Association of Complement C3 Gene Variants with Renal Transplant Outcome of Deceased Cardiac Dead Donor Kidneys
Abstract
Local renal complement activation by the donor kidney plays an important role in the pathogenesis of renal injury inherent to kidney transplantation. Contradictory results were reported about the protective effects of the donor C3F allotype on renal allograft outcome. We investigated the influence of the donor C3F allotype on renal transplant outcome, taking all different donor types into account. C3 allotypes of 1265 donor–recipient pairs were determined and divided into four genotypic groups according to the C3F allotype of the donor and the recipient. The four genotypic groups were analyzed for association with primary nonfunction (PNF), delayed graft function, acute rejection, death-censored graft survival and patient survival. Considering all donor types, multivariable analysis found no association of the donor C3F allotype with renal allograft outcome. Also, for living and deceased brain-dead donors, no association with allograft outcome was found. Post hoc subgroup analysis within deceased cardiac dead (DCD) donors revealed an independent protective association of donor C3F allotype with PNF. This study shows that the donor C3F allotype is not associated with renal allograft outcome after kidney transplantation. Subgroup analysis within DCD donors revealed an independent protective association of the donor C3F allotype with PNF, which is preliminary and warrants further validation.
Abbreviations:
-
- ATG
-
- antithymocyte globulin
-
- DBD
-
- deceased brain death
-
- DCD
-
- deceased cardiac death
-
- DGF
-
- delayed graft function
-
- HLA
-
- human leukocyte antigen
-
- IRI
-
- ischemia-reperfusion injury
-
- Moab
-
- monoclonal antibody
-
- PNF
-
- primary nonfunction
-
- PRA
-
- panel reactive antibody
-
- RA
-
- receptor antagonist
Introduction
Long-term renal allograft survival after kidney transplantation is affected by different variables including donor type and age, kidney preservation methods, ischemia-reperfusion injury (IRI) and acute rejection (1–4). Today, the majority of renal allografts are recovered from deceased brain-dead (DBD) donors whereas donation after cardiac death (DCD) has emerged as an important alternative donor source to enlarge the deceased donor pool. Organs recovered from DBD as well as DCD donors have shown to give inferior transplant outcomes compared to organs recovered from living (un)related donors (5). In DBD donors, local and systemic immune activation occurs and is, at least in part, responsible for the pathogenesis of renal injury of kidney grafts to be. Importantly, a significant part of the brain death induced immune activation can be ascribed to local and systemic complement activation which is associated with reduced renal allograft function (6,7). Notably, significant systemic complement activation has been demonstrated in DCD donors already before withdrawal of treatment (8).
In renal transplantation, it was demonstrated that complement activation plays a substantial role in renal IRI and allograft rejection. Previous studies have shown that complement can also be activated by donor brain death, renal IRI and allograft rejection (6–9). C3 is the central complement component that can be activated by all the three complement pathways (10,11). In mice, it was shown that absence of local renal C3 in the donor kidney significantly improves early postreperfusion injury and late rejection associated allograft survival (12,13). The main producers of C3 in the kidney are tubular epithelial cells and the rejecting transplanted kidney can contribute up to 16% of all circulating C3 compared to 5% in resting kidneys (14). Complement C3 can be activated locally in the kidney at the tubular brush border of proximal tubular epithelial cells, inducing a proinflammatory response, thereby contributing to the pathogenesis of tubulointerstitial injury (15–18).
There are two allelic variants of the C3 allotype, namely, the slow variant (C3S) and the fast variant (C3F) which is based on the ability of C3 to migrate through a gel-electrophoresis system (19,20). The C3F allotype is characterized by a functional single nucleotide polymorphism (C–G) leading to a substitution of glycine (C3F) for arginine (C3S) at position 80. This might affect the ability of C3 to interact with monocyte complement receptors (21,22). The frequency of the C3F allotype is highest amongst Caucasians (20%) and less frequent in blacks (5%) and Asians (1%). In human kidney transplantation, conflicting results about the protective effects of the donor C3F allotype on allograft outcome of deceased donor kidneys have been reported (23,24). The disparity could be explained by an unequal distribution of DBD and DCD donor types among both studies.
