunit, chloroplast | 7.16/47.6 | 7.54/47.2 | 17% | 74 |
| 120c | At1g65930 | Isocitrate dehydrogenase, putative | 6.13/46.1 | 6.52/45.7 | 26% | 100 |
| 121c | At2g35840 | Sucrose-phosphatase 1 (SPP1) | 6.24/47.8 | 6.57/47.8 | 33% | 170 |
| 123 | At4g35650 | Isocitrate dehydrogenase, putative | 7.08/40.3 | 7.49/39.9 | 23% | 72 |
| 124 | At1g24180 | Pyruvate dehydrogenase Ela-like subunit IAR4 | 7.62/43.7 | 8.01/43.3 | 25% | 128 |
| 125 | At4g35260 | Isocitrate dehydrogenase subunit 1 | 8.12/40.0 | 8.27/39.6 | 21% | 104 |
| 128c | At2g47510 | Fumarate hydratase, putative | 7.98/53.4 | 7.98/53.0 | 16% | 72 |
| 131c | At1g48030 | Dihydrolipoamide dehydrogenase 1, mitochondrial | 6.96/54.0 | 7.45/54.0 | 23% | 105 |
| 132c | At5g25880 | Malate oxidoreductase, putative | 6.55/65.0 | 6.98/64.6 | 27% | 177 |
| 133 | At5g63680 | Pyruvate kinase, putative | 6.24/55.6 | 6.62/55.0 | 18% | 79 |
| 134d | At2g07698 | ATP synthase alpha chain, mitochondrial, putative | 6.23/55.3 | 5.23/85.9 | 37% | 229 |
| 141c | At4g35830 | Aconitate hydratase, cytoplasmic | 5.98/98.8 | 6.35/98.1 | 20% | 141 |
| 142c | At2g05710 | Aconitate hydratase, cytoplasmic, putative | 6.72/108.8 | 7.12/108.2 | 16% | 103 |
| 143c | At5g37510 | NADH-ubiquinone dehydrogenase, mitochondrial, putative | 6.24/82.2 | 6.59/81.2 | 31% | 220 |
| 144c | At1g23190 | Phosphoglucomutase, cytoplasmic, putative | 5.82/63.2 | 6.21/63.2 | 20% | 156 |
| 145c | At5g65690 | Phosphoenolpyruvate carboxykinase [ATP], putative | 5.98/69.3 | 6.37/72.9 | 14% | 109 |
| 146c | At2g45290 | Transketolase, putative | 5.63/69.3 | 6.55/79.9 | 12% | 70 |
| 152c | At3g03250 | UTP-glucose-1-phosphate uridylyltransferase, putative | 5.80/51.9 | 5.98/51.7 | 30% | 111 |
| 155c | At5g17310 | UTP-glucose-1-phosphate uridylyltransferase, putative | 5.73/52.1 | 5.79/51.9 | 31% | 100 |
| 156c | At3g03250 | UTP-glucose-1-phosphate uridylyltransferase, putative | 5.80/51.9 | 5.98/51.7 | 37% | 228 |
| 161c | At4g34200 | D-3-phosphoglycerate dehydrogenase, putative | 6.32/63.6 | 6.53/63.3 | 20% | 93 |
| 162c | At3g08590 | 2,3-biphosphoglycerate-independent phosphoglycerate mutase, putative | 5.53/60.9 | 5.62/60.7 | 51% | 261 |
| 163c | At3g08590 | 2,3-biphosphoglycerate-independent phosphoglycerate mutase, putative | 5.53/60.9 | 5.62/60.7 | 32% | 148 |
| 164c | At2g36530 | Enolase (2-phospho-D-glycerate hydroylase) | 5.54/48.0 | 5.51/47.7 | 44% | 189 |
| 165c | At5g08670 | ATP synthase beta chain 1, mitochondrial | 6.52/63.6 | 6.52/59.6 | 62% | 377 |
| 169Ac | At5g08670 | ATP synthase beta chain 1, mitochondrial | 6.18/59.8 | 6.52/59.6 | 48% | 219 |
| 171c | At3g13930 | Dihydrolipoamide S-acetyltransferase, putative | 7.55/58.9 | 7.75/58.4 | 36% | 158 |
| 176c | At1g09780 | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, putative | 5.36/63.0 | 5.20/60.5 | 27% | 119 |
| 181c | At5g08670 | ATP synthase beta chain 1, mitochondrial | 5.41/48.3 | 6.52/59.6 | 21% | 84 |
| 182c | At5g08670 | ATP synthase beta chain 1, mitochondrial | 5.41/48.3 | 6.52/59.6 | 37% | 187 |
| 186 | At1g12240 | Beta-fructosidase (BFRUCT4) | 5.39/73.7 | 5.30/73.8 | 18% | 149 |
| 220c | At2g21870 | Expressed protein | 9.0/25.1 | 6.59/27.6 | 21% | 84 |
| 228 | At1g30120 | Pyruvate dehydrogenase E1 component beta subunit, chloroplast | 5.92/44.7 | 6.31/44.2 | 12% | 68 |
| 241Ac | At5g08670 | ATP synthase beta chain 1, mitochondrial | 6.18/59.8 | 6.52/59.6 | 26% | 98 |
| 241Bc | At5g08680 | ATP synthase beta chain, mitochondrial, putative | 6.06/60.0 | 6.45/59.8 | 26% | 98 |
| 241Cc | At5g08690 | ATP synthase beta chain 2, mitochondrial | 6.18/59.8 | 6.59/59.7 | 26% | 98 |
| 243 | At2g27860 | Expressed protein | 5.49/44.1 | 5.47/43.6 | 14% | 63 |
| 247c | At4g10260 | Pfkb-type carbohydrate kinase family protein | 6.90/35.0 | 7.36/34.6 | 21% | 74 |
| 267c | At5g65690 | Phosphoenolpyruvate carboxykinase [ATP], putative | 5.98/69.3 | 6.37/72.9 | 13% | 100 |
| 268c | At5g13450 | ATP synthase delta chain, mitochondrial, putative | 9.12/26.3 | 9.86/26.3 | 28% | 102 |
| Metabolism (47) | ||||||
| 9 | At1g01050 | Inorganic pyrophosphatase, putative [soluble] | 5.58/26.7 | 5.96/24.5 | 32% | 75 |
| 14 | At5g62530 | Delta-1-pyrroline-5-carboxylate dehydrogenase (P5CDH) | 6.26/62.1 | 6.71/61.7 | 17% | 141 |
| 17c | At1g23730 | Carbonic anhydrase, putative | 6.54/29.2 | 6.99/28.8 | 38% | 112 |
| 21c | At1g23730 | Carbonic anhydrase, putative | 6.54/29.2 | 6.99/28.8 | 31% | 97 |
| 30 | At4g09320 | Nucleoside diphosphate kinase 1 (NDK1) | 6.84/15.8 | 8.47/18.8 | 33% | 70 |
| 38c | At5g26667 | Uridylate kinase, uridine monophosphate kinase (PYR6) | 5.79/22.6 | 5.83/22.5 | 50% | 181 |
| 41c | At1g75270 | Dehydroascorbate reductase, putative | 6.0/23.5 | 6.03/23.4 | 24% | 73 |
| 54c | At2g46860 | Inorganic pyrophosphatase, putative [soluble] | 5.55/25.1 | 5.63/24.9 | 30% | 65 |
