Higher-order repeats in the satellite DNA of the cave beetle Pholeuon proserpinae glaciale (Coleoptera: Cholevidae)
Abstract
The present study characterizes the satellite DNA of the cave beetle Pholeuon proserpinae glaciale which represent about 3–5 % of its genome, and which is composed of monomers of 266 bp and 70.5 % A-T. Concerted evolution seems to act on a higher-order repeat, a dimer, composed of two types of 266-bp monomers that differ in three diagnostic sites. These dimers show a striking nucleotide identity (98.7 % similarity) suggesting strong homogenization processes. The presence of particular mutations shared by several dimers represents an early expansion of these types of repeats as proposed by the molecular drive model. Moreover, evidence of gene conversion tracts in P. proserpinae glaciale, which also could be the result of unequal sister chromatid exchange, would suggest that recombination is involved in the homogenization of stDNA sequences. The presence of a 17-bp-motif repeated six times, along another one of 31-bp repeated twice which also have embedded one 17-bp-motif, suggest that monomers have originated from those basic motifs.
Satellite DNA (stDNA) is a highly repetitive portion of the eukaryotic genome composed of tandem repeat units arranged in large clusters (Elder and Turner 1995). These repeats show striking intraspecific similarities (homogenization of the repeats through a particular stDNA family and fixation through the whole population) but marked interspecific divergences (concerted evolution, Smith 1976). StDNAs described to date vary considerably among taxa in the length of the repeat (usually 100–500 bp), nucleotide composition (mostly A+T rich), number of repeats (from a few thousand to millions) and intra-specific sequence similarity (generally 1–13 %, King and Cummings 1997). In some species the sequence of repeats originated from short motifs (7–9 bp) as observed in Drosophila virilis (Gall and Atherton 1974) and D.melanogaster (Lohe et al. 1993). Other species show even more complex repeats such as the human alphoid stDNA (Willard and Wayne 1987), and the tenebrionid beetle Palorus subdepresus stDNA (Plohl et al. 1998). For instance, in human chromosome 7 there are two higher-order repeats (HORs) based on divergent subfamilies of the ∼171 bp alphoid monomer: a 6-monomer HOR, and a dimer (Willard and Wayne 1987). These HORs show striking sequence identity but within a HOR monomers show substantial sequence divergence because they are composed of divergent subfamilies. In the case of Palorus subdepressus the 144-bp-long repeat sequence is composed of two variants of a 72-bp-long subunit that are themselves composed of two copies of an octanucleotide alternating with 22-nucleotide-long elements of an inverted repeat.
However, evolutionary processes and function of this often called “junk DNA” are still controversial and poorly understood due to the lack of molecular data in most of the taxonomic groups and the discordance of results supporting opposing models. Therefore, we still need new data from a wider range of taxa to shed light on stDNA evolution (e.g. molecular mechanisms and factors involved in homogenization and fixation, and the fundamental unit of concerted evolution). StDNA sequence comparisons for further taxa are also likely to reveal the mechanisms of satellite origination (e.g. from smaller sequence motifs) and persistence of high copy number (major) satellites which seem to be linked to the role of stDNA in the tight condensation of heterochromatin (Ugarkovic and Plohl 2002).
With this in mind, our aim is to characterize the stDNA of Pholeuon proserpinae glaciale JEANNEL (Cholevidae) and hence assess these mechanisms in groups of Coleoptera other than tenebrionids which have been the most widely used study system (see reviews Petitpierre et al. 1995; Ugarkovic and Plohl 2002). It has been difficult to obtain satellite information from other Coleoptera since they do not represent a large fraction of the genome, and only three genera belonging to other beetle families have been studied to date: genus Nicrophorus (Silphidae; King and Cummings 1997), Chrysolina (Chrysomelidae; Lorite et al. 2001), and Cicindela (Cicindelidae, Galian and Vogler 2003). We specifically focussed in P. proserpinae glaciale because the chromosomes of this genus are known to have conspicuous heterochromatic blocks (Buzila and Marec 2000) which might be composed of stDNA sequences, as has been described for tenebrionids (Petitpierre et al. 1995). In such species tandem repeats can be isolated by digesting the genomic DNA with restriction enzymes, as they produce a characteristic ladder of conspicuous oligomeric bands in agarose gels which can easily be cloned and sequenced (Petitpierre et al. 1995).