The aim of this study was to investigate the influence of the donor C3F allotype on renal allograft outcome, taking all different donor types into account.
Methods
Patients and study design
Between March 7, 1993 and February 12, 2008, 1430 patients underwent kidney transplantation at the University Medical Center Groningen, the Netherlands. From this original group, 90 patients were excluded because of three or more kidney transplantations, simultaneous transplantation of other organs (pancreas, liver, lung and intestine) and technical problems during the operation. A total of 4 patients were lost to follow-up, of 65 transplantations, no donor and recipient DNA pairs were available and of 6 patients, no genotype could be assessed (Figure 1). Informed consent was given by all patients. Donor, recipient and transplant characteristics were obtained and documented. First, the whole study group (all donor types, n = 1265) was divided into four genotypic groups according to the presence or absence of the C3F allotype of the donor or the recipient. As the frequency of the C3FF allotype in the general population is low, C3FF and C3FS allotypes were combined and compared with the C3SS allotype in both the donor and recipient. Following this strategy, four groups were defined: (1) SS donor into SS recipient, (2) FS/FF donor into SS recipient, (3) SS donor into FS/FF recipient and (4) FS/FF donor into FS/FF recipient (Figure 1, Tables 1 and 2). Subsequently, the same subdivision was made as described above but now within living, DBD and DCD donor types separately.
Kidney transplantations that were included and excluded from the study.
| Variable | SS donor SS recipient (n = 529) | FS/FF donor SS recipient (n = 255) | SS donor FS/FF recipient (n = 247) | FS/FF donor FS/FF recipient (n = 234) | p-Value1 |
|---|---|---|---|---|---|
| Donor characteristics | |||||
| Age (years)2 | 47 (34–55) | 46 (33–55) | 47 (35–56) | 47 (38–55) | 0.792 |
| Sex no. (%) | |||||
| Male | 269 (51) | 139 (54) | 114 (46) | 120 (51) | 0.313 |
| Female | 260 (49) | 116 (46) | 133 (54) | 114 (49) | |
| Donortype no. (%): | |||||
| Living | 111 (21) | 49 (19) | 52 (21) | 68 (29) | 0.037 |
| DBD | 335 (63) | 161 (63) | 164 (66) | 124 (53) | |
| DCD | 83 (16) | 45 (18) | 31 (13) | 42 (18) | |
| Recipient characteristics | |||||
| Age (years)2 | 51 (39–60) | 48 (37–59) | 48 (38–58) | 48 (39–57) | 0.061 |
| Sex no. (%) | |||||
| Male | 309 (58) | 161 (63) | 137 (56) | 129 (55) | 0.238 |
| Female | 220 (42) | 94 (37) | 110 (44) | 105 (45) | |
| PRA level >5% (%) | 14 | 14 | 20 | 14 | 0.461 |
| Previous transplants no. (%) | |||||
| First transplant | 483 (91) | 225 (88) | 222 (90) | 208 (89) | 0.538 |
| Second transplant | 46 (9) | 30 (12) | 25 (10) | 26 (11) | |
| Primary kidney disease no. (%): | |||||
| Glomerulonephritis | 92 (17) | 45 (18) | 44 (18) | 41 (18) | 0.052 |
| Adult polycystic disease | 68 (13) | 36 (14) | 35 (14) | 28 (12) | |
| Renal vascular disease | 69 (13) | 24 (9) | 19 (8) | 11 (5) | |
| IgA nephropathy | 35 (7) | 15 (6) | 19 (8) | 29 (12) | |
| Pyelonephritis | 47 (9) | 33 (13) | 31 (13) | 35 (15) | |