| 55c | At2g46860 | Inorganic pyrophosphatase, putative [soluble] | 5.55/25.1 | 5.63/24.9 | 43% | 105 |
| 58 | At1g54870 | Short-chain dehydrogenase / reductase (SDR) family protein | 5.92/31.4 | 8.75/36.7 | 23% | 80 |
| 62c | At1g47260 | Bacterial transferase hexapeptide repeat-containing protein | 6.71/30.2 | 7.27/30.0 | 38% | 119 |
| 66c | At5g16510 | Alpha-1,4-glucan-protein synthase (UDP-forming) | 5.06/39.0 | 4.82/38.6 | 27% | 116 |
| 73 | At3g22850 | Expressed protein | 5.84/27.5 | 6.12/27.1 | 27% | 83 |
| 74c | At5g01410 | Stress-responsive protein, putative | 5.79/33.4 | 5.98/33.2 | 25% | 122 |
| 78c | At5g01410 | Stress-responsive protein, putative | 5.79/33.4 | 5.98/33.2 | 24% | 92 |
| 79c | At2g05990 | Enoyl-[acyl-carrier protein] reductase [NADH], chloroplast, putative | 9.19/41.5 | 9.36/41.2 | 40% | 170 |
| 81c | At2g45600 | Expressed protein | 6.15/36.8 | 6.41/36.4 | 38% | 144 |
| 83 | At5g66510 | Bacterial transferase hexapeptide repeat-containing protein | 6.75/28.0 | 7.30/27.8 | 33% | 107 |
| 84 | At5g03300 | Adenosine kinase 2 (ADK2) | 5.14/38.2 | 4.90/37.8 | 31% | 75 |
| 86c | At5g03630 | Monodehydroascorbate reductase, putative | 5.18/47.4 | 4.97/47.4 | 22% | 84 |
| 87Bc | At5g03630 | Monodehydroascorbate reductase, putative | 5.18/47.4 | 4.97/47.4 | 27% | 87 |
| 88c | At3g09820 | Adenosine kinase 1 (ADK1) | 5.29/38.3 | 5.11/37.8 | 72% | 271 |
| 90 | At1g14810 | Semialdehyde dehydrogenase family protein | 5.39/36.9 | 7.00/40.7 | 26% | 96 |
| 95 | At3g17820 | Glutamine synthetase (GS1) | 5.72/38.8 | 5.93/38.6 | 46% | 125 |
| 98c | At3g17940 | Aldose 1-epimerase family protein | 5.88/37.3 | 6.28/37.2 | 62% | 240 |
| 100 | At1g48470 | Glutamine synthetase, putative | 6.20/38.4 | 6.63/38.9 | 32% | 148 |
| 114 | At5g54160 | Quercetin 3-O-methyltransferase 1 | 5.73/40.1 | 5.81/39.6 | 22% | 72 |
| 116c | At1g02500 | S-adenosylmethionine synthetase 1 (SAM1) | 5.50/43.6 | 5.60/43.1 | 39% | 124 |
| 117 | At3g17390 | S-adenosylmethionine synthetase, putative | 5.51/43.2 | 5.60/42.8 | 30% | 103 |
| 118c | At4g01850 | S-adenosylmethionine synthetase 2 (SAM2) | 5.67/43.6 | 5.94/43.2 | 25% | 78 |
| 119c | At2g36880 | S-adenosylmethionine synthetase, putative | 5.76/42.9 | 6.09/42.5 | 46% | 179 |
| 127c | At2g30970 | Aspartate aminotransferase (ASP1) | 8.36/48.1 | 8.34/47.7 | 40% | 143 |
| 135 | At1g23800 | Aldehyde dehydrogenase, mitochondrial (ALDH3) | 6.21/56.8 | 7.35/58.1 | 29% | 185 |
| 138c | At1g53500 | NAD-dependent epimerase/dehydratase family protein | 6.04/75.7 | 6.40/75.2 | 32% | 257 |
| 140c | At5g17920 | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | 6.09/84.6 | 6.47/84.3 | 33% | 224 |
| 147c | At5g17920 | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | 6.09/84.6 | 6.47/84.3 | 16% | 88 |
| 150c | At5g15490 | UDP-glucose-6-dehydrogenase, putative | 5.76/53.7 | 5.82/53.1 | 31% | 185 |
| 153 | At3g29360 | UDP-glucose-6-dehydrogenase, putative | 5.69/53.7 | 5.77/53.1 | 35% | 139 |
| 157 | At1g19920 | Sulfate adenylyltransferase 2 | 6.15/53.7 | 6.58/53.6 | 13% | 74 |
| 159 | At3g06580 | Galactokinase (GAL1) | 5.67/55.1 | 5.81/54.3 | 23% | 123 |
| 160 | At3g29360 | UDP-glucose 6-dehydrogenase, putative | 5.69/53.7 | 5.77/53.2 | 13% | 64 |
| 204c | At5g40370 | Glutaredoxin, putative | 6.71/11.9 | 7.34/117.5 | 62% | 82 |
| 214 | At3g22850 | Expressed protein | 5.84/27.5 | 6.12/27.1 | 22% | 78 |
| 224c | At5g19550 | Aspartate aminotransferase 2 (ASP2) | 6.80/44.5 | 7.32/44.3 | 28% | 81 |
| 225 | At4g31990 | Aspartate aminotransferase, chloroplast | 8.18/50.0 | 8.38/49.8 | 19% | 87 |
| 226c | At5g07440 | Glutamate dehydrogenase 2 (GDH2) | 6.07/45.0 | 6.51/44.7 | 24% | 79 |
| 227c | At4g13930 | Glycine hydroxymethyltransferase, putative | 6.80/52.1 | 7.25/51.7 | 44% | 226 |
| 232 | At4g23100 | Glutamate-cysteine ligase | 6.16/58.9 | 6.52/58.5 | 14% | 100 |
| 234c | At5g57655 | Xylose isomerase family protein | 5.74/53.5 | 7.88/32.4 | 21% | 69 |
| 242 | At5g39320 | UDP-glucose 6-dehydrogenase, putative | 5.60/53.5 | 5.56/53.1 | 19% | 86 |
| 250c | At5g15490 | UDP-glucose 6-dehydrogenase, putative | 5.76/53.7 | 5.82/53.1 | 13% | 61 |
| 253c | At5g15490 | UDP-glucose 6-dehydrogenase, putative | 5.76/53.7 | 5.82/53.1 | 13% | 81 |
| 254 | At1g79440 | Succinate-semialdehyde dehydrogenase (SSADH1) | 6.10/55.6 | 6.90/56.5 | 39% | 175 |
| 255 | At5g35360 | Acetyl-coa carboxylase, biotin carboxylase subunit (CAC2) | 8.26/54.6 | 7.28/58.4 | 24% | 143 |
| 269 | At3g22850 | Expressed protein | 5.84/27.5 | 6.12/27.1 | 27% | 76 |
| 272c | At5g17920 | 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | 6.09/84.6 | 6.47/84.3 | 16% | 125 |
| 273c | At2g17420 | Thioredoxin reductase 2 | 6.26/40.2 | 5.81/54.3 | 22% | 77 |
| 276 | At3g22200 | 4-aminobutyrate aminotransferase | 6.33/51.3 | 8.07/55.2 | 17% | 85 |
| Protein fate (25) | ||||||