Material and methods
Sampling, genomic DNA isolation, and isolation, cloning and sequencing of satellite DNA
Pholeuon proserpinae glaciale specimens were collected in the cave Avenul din Sesuri (Romania). Total DNA was isolated from ethanol preserved adult specimens using the DNeasy Tissue Kit (Qiagen). 1–2 micrograms of genomic DNA were digested with restriction enzymes (Alu I, Bfa I, Eco RI, Eco RV, Hind III, Hinf I, Not I, Rsa I, Sau 3A, Xba I) according to the instructions of the manufacturer (New England). Fragments were electrophoresed on 1.5 % agarose gels containing ethidium bromide. DNA from agarose block was purified with the Qiaquick Gel Extraction Kit (Qiagen), and cloned using the pMOSBlue blunt ended cloning kit (Amersham Pharmacia Biotech). Recombinant clones were sequenced on both strands by the dideoxy sequencing method using the Big Dye™ Terminator Cycle Sequencing Kit and a ABI PRISM™ 3700 DNA Analyzer (Applied Biosystems).
Southern hybridizations
The genomic restriction fragments analyzed on agarose gels were denatured, transferred onto nylon membranes (Osmotics), and cross-linked to the membrane by exposure to ultraviolet light. Digoxigenin labeling of the probe, filter hybridizations, and detection of the hybridization signals were performed as described in the manual for the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche). Stringency washes were performed to allow hybridization at the level of sequence similarity of 80–75 % (2×5 min in 2×SSC and 0.1 % SDS at RT, and 2×15 min in 0.1×SSC and 0.1 % SDS at 37 ° C).
Estimation of stDNA percentages
The digitalization and densitometric measurements of the gel were performed with Quantitative One software v. 4.2.1 (BIO-RAD). Thus, a conspicuous band of about 550 bp corresponding to stDNA could be directly compared to the total DNA signal (Pons et al. 2002).
Sequence analysis
Multiple sequence alignment was performed using the default parameters of Clustal W available in the Institute Pasteur's web page. Parsimony trees were obtained after 1000 random addition searches with TBR branch swapping and under the “Multrees” option in PAUP* 4.05 (Swofford 2002). All nucleotide positions in the matrix, including gap positions, were considered equivalent and weighted equally. Distance trees were built by the Neighbor-Joining method, and sequence divergences were calculated according the maximum likelihood model of nucleotide evolution F81+G selected by hLRT in Modeltest v. 3.06 (Posada and Crandall 1998). Bootstrap values were calculated on 1000 replicates in PAUP* 4.05 (Swofford 2002). The nucleotide composition and homogeneity of base frequencies across repetitive sequences were also estimated using χ2 tests in PAUP* 4.05. DNA polymorphism, DNA divergence, and putative gene conversion events were estimated using DnaSP v. 3.51 (Rozas and Rozas 1997). The search of substructures within the sequences was performed using the following software available in the Institute Pasteur's web page: dotmatcher, repeats, einverted, etandem, fuzznuc, palindrome, and equicktandem.
Results
Detection, cloning and Southern blot of stDNAs
The genomic DNA of P. proserpinae glaciale was digested with several endonucleases and the restriction fragments obtained were analyzed on agarose gels. Only Alu I induced a band of about 550 bp (Fig. 1a). The band was excised from the gel and the purified DNA was then labeled with digoxigenin. The partial digestion of the genomic DNA of P. proserpinae glaciale with Alu I, and its subsequent Southern hybridization using the labeled band as probe revealed a ladder of oligomers (Fig. 1b). Densitometric analysis of P. proserpinae glaciale genomic DNA digested with Alu I estimated that the band of about 550 bp constitutes about 3–5 % of the total DNA. The same band was cloned and 18 randomly selected clones were sequenced (named pPHO, acc. num. AJ535467-84). These sequences retrieved no significant Blast matches with any sequence in Genbank.
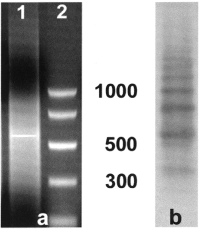
(a) Agarose gel electrophoresis of P. proserpinae glaciale genomic DNA digested with Alu I (lane 1). Lane 2 shows the DNA molecular standard whose bands range from 1 Kb to 150 bp. (b) Southern blot of P. proserpinae glaciale genomic DNA partially digested with Alu I and subsequent electrophoresed on agarose gel. The band of about 550 bp excised from gel (a) was used as a probe.
Analysis of stDNA sequences
The cloned sequences are 532 bp long, except for the sequence pPHO10 with a single base insertion at position 289, and sequence pPHO11 with two deletions (one base at position 288, and a 12 bp deletion corresponding to positions 498–509). They are A+T rich (A=32.3 %, C=16.5 %, G=12.8, and T=38.4 %). The 18 repeats studied have a striking sequence similarity (nucleotide diversity 1.3±0.1 %), and the variation seems to be evenly distributed throughout the repeat. Half of the nucleotide substitutions (15 out of 31) are single mutations of a particular monomer (autapomorphies). The remaining ones are particular nucleotide substitutions shared across several sequences within a particular site (synapomorphies), e.g. six sequences share a G in the position 63, and eight other sequences share a T in the position 365.