| Diabetes | 27 (5) | 9 (4) | 8 (3) | 7 (3) | |
| Chronic | 71 (13) | 35 (14) | 30 (12) | 31 (13) | |
| Other | 120 (23) | 58 (23) | 61 (25) | 52 (22) | |
| Initial immunosuppression no. (%): | |||||
| Corticosteroids | 496 (94) | 240 (94) | 238 (96) | 221 (94) | 0.716 |
| Mycophenolic acid | 368 (70) | 189 (74) | 179 (73) | 166 (71) | 0.703 |
| Cyclosporin | 454 (86) | 218 (86) | 215 (87) | 193 (83) | 0.238 |
| Azathioprine | 28 (5) | 12 (5) | 14 (6) | 18 (8) | 0.204 |
| Tacrolimus | 38 (7) | 18 (7) | 20 (8) | 20 (9) | 0.507 |
| ATG | 36 (7) | 22 (9) | 24 (10) | 20 (9) | 0.397 |
| Anti-CD3 moab | 8 (2) | 4 (2) | 1 (0) | 6 (3) | 0.324 |
| Interleukin-2 RA | 87 (16) | 35 (14) | 40 (16) | 37 (16) | 0.834 |
| Sirolimus | 14 (3) | 6 (2) | 8 (3) | 9 (4) | 0.366 |
| Transplant characteristics | |||||
| Cold ischemia time (h)2 | 18 (12–23) | 17 (13–23) | 18 (10–23) | 17 (3–23) | 0.431 |
| HLA no. of 0 mismatches (%) | 101 (23) | 47 (22) | 50 (24) | 42 (23) | 0.991 |
- DBD = deceased brain death; DCD = deceased cardiac death; PRA = panel reactive antibody; ATG = antithymocyte globulin; moab = monoclonal antibody; RA = receptor antagonist.
- 1All p-values are two-sided. Kruskal–Wallis test for continuous variables, and chi-square test for categorical variables.
- 2Median (interquartile range).
| Variable | SS donor SS recipient (n = 529, reference group) | FS/FF donor SS recipient (n = 255) | SS donor FS/FF recipient (n = 247) | FS/FF donor FS/FF recipient (n = 234) | p-Value |
|---|---|---|---|---|---|
| Posttransplant follow up (years)1 | 6.89 | 7.01 | 7.09 | 7.08 | 0.7892 |
| (6.68 – 7.12) | (6.71 – 7.30) | (6.80 – 7.38) | (6.78 – 7.38) | ||
| Death censored graft survival3 | 1.00(1) | 0.78 | 0.87 | 0.85 | 0.793 |
| 1.00(2) | (0.52–1.18)(1) | (0.58–1.30)(1) | (0.56–1.31)(1) | ||
| 0.78 | 0.85 | 0.87 | 0.657 | ||
| (0.51–1.19)(2) | (0.56–1.30)(2) | (0.56–1.35)(2) | |||
| Patient survival3 | 1.00(1) | 0.93 | 1.03 | 0.72 | 0.518 |
| 1.00(2) | (0.61 –1.42)(1) | (0.68–1.55)(1) | (0.45–1.15)(1) | ||
| 0.99 | 1.11 | 0.71 | 0.501 | ||
| (0.63–1.56)(2) | (0.72 –1.72)(2) | (0.42–1.21)(2) | |||
| Primary nonfunction3 | 1.00(1) | 0.59 | 0.82 | 0.57 | 0.369 |
| 1.00(2) | (0.28–1.25)(1) | (0.42–1.63)(1) | (0.26–1.26)(1) | ||
| 0.52 | 0.78 | 0.54 | 0.351 | ||
| (0.23–1.19)(2) | (0.36–1.67)(2) | (0.22–1.38)(2) | |||
| Delayed graft function3 | 1.00(1) | 1.00 | 0.81 | 0.90 | 0.597 |
| 1.00(2) | (0.73–1.38)(1) | (0.59–1.13)(1) | (0.65–1.25)(1) | ||
| 1.08 | 0.83 | 1.04 | 0.627 | ||
| (0.76–1.55)(2) | (0.57–1.21)(2) | (0.70–1.53)(2) | |||
| Biopsy proven acute rejection | 1.00(1) | 1.27 | 1.18 | 1.03 | 0.447 |
| (1st year)3 | 1.00(2) | (0.92–1.75)(1) | (0.85–1.63)(1) | (0.74–1.44)(1) | |
| 1.09 | 1.03 | 1.06 | 0.967 | ||
| (0.75–1.58)(2) | (0.71–1.51)(2) | (0.71–1.58)(2) | |||
| Acute rejection classification no. (%) | |||||
| Overall | 155 (29) | 88 (35) | 81 (33) | 70 (30) | 0.4464 |
| Borderline | 49 (9) | 26 (10) | 27(11) | 20 (9) | 0.8084 |
| Banff IA or IB | 64 (12) | 39 (15) | 34(14) | 31 (13) | 0.6624 |
| Banff IIA, IIB, III | 42 (8) | 23 (9) | 20 (8) | 19 (8) | 0.9644 |
- DCD = deceased cardiac death.