| 2c | At5g42020 | Luminal binding protein 2 (BiP-2) (BP2) | 5.11/73.8 | 4.84/73.5 | 31% | 273 |
| 25 | At5g48580 | FK506-binding protein 2-2 (FKBP15-2) | 4.90/17.8 | 5.05/17.6 | 22% | 64 |
| 42 | At4g31300 | 20S proteasome beta subunit A (PBA1) (PRCD) | 5.31/25.2 | 5.24/25.1 | 33% | 89 |
| 50 | At3g60820 | 20S proteasome beta subunit F1 (PBF1) | 6.95/24.9 | 7.49/24.6 | 53% | 154 |
| 52 | At1g18840 | Lactoylglutathione lyase, putative | 5.11/32.0 | 9.75/62.7 | 25% | 79 |
| 65 | At5g42790 | 20S proteasome alpha subunit F1 (PAF1) | 4.99/30.7 | 4.73/30.5 | 45% | 94 |
| 97-2c | At2g47470 | Thioredoxin family protein | 5.52/29.5 | 5.86/39.5 | 35% | 84 |
| 136c | At3g11830 | Chaperonin, putative | 6.03/60.2 | 6.29/59.8 | 15% | 73 |
| 149c | At3g02090 | Mitochondrial processing peptidase beta subunit, putative | 6.29/59.2 | 6.76/59.1 | 37% | 190 |
| 154c | At5g20890 | Chaperonin, putative T-complex protein 1, beta subunit | 5.59/57.8 | 5.66/57.3 | 29% | 130 |
| 158 | At2g24200 | Cytosol aminopeptidase | 5.66/54.8 | 5.72/54.5 | 25% | 157 |
| 166c | At5g28540 | Luminal binding protein 1 (bip-1) (BP1) | 5.08/73.9 | 4.81/73.6 | 11% | 62 |
| 167c | At5g28540 | Luminal binding protein 1 (bip-1) (BP1) | 5.08/73.9 | 4.81/73.6 | 17% | 93 |
| 168c | At5g02500 | Heat shock cognate 70 kDa protein 1 (HSC70-1) (HSP70-1) | 5.03/71.7 | 4.75/71.3 | 20% | 128 |
| 169Bc | At5g28540 | Luminal binding protein 1 (bip-1) (BP1) | 5.08/73.9 | 4.81/73.6 | 20% | 97 |
| 173c | At3g23990 | Chaperonin (CPN60) (HSP60) | 5.66/61.6 | 5.41/61.3 | 28% | 125 |
| 174c | At5g09590 | Heat shock protein 70/HSP70 (HSP70-5) | 5.63/73.2 | 5.44/73.0 | 11% | 74 |
| 178c | At1g51980 | Mitochondrial processing peptidase alpha subunit, putative | 5.94/54.5 | 6.26/54.4 | 35% | 215 |
| 180 | At3g13860 | Chaperonin, putative | 7.57/46.0 | 5.89/60.4 | 11% | 68 |
| 184 | At5g58290 | 26S proteasome AAA-ATPase subunit (RPT3) | 5.42/45.9 | 5.25/45.7 | 21% | 95 |
| 191 | At3g05530 | 26S proteasome AAA-ATPase subunit (RPT5a) | 4.91/47.7 | 4.65/47.5 | 37% | 132 |
| 212 | At2g27020 | 20S proteasome alpha subunit G (PAG1) (PRC8) | 5.77/27.5 | 6.25/27.3 | 29% | 92 |
| 218 | At1g11910 | Aspartyl protease family protein | 5.29/53.0 | 5.20/54.6 | 17% | 101 |
| 219 | At5g01660 | Ferritin 1 (FER1) | 5.73/28.2 | 4.79/68.5 | 13% | 74 |
| 235c | At1g21750 | Protein disulfide isomerase, putative | 4.81/55.9 | 4.54/55.6 | 54% | 301 |
| 239A | At2g05850 | Serine carboxypeptidase S10 family protein | 5.46/55.2 | 5.55/54.7 | 17% | 65 |
| 288 | At2g05840 | 20S proteasome alpha subunit A2 (PAA2) | 5.75/27.4 | 5.91/27.3 | 30% | 108 |
| Protein synthesis (2) | ||||||
| 67c | At3g09200 | 60S acidic ribosomal protein P0 (RPP0B) | 5.0/34.2 | 4.69/34.1 | 27% | 63 |
| 68c | At3g09200 | 60S acidic ribosomal protein P0 (RPP0B) | 5.0/34.2 | 4.69/34.1 | 30% | 87 |
| 99c | At4g02930 | Elongation factor Tu, putative/EF-Tu, putative | 6.25/49.6 | 6.68/49.4 | 39% | 195 |
| Signal transduction (11) | ||||||
| 4c | At1g35720 | Annexin 1 (ANN1) | 5.21/36.1 | 5.01/36.2 | 41% | 124 |
| 10A | At1g15470 | Transducin family protein / WD-40 repeat family protein | 5.72/36.7 | 6.07/36.3 | 42% | 107 |
| 10B | At5g65020 | Annexin 2 (ANN2) | 5.76/36.4 | 5.96/36.3 | 50% | 154 |
| 18c | At5g63400 | Adenylate kinase | 6.91/27.1 | 7.45/26.9 | 41% | 119 |
| 104c | At5g16760 | Inositol 1,3,4-Trisphosphate 5/6 kinase | 5.89/36.4 | 6.43/36.2 | 36% | 119 |
| 105c | At5g16760 | Inositol 1,3,4-Trisphosphate 5/6 kinase | 5.89/36.4 | 6.43/36.2 | 39% | 135 |
| 112 | At2g46280 | Eukaryotic translation initiation factor 3 subunit 2 | 6.50/36.7 | 6.99/36.4 | 38% | 198 |
| 139 | At1g50480 | 10-formyltetrahydrofolate synthetase (THFS) | 6.26/68.3 | 6.69/67.8 | 42% | 302 |
| 172c | At5g09550 | Rab GDP dissociation inhibitor, putative | 5.11/50.0 | 4.91/49.5 | 34% | 121 |
| 246 | At1g48630 | Guanine nucleotide-binding family protein | 7.07/36.3 | 7.12/35.8 | 34% | 131 |
| 270c | At1g30580 | Expression protein,putative GTP-binding protein | 6.35/44.7 | 6.79/44.5 | 50% | 215 |
| 283 | At4g34490 | Cyclase-associated protein (cap1) | 6.23/51.5 | 6.62/50.9 | 14% | 66 |
| Storage protein (2) | ||||||
| 97-1c | At1g07750 | Cupin family protein | 5.83/38.5 | 6.06/38.3 | 20% | 62 |
| 103c | At2g28680 | Cupin family protein | 6.25/38.7 | 6.68/38.4 | 31% | 88 |
| Subcellular localization (1) | ||||||
| 239B | At3g23940 | Dehydratase family | 5.85/65.6 | 6.15/64.9 | 23% | 123 |
| Transcription (2) | ||||||
| 32 | At2g01860 | Pentatricopeptide (PPR) repeat-containing protein | 9.25/56.0 | 9.66/55.6 | 17% | 72 |
| 93Bc | At4g12130 | Glycine cleavage T family protein/aminomethyl transferase family protein | 6.30/43.6 | 6.69/43.5 | 30% | 75 |
| Transport (7) | ||||||
| 36c | At4g16160 | Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein | 6.97/18.7 | 7.89/18.6 | 64% | 146 |