The analysis for the presence of substructures within the 532-bp-long sequences retrieved several short direct and inverted subrepeats, but surprisingly also revealed two direct repeats of 266 bp in length (Fig. 2). The duplicated pattern of the restriction sites also indicates that the 532-bp-long sequences are dimers composed of two very similar subunits or monomers (Fig. 3). Interestingly, the 266 bp-subunit harbours three direct and another three inverted repeats of a 17 bp-motif (TAATACATGCGTTTTTT) differing from the basic motif by two to six residues (motifs α1–α6, Fig. 4). Moreover, the 266 bp-monomer is flanked by two direct repeats of 31 bp, differing in 10 bp, that include one very conserved motif of 17 bp (motifs a1–a2, Fig. 4).

Dot plot of the 532-bp-long consensus satellite DNA sequence of P. proserpinae glaciale compared to itself. Plots were generated with the program Dotmatcher using a 10-bp window with a threshold of 17.
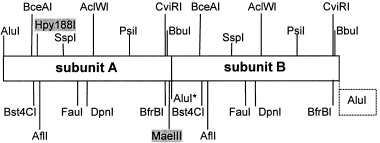
Schematic restriction map (single cutters only) of the 532-bp-long consensus satellite DNA sequence of P. proserpinae glaciale. Note the existence of two identical subunits (A and B) both 266 bp long and identical restriction map, except for Alu I, Hpy 188I and Mae III sites that are specific of subunit A. When genomic DNA is digested with Alu I, and since this site is mutated in subunits B (Alu I*), both subunits are isolated as a single fragment. The Alu I restriction site of the next repeat is highlighted in a dim-line box.

Consensus sequence of the 266-bp subunit (monomer) of P. proserpinae glaciale. Direct and inverse repeats of a basic 17 bp-motif (α1, TAATACATGCGTTTTTT) are indicated by arrows, and nucleotides substitutions relative to basic motif are indicated in brackets. Moreover, another direct repeat of 31 bp (a) flanks the monomer, which differ in 10 nucleotide substitutions and harbour two very conserved α repeats.
Monomers were analysed phylogenetically based on parsimony. The monomers exhibiting the Alu I restriction site were named pPHO-A, and those lacking the site were called pPHO-B. Parsimony searches on the monomers retrieved 30 equally most parsimonious trees of 54 steps. All of them showed the two types of monomers as monophyletic group with respect to each other, supported by high bootstrap values (Fig. 5). The Neighbor-Joining tree based on sequence divergence retrieved similar results (not shown). Within the two types the sequences have high sequence similarity (nucleotide diversity 1.2±0.2 % and 1.4±0.2 %, respectively, for pPHO-A and pPHO-B), which is similar to that between dimers (1.3±0.1 %). The level of sequence divergence between both types of monomers is slightly higher (Dxy divergence 3.4±0.3 %). Despite their striking similarities the sequences of each type can be distinguished due to the presence of three diagnostic sites (positions 1, 52, and 56). The presence of diagnostic nucleotide substitutions between types allows to test for the presence of gene conversion tracts in specific sequences. This analysis showed that both monomers 13a and 16a have one tract with the diagnostic mutations of the other type possibly caused by gene conversion (positions 62–68).
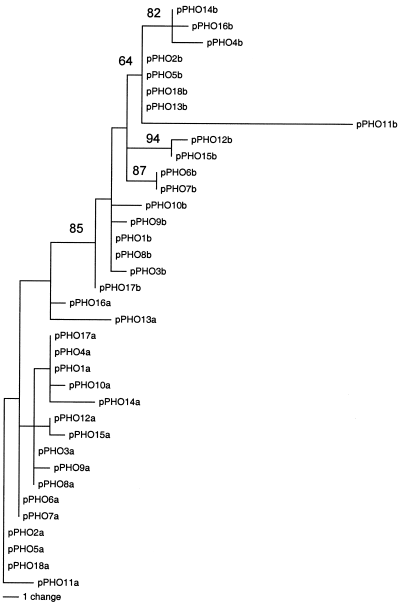
One of 30 shortest trees (54 steps) showing the relationships among the 36 P. proserpinae glaciale subunits studied (266 bp long). The subunits exhibiting the Alu I restriction site were called pPHO-A, and those lacking that restriction site were called pPHO-B. Numbers at each node indicate bootstrap proportions (>50 %) from 1000 bootstrap replicates.