- 1Mean estimate (95% confidence interval).
- 2Log-rank test.
- 3Hazard ratios (95% confidence interval).(1)Crude and (2)adjusted for donor and recipient gender and age, donor type, cold ischemia time, HLA-A, -B and -DR mismatch, transplant number, recipient primary renal disease.
- 4Chi-square test.
DNA isolation and genotyping
DNA was extracted from peripheral blood samples or splenocytes from deceased donors using a commercial kit following the manufacturer's instructions. Genotyping of the selected C3 single nucleotide polymorphism (rs2230199) was performed using the Illumina VeraCode GoldenGate Assay kit (Illumina, San Diego, CA, USA), according to the manufacturer's instructions. Genotype clustering and calling were performed using BeadStudio Software (Illumina). The overall genotype success rate was 99.5% and six samples with a high missing call rate were excluded from subsequent analyses.
Study end-points
The primary end-points in this study were: primary nonfunction (PNF, defined as nonfunctioning of the allograft from transplantation on), delayed graft function (DGF, defined as the requirement for dialysis within the first week after transplantation), biopsy proven acute rejection (all biopsies were re-evaluated according to the Banff 2007 classification) during the first year after transplantation, death censored graft survival (defined as the need for dialysis or re-transplantation) and patient survival.
Statistical analysis
Statistical analyses were performed using SPSS (version 18.0; SPSS Inc., Chicago, IL, USA) and two-sided p-values under 0.05 were considered to indicate statistical significance. To compare demographics between the four genotypic groups, the Kruskal–Wallis test was performed for continuous variables and the Chi-square test for categorical variables. The Mann–Whitney U test was applied to compare continuous variables between two genotypic groups.
First, univariate analyses were performed on the whole study cohort, for the association of the four genotypic groups with all transplant outcome parameters. Subsequently, same analyses were performed for the three donor types separately (living, DBD, DCD). Kaplan–Meier survival curves and log-rank tests were performed between the genotypic groups to assess the difference in death-censored graft survival rates and patient survival. To assess difference in PNF, DGF and acute rejection between the genotypic groups, chi-square tests were performed.
Second, multivariable logistic-regression models were constructed to find independent risk factors for PNF, DGF and acute rejection. Furthermore, Cox proportional hazards models were used to identify independent risk factors for death-censored graft failure and patient survival.
Results
No appreciable difference was observed between baseline characteristics of the study group compared to the original group (Supporting Information, Table S1). In donors, the frequency of the C3F allotype was 0.21 and 0.22 in recipients, respectively. These are comparable frequencies as reported by others and in accordance with the Hardy–Weinberg equilibrium.