| 37c | At4g16160 | Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein | 8.09/16.9 | 7.89/18.6 | 25% | 62 |
| 75 | At2g30050 | Transducin family protei/WD-40 repeat family protein | 5.42/32.8 | 5.53/32.6 | 30% | 79 |
| 129c | At3g24170 | Glutathione reductase, putative | 6.36/54.3 | 6.78/53.8 | 18% | 64 |
| 130c | At3g24170 | Glutathione reductase, putative | 6.36/54.3 | 6.78/53.8 | 36% | 175 |
| 151c | At1g80460 | Glycerol kinase, putative | 6.11/53.0 | 6.20/56.4 | 23% | 69 |
| 170c | At1g78900 | Vacuolar ATP synthase catalytic subunit A | 5.11/69.1 | 4.86/68.8 | 19% | 117 |
| 188c | At4g38510 | Vacuolar ATP synthase subunit B, putative | 5.03/54.4 | 4.77/54.3 | 35% | 146 |
| 189c | At1g20260 | Vacuolar ATP synthase subunit B, putative | 5.11/54.4 | 4.55/36.3 | 53% | 235 |
| Unclassified protein (4) | ||||||
| 31 | At2g10360 | Hypothetical protein | 7.74/15.0 | 8.18/14.8 | 17% | 68 |
| 43 | At2g31985 | Expressed protein | 5.79/26.9 | 6.03/26.7 | 33% | 72 |
| 183 | At1g72880 | Acid phosphatase survival protein surE, putative | 5.29/38.3 | 5.13/40.6 | 21% | 75 |
| 207 | At1g10590 | DNA-binding protein-related | 6.60/15.5 | 7.36/15.4 | 32% | 63 |
- aTotal protein spots identified from mature pollen and pollen tubes gels. The annotation of spot number with ‘A’, ‘B’ or ‘C’ indicates that two or three different proteins were identified from one protein spot.
- bFunctional category analysis was conducted using the website tools provided by Munich Information Center for Protein Sequences (MIPS), Bio-Array Resource (BAR), protein information resource (PIR), and Plant Energy Biology.
- cProteins reported by other pollen proteomic researches (Holmes-Davis et al. 2005; Noir et al. 2005; Sheoran et al. 2006).
- dThe theoretical pI and molecular weight (MW) of each protein was from the data of The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/index.jsp).
- AGI, Arabidopsis Genome Initiative.
It should be noted that the identified proteins did not always have a one-to-one correlation with the spots on the gels according to other proteomic analyses, which may have resulted from polypeptide variants that were present in different spots on the gel, but encoded by the same gene (Kerim et al. 2003; Holmes-Davis et al. 2005; Noir et al. 2005). In our experiments, there were 24 proteins detected from different spots (Table 1). There may be several possibilities for differential migration of the same protein. One possibility is a difference in post-translational modification of the proteins in vivo, such as phosphorylation, glycosylation, or acetylation (Krishna and Wold 1993; Jensen 2004). In most cases, these modifications do not significantly affect the molecular weight of a protein, but may induce a pI shift on the gel (Holmes-Davis et al. 2005; Noir et al. 2005). Another possibility is alternative splicing of mRNAs during translation (Smith et al. 1989; Brett et al. 2002) or chemical modification of the proteins during sample preparation.
Among the 303 protein spots analyzed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS), 213 protein spots representing 189 proteins were identified. This is the greatest number of identified proteins in Arabidopsis pollen and pollen tubes so far. One previous study (Holmes-Davis et al. 2005) identified a total of 135 proteins from the Arabidopsis mature pollen proteome, and 60 of them were identified in the present study. Another study identified 121 proteins from the Arabidopsis mature pollen proteome (Noir et al. 2005), and 76 of them were detected in our study. Compared with the proteomic analysis of Arabidopsis Landsberg ecotype pollen (Sheoran et al. 2006), 39 out of 95 identified proteins in the previous study were common in the present study. These differences may be attributed to the methods of protein extraction, choices of immobilized pH gradient (IPG) strips with different pH ranges, spots selection for analysis and the ecotype used (Sheoran et al. 2006). In our study, 75 new proteins were identified that were not reported in the previous studies (Table 1). Most of the newly identified proteins were classified into metabolism, energy and protein fate. Especially, we found that spots 42, 50, 65, 184, 191, 212, 288 were proteasome-related proteins. The ubiquitin/proteasome pathway represents one of the most important proteolytic systems in eukaryotes and has been proposed as being involved in pollen tube growth (Sheng et al. 2006). Inhibition of proteasome activity can significantly prevent pollen tube development and markedly alter tube morphology (Sheng et al. 2006). The pre-synthesized proteasome-related proteins in mature pollen indicate their important roles for rapid pollen germination and tube growth. Some other newly identified proteins (spots 3, 6, 7, 9, 20, 31, 32, 33, 73, 214, 269) are differently expressed between mature pollen and pollen tubes.