Discussion
The ladder of oligomers obtained after the separation of genomic restriction fragments unveiled the existence of stDNA sequences in the cave beetle Pholeuon proserpinae glaciale. However, they constitute a smaller fraction of the genome than in darkling beetles (Ugarkovic et al. 1995) or nematodes (Elder and Turner 1995) where stDNA constitutes, in some species, more than 50 % of the genome. Interestingly, the nucleotide composition of P. proserpinae glaciale monomers is remarkably A-T rich compared with most of the stDNAs sequences described to date (Elder and Turner 1995; King and Cummings 1997).
It is clear that the 266-bp-long monomer is the repeat unit with regard to DNA similarity. If the monomer was also the unit that undergoes converted evolution, then dimers (two adjacent monomers) should randomly link monomers bearing different mutations, as described in the darkling beetle Tenebrio molitor (Plohl et al. 1992). This particular scenario is not applicable to P. proserpinae glaciale because its dimers are always composed of one monomer type A and one type B. Hence, this pattern of variation suggests that the dimer is the repetitive unit undergoing concerted evolution in P. proserpinae glaciale as described for the human alphoid stDNA of some chromosomes (Willard and Wayne 1987). In addition, the presence of 17-bp and 31 bp-motifs, along with the 266-bp subunits, suggests a trend where the actual repetitive sequence of P. proserpinae glaciale, the dimer, has originated from successive fusion of smaller motifs. Additional motifs may not be retrieved in the searches because mutation may have changed sequence motifs beyond any recognition. The formation of longer repeat units from small motifs also has been described in Mus musculus (Horz and Altenburger 1981), bovids (Lee et al. 1997), parasitic wasps (Rojas-Rousse et al. 1993) and fish of the genus Diplodus (Kato 1999). Theoretical simulations on short tandem repeats indicate that low ratios of unequal crossing over to mutation, common in heterochromatic regions, tend to form longer and complex repetitive units (Stephan and Cho 1994). Hence, the linking of divergent repeats would lead to the preservation of sequence identity, now between HORs but not within them, which is a prerequisite for recombination to occur. Otherwise, the coexistence of unlinked divergent sequences, which are not longer recognized by the recombination machinery, would lead to the extinction of such repetitive sequences.
P. proserpinae glaciale repeats exhibit a higher intra-specific sequence identity than most of the stDNA described in other beetles (1.2–20 % in tenebrionids, Petitpierre et al. 1995, Pons et al. 2002; 5 % in the leaf beetle Chrysolina americana, Lorite et al. 2001), and in other insects (1–13 %, King and Cummings 1997). This high nucleotide similarity suggests P. proserpinae glaciale HOR undergo very efficient homogenization. The presence of particular mutations shared by several HORs (synapomorphies) represents the result of an early expansion of these types of HORs (Strachan et al. 1985; Dover 2002). Further modification recognizable as nucleotide changes confined to a single repeat unit apparently has not spread widely by concerted evolution mechanisms. The molecular drive model predicts that gradual processes of homogenization will eventually result in the expansion of novel variants throughout the genome (Dover 2002), as has been demonstrated experimentally in a few cases (Strachan et al. 1985; Bachmann and Sperlich 1993; Pons et al. 2002). Nevertheless, these mutated sequences usually are lost due to homogenization because the rate of random shuffling of chromosomes carrying them in a large sexual population is substantially higher than their rate of spreading by molecular mechanisms of non-equal recombination (Dover 2002). Moreover, evidence of gene conversion tracts in P. proserpinae glaciale, which also could be the result of unequal sister chromatid exchange, would suggest that recombination is involved in the homogenization of stDNA sequences.
In summary, stDNA sequences of P. proserpinae glaciale exhibit similar characteristics to those described previously in other Coleoptera and other insects, but also reveal an interesting discrepancy regarding the repeat unit with respect to DNA similarity (monomers) and to processes of concerted evolution (dimers). Both detection of putative recombination events and mutations shared in several units suggest that concerted evolution of Pholeuon repeats is better explained by the molecular drive model (Dover 2002) rather than through extra-chromosomal transposition by rolling-circle replication as has been suggested in the rodent genus Ctenomys (Rossi et al. 1990). Finally, the presence of smaller motifs embedded in the P. proserpinae glaciale sequence suggests a trend towards increasing repeat length, first forming the 266-pb-monomer from a 17-pb-motif, and then linking two monomers to form the actual 532-bp-HOR.
Acknowledgements
We acknowledge funding through NERC grant NER/S/A/000674 to APV, JP and Tim Barraclough, and a grant of the European Commission's IHP Access to Research Infrastructures Programme to BR to visit the Natural History Museum.