First, the whole study group (all donor types, n = 1265) was divided into four genotypic groups, as previously described. Baseline characteristics between the four genotypic groups did not significantly differ (Table 1). Considering the whole study group, univariate and multivariable analysis showed that neither the C3 allotype of the donor nor that of the recipient was associated with PNF, DGF, acute rejection, death-censored graft survival and patient survival (Figure 2A, Table 2).
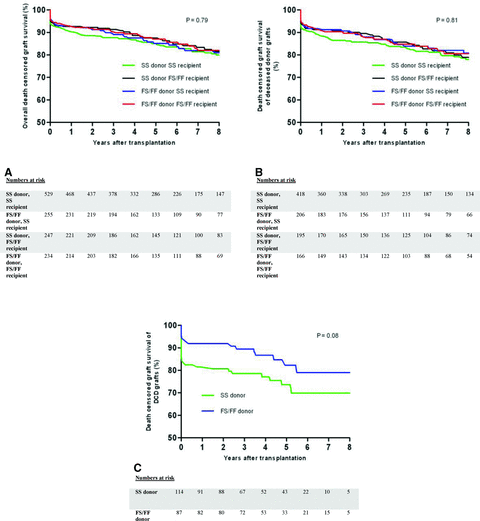
Death-censored graft survival among the four genotypic groups according to the presence or absence of the donor and recipient C3F allotype considering all donor types together (A) or only deceased donor types (B). Graft survival was not found to be significantly different between the four genotypic groups. In recipients of a deceased cardiac dead-donor kidney, superior graft survival of C3F donor kidneys can be fully attributed to the high incidence of primary nonfunction in recipients of a C3SS donor kidney (C).
As previously indicated, the disparity between the studies on C3 allotyping and renal allograft outcome might be explained by an unequal distribution of DBD and DCD donors among the two study cohorts. Therefore, similar analyses were also performed for living, DBD and DCD donor types separately. For living or DBD donors, no association was found between C3 donor and recipient allotype on all transplant outcome parameters (data not shown). Also when analyses were performed for deceased donors (DBD and DCD) separately, no effect of the donor C3F allotype on transplant outcome was found (Figure 2B). As the presence of the donor C3F allotype of kidneys recovered from deceased donors has been shown to give superior allograft survival rates when transplanted into C3SS recipients, we specifically analyzed this genotypic combination (24). Transplantation of a C3F allelic donor kidney into C3SS recipients was not associated with superior transplant outcome compared to C3SS into C3SS recipients for all transplant outcome parameters. Also, when analyses were performed for C3FF allotypes separately compared to all other allotypes, no differences were found for all transplant outcome parameters (data not shown).
However, in the subgroup of only DCD donors (n = 201), univariate analysis showed that transplantation of C3F allotypic DCD donor kidneys was significantly associated with lower PNF rates (p = 0.019, Table 3). Baseline characteristics did not significantly differ between the donor groups according to the presence of the C3F allotype (Table 4). Subsequently, multivariable logistic-regression analysis was performed with covariates that were found to be significantly associated with PNF by univariate analysis and the factors known from literature influencing graft outcome. These included donor and recipient gender and age, donor type, cold ischemia time, HLA-A, -B and -DR mismatch, transplant number, PRA level and the first warm ischemic time. Because it was commented that the disparity in the results between the studies by Brown et al. and Varagunam et al. might be due to an unequal distribution of primary kidney disease among the genotypic groups, we also included primary recipient kidney disease in our multivariable analysis (25). Multivariable analysis revealed that the C3F allotype of DCD donor kidneys was independently associated with a lower incidence of PNF (odds ratio 0.13, 95% CI 0.03–0.67; Table 5). Interestingly, the association with PNF was strongest among C3SS recipients receiving a C3SS DCD donor kidney (18%). Moderate or severe vascular rejection (Banff IIA, IIB, III) was the main cause of PNF in these patients (44%). Also, the incidence of acute vascular rejection was significantly higher in the first year after transplantation in recipients of a C3SS compared to C3F allelic donor kidneys (Table 3).