According to the gene sequences and a homological comparison with other known proteins, the identified proteins were classified into 15 different functional categories (Table 1; Figure 4A), including energy (31.2%), metabolism (24.9%), protein fate (13.2%), protein synthesis (1.1%), cell structure (6.3%), signal transduction (5.8%), defense/stress responses (4.8%), transport (3.7%), cell fate (2.6%), storage protein (1.1%), subcellular localization (0.5%), cell type differentiation (1.1%), development (0.5%), transcription (1.1%), and unclassified proteins (2.1%). Among the 189 proteins identified, approximately 70% of them were classified into three categories, including energy (31.2%), metabolism (24.9%) and protein fate (13.2%). This result may indicate the special requirement of these categories of proteins for pollen development or pollen germination. There were only four proteins (spots 31, 43, 183, 207) out of the 189 that could not be functionally classified as they were not observed to contain any known conserved domains.
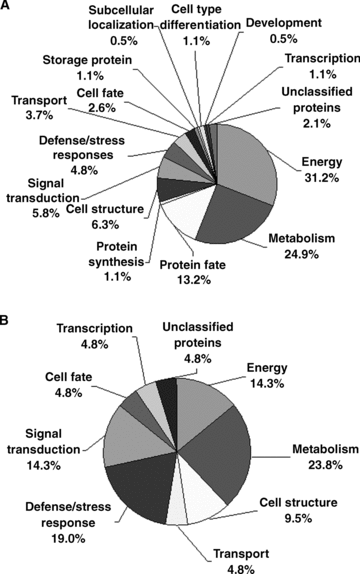
Functional categories of the total and differentially expressed proteins identified in Arabidopsis pollen grains and pollen tubes. Each different portion represents the percentage of proteins in the corresponding functional category. The putative functions of these proteins are listed in Table 1.(A) Functional categories of total identified proteins.(B) Functional categories of differentially expressed proteins.
Changes in protein expression profiles during the transition from the mature pollen grains to the germinated pollen
Based on the analysis of the reproducible protein spots of mature pollen grains and pollen tubes, 40 protein spots in total exhibited significant and reproducible changes during the transition from the mature pollen grains to germinated pollen materials (2, 3 and 5). There were 23 protein spots representing 21 proteins identified by MALDI-TOF MS (Table 2). Database analysis revealed that most of these proteins have either an unknown function, or are not yet characterized for their potential function in pollen development or germination. Further detailed functional characterization of these proteins may expand our understanding of complex mechanisms in regulation of pollen germination and pollen tube growth. Analysis of the subcellular locations (http://www.arabidopsis.org) of these proteins showed that most of these proteins are targeted to, or associated with, the cytoplasm and endomembranes. Further functional characterization of these proteins may reveal their potential roles in the regulation of pollen germination.
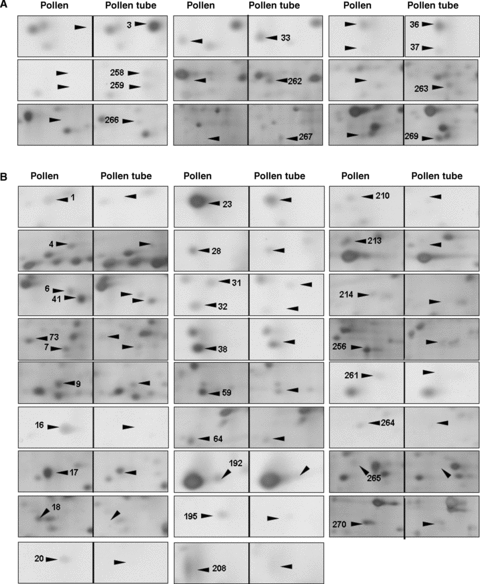
An enlarged view of the differentially expressed protein spots on the 2-D electrophoresis (2-DE) gels between pollen and pollen tubes.(A) Upregulated protein spots.(B) Downregulated protein spots.
| Spot No. | AGI name | Fold change (n= 3)a in proteome | Pollen specificityb | Reported in other proteome studies | Subcellular localizationc |
|---|---|---|---|---|---|
| Upregulated protein | |||||
| 3 | At1g60740 | Appeared | No | No | Unknown |
| 3 | At1g65970 | Appeared | No | No | Cytosol |
| 267 | At5g65690 | Appeared | No data | Yes | Unknown |
| 33 | At1g08480 | ↑2.0 ± 0.3 | No | No | Mitochondrion |
| 36 | At4g16160 | ↑2.0 ± 0.2 | No | Yes | Plastid outer membrane |
| 37 | At4g16160 | ↑3.7 ± 0.4 | No | Yes | Plastid outer membrane |
| 269 | At3g22850 | ↑3.0 ± 0.3 | No | No | Unknown |
| Downregulated protein | |||||
| 18 | At5g63400 | Disappeared | No | Yes | Mitochondrion |
| 20 | At1g48020 | Disappeared | No data | No | Endomembrane system |
| 214 | At3g22850 | Disappeared | No | No | Unknown |
| 1 | At3g52300 | ↓6.4 ± 0.3 | No | Yes | Mitochondrion, cytoplasm |
| 4 | At1g35720 | ↓2.6 ± 0.4 | No | Yes | Cytosol, membrane |
| 6 | At2g30870 | ↓2.3 ± 0.1 | No | No | Cytoplasm |
| 7 | At4g08390 | ↓2.6 ± 0.3 | No | No | Mitochondrion |
| 9 | At1g01050 | ↓3.1 ± 0.6 | No | No | Cytoplasm, nucleus |
| 17 | At1g23730 | ↓2.2 ± 0.3 | No | Yes | Cytoplasm |
| 23 | At4g24640 | ↓2.8 ± 0.5 | Yes | Yes | Endomembrane system |
| 28 | At1g26630 | ↓3.5 ± 1.2 | No | Yes | Unknown |
| 31 | At2g10360 | ↓3.0 ± 0.1 | No data | No | Endomembrane system |
| 32 | At2g01860 | ↓5.1 ± 0.4 | No | No | Chloroplast |
| 38 | At5g26667 | ↓2.4 ± 0.2 | No | Yes | Unknown |
| 41 | At1g75270 | ↓2.0 ± 0.2 | No | No | Unknown |
| 73 | At3g22850 | ↓2.2 ± 0.2 | No | No | Unknown |
| 270 | At1g30580 | ↓4.0 ± 1.2 | No | Yes | Unknown |
- aThe fold change values were determined by comparing the values between pollen tube and pollen.