| Variable | SS donor (n = 114, reference group) | FS/FF donor (n = 87) | p Value |
|---|---|---|---|
| Posttransplant follow-up time (years)1 | 6.07 (5.47–6.68) | 6.88 (6.33–7.42) | 0.0782 |
| Death censored graft survival3 | 1.00(1) | 0.58 (0.30–1.09)(1) | 0.079 |
| 1.00(2) | 0.49 (0.25–0.97)(2) | 0.041 | |
| Patient survival3 | 1.00(1) | 0.43 (0.17–1.12)(1) | 0.084 |
| 1.00(2) | 0.14 (0.04–0.53)(2) | 0.003 | |
| Primary nonfunction3 | 1.00(1) | 0.22 (0.06–0.78)(1) | 0.019 |
| 1.00(2) | 0.12 (0.02–0.73)(2) | 0.021 | |
| Delayed graft function3 | 1.00(1) | 0.82 (0.40–1.70)(1) | 0.599 |
| 1.00(2) | 0.75 (0.31–1.79)(2) | 0.512 | |
| Acute rejection3 | 1.00(1) | 0.69 (0.36–1.31)(1) | 0.255 |
| 1.00(2) | 0.91 (0.43–1.96)(2) | 0.812 | |
| Acute rejection classification no. (%) | |||
| Overall | 33 (29) | 19 (22) | 0.2534 |
| Borderline | 14 (12) | 7 (8) | 0.3284 |
| Banff IA or IB | 11 (10 | 11 (13) | 0.5014 |
| Banff IIA, IIB, or III | 8 (7) | 1 (1) | 0.0434 |
- DCD = deceased cardiac death.
- 1Mean estimate (95% confidence interval).
- 2Log-rank test.
- 3Hazard ratios (95% confidence interval).(1)Crude and (2)adjusted for donor and recipient gender and age, donor type, cold ischemia time, HLA-A, -B and -DR mismatch, transplant number, recipient primary renal disease, PRA level and the first warm ischemic time.
- 4Chi-square test.
| Variable | SS donor (n = 114) | FS/FF donor (n = 87) | p-Value1 |
|---|---|---|---|
| Donor characteristics | |||
| Age (years)2 | 46 (35–55) | 44 (33–52) | 0.233 |
| Sex no. (%) | |||
| Male | 60 (53) | 55 (63) | 0.133 |
| Female | 54 (47) | 32 (37) | |
| Recipient characteristics | |||
| Age (years)2 | 52 (44–61) | 52 (42–62) | 0.977 |
| Sex no. (%) | |||
| Male | 57 (66) | 73 (64) | 0.828 |
| Female | 30 (34) | 41 (36) | |
| PRA level >5% (%) | 7 | 4 | 0.802 |
| Previous transplants (% second) | 7 | 10 | 0.401 |
| Primary kidney disease no. (%) | |||
| Glomerulonephritis | 21 (18) | 16 (18) | 0.319 |
| Adult polycystic disease | 18 (16) | 10 (12) | |
| Renal vascular disease | 10 (9) | 10 (12) | |
| IgA nephropathy | 11 (10) | 7 (8) | |
| Pyelonephritis | 7 (6) | 11 (13) | |
| Diabetes | 10 (9) | 4 (5) | |
| Chronic | 17 (15) | 8 (9) | |
| Other | 20 (18) | 21 (24) | |
| Initial immunosuppression no. (%): | |||
| Corticosteroids | 107 (94) | 81 (93) | 0.829 |
| Mycophenolic acid | 100 (88) | 75 (86) | 0.752 |
| Cyclosporin | 104 (91) | 78 (90) | 0.706 |
| Azathioprine | 5 (4) | 3 (3) | 0.737 |
| Tacrolimus | 3 (3) | 1 (1) | 0.457 |
| ATG | 5 (4) | 3 (3) | 0.737 |
| Anti-CD3 moab | 1 (1) | 1 (1) | 0.848 |
| Interleukin-2 RA | 43 (38) | 24 (28) | 0.132 |
| Sirolimus | 1 (1) | 1 (1) | 0.848 |
| Transplant characteristics | |||
| First warm ischemia time (min)2 | 18 (15–23) | 19 (15–23) | 0.935 |
| Cold ischemia time (h)2 | 18 (15–21) | 17 (14–21) | 0.444 |
| HLA no. of 0 mismatches (%) | 10 (9) | 7 (8) | 0.461 |
- DCD = deceased cardiac death; PRA = panel reactive antibody; ATG = antithymocyte globulin; moab = monoclonal antibody; RA = receptor antagonist.
- 1All p-values are two-sided. Mann–Whitney test for continuous variables, and chi-square test for categorical variables.
- 2Median (interquartile range).
| Variable | Odds ratio (95% CI) | p-Value |
|---|---|---|
| C3FS/FF vs. C3SS donor | 0.13 (0.03–0.67) | 0.014 |
| Donor age | 1.05 (1.00–1.11) | 0.045 |
| Donor gender | 1.30 (0.36–4.74) | 0.691 |
| Recipient age | 1.03 (0.98–1.09) | 0.243 |
| Recipient gender | 3.69 (0.73–18.72) | 0.122 |
| Second vs. first transplantation | 2.41 (0.12–47.16) | 0.561 |
| Recipient primary kidney disease | 1.15 (0.89–1.48) | 0.298 |
| PRA percentage | 0.95 (0.70–1.29) | 0.757 |
| First warm ischemia time | 1.13 (1.02–1.24) | 0.016 |
| Cold ischemia time | 1.00 (1.00–1.00) | 0.004 |
| HLA mismatch | 1.70 (1.03–2.82) | 0.040 |
- DCD = deceased cardiac death; CI = confidence interval; PRA = panel reactive antibody. A logistic-regression model was used to determine the odds ratio for PNF.
Death-censored graft survival and patient survival were also superior in recipients of a C3F allotypic donor kidney, whereas no association was found with acute rejection and DGF (Table 3). The difference in graft survival is likely to be attributed to the high incidence of PNF in recipients of a C3SS DCD donor kidney as graft survival rapidly declines shortly after transplantation (Figure 2c). To investigate this hypothesis, we analyzed the difference in graft survival, excluding patients suffering from PNF. In agreement with what we expected, no significant difference was found in graft survival between DCD donor kidneys based on the C3 genotype, excluding patients with PNF (p = 0.864). However, the difference in patient survival could not be explained by the high rate of PNF (all patients died with a functioning transplant).
Discussion
Complement activation has been shown to play an important role in the pathogenesis of renal injury after kidney transplantation. C3 is the central complement component that is activated by all complement activation pathways and is, therefore, essential for complement function. Two polymorphisms, C3S and C3F, of the C3 gene have been described, resulting in a substitution of glycine (C3F) for arginine (C3S). Recently, conflicting results regarding the protective effect of the donor C3F allele on renal allograft survival of deceased donor kidneys have been published (23,24). In our study, we confirm a recent report that the C3F allotype of the donor and recipient is not associated with renal allograft outcome after kidney transplantation (23). However, post hoc subgroup analysis within DCD donors revealed an independent protective association of the C3F allotype with PNF.
Earlier studies demonstrated that complement activation has detrimental effects on allograft function at different time-points during transplantation. It was demonstrated in several rodent models that activation of the complement system is an important mediator of renal IRI (9,26). Moreover, in a mouse allograft model, donor kidneys from C3 deficient animals showed superior graft survival rates compared to wild type donor kidneys (12). In contrast, when kidneys from wild-type mice were transplanted into C3 deficient mice, no such protection against transplant injury was found (13). These results indicate that not circulating C3 in the recipient but local C3 synthesis by the donor kidney is detrimental for renal transplant outcome. Besides, local renal complement induction and activation can be a direct result of brain death in the donor which is associated with impaired renal allograft function in the recipient (6,7).