- bPollen specificity determined according to Genevestigator (https://www.genevestigator.ethz.ch/).
- cSubcellular localization determined by using The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org).
- AGI, Arabidopsis Genome Initiative.
These differentially expressed proteins between mature pollen grains and pollen tubes were grouped into nine functional categories, including metabolism, energy, defense/stress responses, signal transduction, transport, and cell structure (Figure 4B). It is known that many processes, such as cell signaling, vesicle transport and fusion, expansion of the cell wall and plasma membrane, and actin dynamics, play essential roles in pollen germination and tube growth (Krishna and Wold 1993; Mascarenhas 1993; Franklin-Tong 1999a; Brett et al. 2002). Previous pollen transcriptome analyses have shown that a high proportion of genes in signal transduction, cell wall biosynthesis, and cytoskeletal dynamics are preferentially and selectively expressed in pollen (Pina et al. 2005). The present proteomic analysis demonstrated that approximately 29% of the proteins that were differentially expressed during the transition from mature pollen to germinated pollen belong to these functional categories. The proteins associated with energy and metabolism accounted for 38%, and the stress/defense responses proteins accounted for 19% of the differentially expressed proteins during the transition from mature pollen to germinated pollen.
Discussion
Proteins involved in cell structure
Pollen tube growth is a typical tip growth process. Thus, the apical cell wall must be plastic for stretching and for the incorporation of newly synthesized wall materials, and must be also rigid to withstand high internal turgor pressures (Bosch and Hepler 2005). The apical cell wall of a pollen tube is a pectic network, and pectin methylesterases (PMEs) act as key regulators of pollen tube growth (Bosch et al. 2005; Bosch and Hepler 2005). In the present proteomic analysis, two proteins (At1g48020, spot 20 in Figure 5B and At4g24640, spot 23 in Figure 5B) that match with pectin methylesterase inhibitor (PMEI) were identified. The At1g48020 had disappeared and At4g24640 was dramatically decreased in pollen tubes, as compared with mature pollen grains. Wolf et al. (2003) reported that At1g48020 (named as AtPMEI1) inhibited PME activity in flowers and siliques, and AtPMEI1 was preferentially expressed in pollen. Our results, together with the previous reports, suggest that PMEI may play an important role in loosening the cell wall during pollen tube growth. Further functional characterization of the PMEI may increase our understanding of cell wall construction during pollen tube growth.
Signaling proteins
It is well known that Ca2+ is an essential messenger and that Ca2+-regulated proteins serve as key signal transducers in the regulation of pollen tube growth. In plants, members of the annexin family are considered to be a group of calcium sensors involved in a number of signaling processes, such as interactions with the actin cytoskeleton and Golgi-mediated vesicle secretion (Clark et al. 2005). These annexin-associated processes also occur in a growing pollen tube (Franklin-Tong 1999a; Hepler et al. 2001; Robinson and Messerli 2002). In the present study, 2-DE gels analysis revealed that the expression of annexin 1 (AnnAt1, At1g35720, spot 4 in Figure 5B) was significantly decreased during the transition from mature pollen grains to germinating pollen. AnnAt1 is a 317-amino acid residue protein with a molecular weight of 37 kDa (pI 5.0). This protein mainly exists in a soluble form in the cytosol, but is also associated with the membranes (Lee et al. 2004). AnnAt1 protein has been reported to be involved in plant responses to NaCl, abscisic acid (ABA), osmotic, and oxidative stress (Lee et al. 2004; Gorecka et al. 2005). It is known that these stress responses are closely related to Ca2+-signaling events (Xiong et al. 2002). Our proteome analysis showed that AnnAt1 was significantly downregulated, with about 2.6-fold change, from mature pollen grains to pollen tubes, suggesting its potential function in the regulation of pollen tube growth. Another protein (At1g30580, spot 270 in Figure 5B) identified in mature pollen grains matched a predicated guanosine triphosphate (GTP)-binding protein with an unknown function. GTP-binding proteins have been reported to play important roles in pollen germination and pollen tube growth (Ma et al. 1999; Zheng and Yang 2000). The novel GTP-binding protein (At1g30580) identified in this study was significantly downregulated during pollen germination. It is worthwhile to further characterize this protein for its potential function in the regulation of pollen tube growth.
The ADK1 (At5g63400, spot 18 in Figure 5B) protein was only detected in the mature pollen samples. Adenosine kinase (ADK) is an enzyme involved in the adenylate metabolic network, by which adenosine (Ado) is converted into adenosine 5′-monophosphate (AMP) using one molecule of adenosine 5′-triphosphate (ATP) (Moffatt et al. 2000). In animal cells, ADK has been reported to modulate methyltransferase activity, the production of polyamines and secondary compounds, and cell signaling (Young et al. 2006). In plants, an analysis of a mutation in ADK1 has suggested that the function of ADK was to modulate root cap morphogenesis and gravitropism (Young et al. 2006). It is also known that the expression level of ADK1 is low in leaves, but very high in flowers, stems, and roots (Moffatt et al. 2000). Our results indicate that ADK1 is only expressed in mature pollen grains and not in pollen tubes, which may indicate a possible involvement of ADK1 in pollen maturation development.
Proteins associated with transport events
It is well known that the transport of various ions, such as Ca2+, K+ and Cl−, is extremely important for pollen germination and tube growth (Hepler et al. 2001; Robinson and Messerli 2002), and many transporter proteins have been reported to regulate pollen germination and tube growth (Bock et al. 2006). Among the differentially expressed proteins between the mature pollen grains and the pollen tubes, one protein (At4g16160, spots 36 and 37 in Figure 5A) was categorized for potential functions in membrane transport. At4g16160 was detected in different locations on the gels (spots 36 and 37 in Figure 5A), and its expression was upregulated during the transition from mature pollen grains to growing pollen tubes. This protein has been previously suggested to be a transporter protein (named AtOEP16-S) because it shares a 32% identity with pea OEP16 that was the first chloroplast outer membrane channel protein to be isolated (Pohlmeyer et al. 1997; Drea et al. 2006). The β-glucuronidase (GUS) expression analysis of AtOEP16-S demonstrated that pollen grains and seeds contain a high level of GUS activity, which is consistent with the results of in situ hybridization, and that the AtOEP16-S promoter appeared to be most active during the maturation phase in both pollen and seed (Pohlmeyer et al. 1997; Drea et al. 2006). In addition, the expression of AtOEP16-S in siliques is related to the ABA signaling pathway because it requires the transcription factors ABI3 and ABI5 (Drea et al. 2006). The increased expression of AtOEP16-S in pollen tubes in the present study (spots 36 and 37 in Figure 5A) may indicate that this protein functions as a potential transporter for facilitating ion and/or metabolite fluxes during pollen tube growth.