Analysis of C3 polymorphisms in both the donor and recipient is an elegant, noninvasive, method to study the relevance of complement in human kidney transplantation. Brown et al. were the first to analyze the effect of the C3 allotype and found superior graft survival rates of C3F allotypic donor kidneys when transplanted into C3SS recipients (24). In a separate study by Varagunam et al., comprising a much larger transplant cohort, these results could not be confirmed (23). Unfortunately, it is difficult to compare both studies since Brown et al. included only 478 kidney transplants, of which, approximately 75% were derived from deceased donors. Importantly, the protective effect of the donor C3F allotype was only found in grafts recovered from deceased donors, not from living donors. In contrast, the study by Varagunam et al. included solely deceased kidney transplants of 1147 transplantations. Moreover, their study had follow-up data available, 8 years after transplantation, of 35% compared to 7% in the study by Brown et al. Hence, the study by Brown et al. is likely to be underpowered and, therefore, a final study was required to validate the findings by Varagunam et al.
Our study comprises the largest transplant cohort of 1265 kidney transplantation. Follow-up data was available 8 years after transplantation of 30% when all donor-types are considered and 26% among deceased donor types. Therefore, results from our cohort are comparable to the study cohort of Varagunam et al. Among recipients of a living donor kidney, we confirmed the study of Brown et al. that no effect of the C3F allotype on graft outcome is seen. Considering deceased donor kidneys, we validated the study by Varagunam et al. that the donor and recipient C3 allotype does not influence PNF, DGF, acute rejection, death-censored graft survival and patient survival.
However, when subgroup analyses were performed for DCD donor types separately, the donor C3F allotype was independently associated with lower rates of PNF in grafts derived from DCD donors. Also, an association of the donor C3 allotype with graft survival was found, however, these results could be fully attributed to the high incidence of PNF in recipients of a C3SS donor kidney. We are aware that the association found within DCD donors is preliminary and needs to be validated in other study cohorts. Our findings of an association within DCD donors might, however, explain the disparity in findings between the previous studies by Brown et al. and Varagunam et al. Although both studies performed analyses within deceased donors, no differentiation was made between DBD and DCD donor kidneys. Taking our findings into account, a significant difference between the proportion of DBD and DCD donors in both study cohorts might have influenced their results.
We envision that, particularly in DCD grafts experiencing PNF, the contribution of the C3F allotype is likely to be most significant. It is well-known that DCD kidneys that develop PNF are allografts that have experienced the worst type of renal injury. Recently, it has been demonstrated that renal injury and subsequent necrosis of tubular cells can activate the complement system (27). Therefore we hypothesize that, in this particular donor-type, the C3 allotype might be a risk factor for PNF. Obviously, this should be considered in the context of other well-known risk factors for PNF such as prolonged cold and first warm ischemia time, and a high number of HLA mismatches. This risk profile might help to reveal high-risk patients who require intensive surveillance or immunosuppression in the recipient.
In conclusion, this study shows that the C3F allotype of the donor and recipient is not associated with renal allograft outcome after kidney transplantation. Subgroup analysis within DCD donors revealed an independent protective association of the C3F allotype with PNF, which is preliminary and warrants further validation in other study cohorts.
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Acknowledgment
Funding source: This project has been supported by the foundation “De Drie Lichten” in the Netherlands.
Authors’ Contributions
J. Damman, M.R. Daha and M. A. Seelen designed and performed experiments, analyzed data and wrote the paper, H. Snieder analyzed data and edited the paper, M. C. R. F. van Dijk provided biopsy data, edited and approved the final manuscript, H. G. D. Leuvenink, H. van Goor, J. L. Hillebrands, B. G. Hepkema, J. van den Born, M. H. de Borst, S. J. L. Bakker, G. J. Navis and R. J. Ploeg edited and approved the final manuscript.