Proteins related to stress responses
Pollen germination begins with pollen hydration, and pollen tube growth requires a relatively high turgor pressure at the pollen tube tip (Bosch et al. 2005). During this continuous growth process, pollen and pollen tubes appear to use a series of strategies against various stresses (Zonia and Munnik 2004). In the present study, four proteins (At1g60740, At1g65970, At2g30870 and At4g08390) were identified and they may be related to pollen responses to stress.
The proteins At1g60740 and At1g65970 both matched with spot 3 (Figure 5A) and only expressed in pollen tube samples. These two proteins are predicted to be putative type 2 peroxiredoxin (PRXII) and are named as AtPRXII-D and AtPRXII-C. They each have an open reading frame (ORF) encoding 162 amino acids and have a high degree of similarity. It has been reported that the GUS activity in AtPRXII-D::GUS plants was not only detected in mature pollen (similar to AtPRXII-C), but also in germinating pollen, pollen tubes as well as fertilized ovules (Bréhélin et al. 2003). However, the corresponding spot on the gel for these two proteins was exclusively and remarkably detected in pollen tube samples in this study, which indicates that these two proteins were either synthesized during pollen germination or present in the mature pollen grains in an undetectably low abundance. The peroxiredoxin (PRX) family includes a group of alkyl hydroperoxide reductases, consisting of antioxidant enzymes that protect macromolecules from damage caused by reactive oxygen species (ROS), and the donor molecules of these reducers are all thio-containing substances such as thioredoxin (TRX) and glutaredoxin (GRX) (Bréhélin et al. 2003). The significant expression of AtPRXII-D and AtPRXII-C in growing pollen tubes may protect some required components for pollen tube growth, such as the TRX protein (spot 98 in Figure 1), which was detected without changes in expression in different materials in this study.
The expressions of two other stress-related proteins (At2g30870, At4g08390) were downregulated in pollen tubes (spot 6 and spot 7 in Figure 5B). At2g30870 is a dehydration early-response gene (ERD13) that has been reported as a stress-induced gene (Bianchi et al. 2002). At4g08390 encodes an ascorbate peroxidase that should localize to the chloroplast stroma and the mitochondria, according to database analysis (http://www.arabidopsis.org). As one of the ROS-scavenging enzymes, it plays a major role in the removal of H2O2 in living cells. Although both proteins were detected in the mature pollen samples, as shown in this study, the transcription of these two genes was very low in mature pollen (Honys and Twell 2004; Pina et al. 2005). These results may suggest that these two proteins may accumulate in mature pollen grains for protecting pollen from dehydration and oxidation damage during the pollen late maturation phase.
Proteins involved in energy and metabolism
It is known that pollen tube development and growth has large energetic and biosynthetic requirements (Hepler et al. 2001). In order to meet these demands, mature pollen grains contain abundant carbohydrates as energy sources, and pollen germination and tube growth are accompanied by high metabolic activity (Tagede and Kuhlemeier 1997). In the present study, eight differentially expressed proteins (At5g65690, At1g08480, At3g22850, At3g52300, At1g01050, At1g23730, At5g26667, At1g75270) were identified and categorized as energy and metabolism proteins, and account for 38% of all differentially expressed proteins. According to a database analysis, these proteins are closely related to many processes in metabolism and energy production, such as oxidative phosphorylation, carbohydrate metabolism, and sugar metabolism. However, no detailed information is available for their specific function so far. Further detailed functional characterization of these proteins is required to reveal their potential functions in pollen germination and tube growth.
Materials and Methods
Pollen grain collection and in vitro pollen germination
Arabidopsis plants (ecotype, Columbia) were grown under the same condition described by Wang et al. (2008). Mature pollen grains were collected from freshly anther-dehisced flowers using a vacuum cleaner as described previously (Johnson-Brousseau and McCormick 2004) and were immediately frozen in liquid nitrogen for protein extraction. The “thin liquid layer” germination methods were used for collection of a large quantity of pollen tubes (Wang et al. 2008). After incubation at 25°C for 3.5 h, the averaged pollen tube length was about 140 μm. The pollen tube samples were collected and centrifuged at 1 500 ×g at 4°C for 3 min. The supernatants were discarded and the pelleted pollen tubes were resuspended in double-distilled water supplemented with 1 mM phenylmethanesulfonyl fluoride (PMSF) and quickly frozen in liquid nitrogen for protein extraction.
Protein extraction
The proteins were extracted following the methods described previously with slight modifications (Damerval et al. 1986; Natera et al. 2000). The frozen samples of pollen grains or pollen tubes were ground in liquid nitrogen into fine powder and collected in a sterilized 2 mL centrifuge tube. The homogenate was precipitated with cold protein extraction buffer containing 10% trichloroacetic acid (TCA) (w/v) and 0.07%β-mercaptoethanol (v/v) in acetone for 1 h at −20°C, and then was centrifuged at 15 000g for 25 min at 4°C. The precipitate was washed three times with the same cold protein extraction buffer without 10% TCA (w/v), followed by a 1 h incubation at −20°C and subsequent centrifugation at 15 000g for 25 min at 4°C for each wash. The pellets were vacuum-dried, weighed, and dissolved in a lysis buffer containing 7 M urea, 2 M thiourea, 4% CHAPS (w/v), 50 mM dithiothreitol (DTT), and 0.5% carrier ampholytes (pH 3–10). After 1 h of lysis at room temperature with vortexing every 10 min, the homogenate was centrifuged at 15 000g for 20 min. The supernatant was transferred to a 1.5 mL centrifuge tube and stored in aliquots at −80°C. Protein samples extracted from four collections of pollen or pollen tubes were combined together for further analysis. The protein concentration of the supernatant was determined by Bradford assay, with a range of known concentrations of bovine serum albumin (BSA) as standard (Bradford 1976).
2-D gel electrophoresis and silver staining
Protein samples (300 μg) were diluted in a rehydration buffer containing 8 M urea, 2% CHAPS (w/v), 50 mM DTT, 0.5% pH 3–10 carrier ampholytes (v/v), and 0.01% bromophenol blue (w/v) for gel electrophoresis using a 17 cm IPG strip (pH 5–8, Bio-Rad, Hercules, CA, USA). After active rehydration (20°C, 50 V) for 12 h in a Bio-Rad PROTEAN IEF Cell, the IEF (isoelectric focusing) was performed at 20°C following the manufacturer's protocols. After IEF, the IPG strips were treated in a equilibration buffer containing 6 M urea, 0.375 M Tris-HCl (pH 8.8), 2% sodium dodecyl sulfate (SDS) (w/v), 20% glycerol (v/v), and 2% DTT (w/v) for 15 min, and subsequently, in the same buffer containing 2.5% iodoacetamide (w/v) without DTT for another 15 min. The second-dimension separations were carried out on 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels with a running buffer in a PROTEAN Plus Dodeca cell (Bio-Rad) electrophoresis tank at 16°C. The gels were run with a constant voltage at 100 V for 1 h, and then at 200 V until the tracking dye reached the bottom of the gels (approximately 6 h).
After electrophoresis, a silver-staining procedure compatible with mass spectrometric analysis was carried out with a slight modification (Yan et al. 2000). The gels were fixed in 40% ethanol (v/v) and 10% acetic acid (v/v) for 1 h, washed twice for 10 min in Milli-Q water, and then sensitized with a 30% ethanol (v/v), 6.8% sodium acetate (w/v), and 0.2% sodium thiosulfate (w/v) solution for 30 min. The gels were then rinsed with Milli-Q water three times for 10 min each. The gels were incubated in 0.25% (w/v) silver nitrate for 20 min, rinsed twice with Milli-Q water for 1 min, and then developed in a solution of 2.5% sodium carbonate (w/v) with formaldehyde (37%,w/v) added (400 μL/L) before use. When the desired intensity of staining was achieved, development was stopped with 1.46% ethylenediaminetetraacetic acid (EDTA) ·Na2·2H2O (w/v) for 10 min. The gels were stored in 1% acetic acid (v/v) at 4°C until further analysis. Three representative gels per sample were used for analysis.
Gel scanning and image analysis
The gels were scanned by ScanMaker 6000 (Microtek, Shanghai, China). Image analysis was carried out with PDQuest 6.2 software (Bio-Rad), including the quantitative analysis. After background subtraction and spot detection, the volume of each spot from three replicate gels was normalized against the total valid spots. Protein spots with reproducible and statistically significant changes in intensity (greater than twofold, paired t-test, P < 0.05) were considered to be differentially expressed proteins. Except for the differentially expressed protein spots, the best focused 263 spots present in both pollen and pollen tube samples were picked and subjected to detailed proteomic analysis.
In-gel digestion and mass spectrometry
In-gel digestion of proteins for MALDI-TOF MS was carried out according to the method described by Gharahdaghi et al. (1999) with some modifications. Before digestion, the protein spots were cut from the gels and rinsed twice with Milli-Q water, and then destained twice for 15 min using 200 μL of a freshly prepared 1:1 solution of 100 mM Na2S2O3 and 30 mM K3Fe(CN)6 by slow vortexing. After being rinsed twice for 5 min with Milli-Q water, the samples were dehydrated twice for 10 min in 200 μL of dehydration solution containing 25 mM NH4HCO3 and 50% acetonitrile (ACN) (v/v), shrunk for 10 min in 100 μL of ACN, and then completely dried under vacuum. For digestion, the samples were incubated in 10 μg/mL Roche sequencing-grade modified trypsin buffer (containing 1 mM CaCl2, 25 mM NH4HCO3, pH 8.3) for 45 min on ice. The remaining solution was then removed and replaced with 10 μL of 25 mM NH4HCO3 for digestion at 37°C for 12 h. The digestion solution was transferred to a 0.5 mL centrifuge tube and the digested samples were extracted using 40 μL of 0.1% trifluoroacetic acid (TFA) (v/v) by vortexing 20 min at room temperature, followed by 30 μL of a 70% ACN (v/v) and 5% TFA (v/v) solution twice by vortexing for 20 min. The digested samples from different tubes of the same original material were combined and vacuum-dried. A piece of the gel was cut from a protein-free region and processed in parallel with the samples as a control.
To acquire the PMF data, the vacuum-dried samples were dissolved in 10 μL of a 70% ACN (v/v) and 0.1% TFA (v/v) solution. Then the samples were spotted onto the Anchorchip target plate (600 μm, Bruker Daltonics, Germany) (1.0 μL) twice and 0.3 μL of 4 mg/mL α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution dissolved in 70% ACN (v/v), and 0.1% TFA (v/v) once. The sample spot was desalted with 1 μL of 0.1% TFA (v/v) twice and completely dried. The tryptic peptide masses were generated using a MALDI-TOF/TOF mass spectrometer (AUTOFLEX II TOF-TOF; Bruker Daltonics, Germany). The peptide calibration standard mono (Bruker Daltonics) was used as an external calibration to ensure the accuracy of protein identification.
Data analysis
The PMF data searches were carried out using MASCOT search tools (http://www.matrixscience.com) in the NCBI non-redundant public protein database. Arabidopsis thaliana (thale cress) was selected for the taxonomic category. All of the peptide masses were assumed to be monoisotopic and [M+H]+. Carbamidomethylcysteine was considered to be a fixed modification, while oxidation of methionine was considered to be a variable modification. The mass accuracy was set to ±100 ppm, and the maximum number of missed cleavages was set at one. The identified proteins should have more than four matched peptides, and the percentage of sequence coverage should be greater than 10%. All of the positive protein identification scores were significant (P < 0.05, mascot score > 60). Functional categories were assigned according to the data from Munich Information center for Protein Sequences (MIPS) (http://mips.gsf.de/proj/funcatDB/search_main_frame.html), Bio-Array Resource (BAR) (http://bar.utoronto.ca/), plant energy biology (http://www.plantenergy.uwa.edu.au/applications/suba/flatfile.php), and protein information resource (PIR, http://pir.georgetown.edu/). The lowest P-value was selected when multiple functional categories were allotted to one protein. The subcellular location and preferential expression pattern of the proteins were deduced by using The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org/) and Gene Atlas tool of Genevestigator (https://www.genevestigator.ethz.ch/).
(Handling editor: Weicai Yang)
Acknowledgements
We thank Dr Heven Sze (Department of Cell Biology & Molecular Genetics, University of Maryland) and Dr Tai Wang (Institute of Botany, Chinese Academy of Sciences, China) for critical reading of this article. We also thank Mr Jidong Feng (State Key Laboratory of AgroBiotechnology, China Agricultural University) for his assistance in mass spectrometry analysis.




