Matching T-Cell Receptors Identified in Renal Biopsies and Urine at the Time of Acute Rejection in Pediatric Renal Transplant Patients
Abstract
Urinary monitoring of kidney allograft function has been used for many years. More recently, molecular identification of cytotoxic T-cell products has been used as a diagnostic tool in acute rejection. Monitoring of T-cell infiltrates by analysis of the T-cell receptor (TcR) gene usage has been performed on biopsies with acute and chronic rejection, but not on urine samples. The aim of this study was to identify and compare TRBV gene usage assessing the CDR3 (Complementarity Determining Region 3) length distribution and sequence in urine and biopsies of pediatric renal allograft patients at the time of acute rejection and compare them with peripheral blood. We studied four pediatric renal transplant recipients with acute cellular rejection. We identified restricted and matched TRBV CDR3 spectratypes with overexpressed TRBV families and show identical, clonally expanded TRBV CDR3 sequences in all four patients present in the urine and renal allograft. We demonstrate that urinary monitoring can detect graft-infiltrating lymphocytes in acute rejection and may have a role in the monitoring of renal transplants.
Introduction
Kidney transplantation is now the standard of care for patients with end stage of renal disease. The success of transplantation is significantly influenced by immune rejection. The rate of renal allograft failure in the first-year posttransplant has been reported as 18% for cadaver transplants and 6% for living-donor transplants (1). Acute rejection has occurred in up to 35% of recipients in the first-year posttransplant and is a major risk factor for allograft failure (2). Transplant rejection is dependent on T-cell-mediated allorecognition (3–5). The rejection of an allograft involves the activation of T cells by alloantigen either directly or indirectly (6,7). A large proportion of the T-cell population is activated by an allograft reflecting high levels of T-cell receptor crossreactivity with mismatched MHC molecules. Alloreactive T-cell frequencies vary 30-fold from 0.71 ± 0.31% to 21.05 ± 3.62% depending on different conditions in vivo (8).
The T-cell receptor (TcR) is the antigen-specific receptor on the surface of T lymphocytes. Over 95% of human peripheral blood T cells express the αβ TcR complex (9). TRBV repertoires reflect the diversity of the T-cell repertoire used by alloreactive T cells to recognize foreign tissue in transplantation. Altered TRBV repertoires have been described in studies of allograft rejection (10–12). Restricted TRBV repertoires were observed in patients with both acute and progressive chronic rejection (10). A highly altered TRBV repertoire and restricted CDR3 spectratypes have been shown in chronically rejected human kidney allografts with acute histological changes (13). Restricted usage has also been shown in peripheral blood mononuclear cells of patients with chronic renal rejection (14). The presence of an alloreactive clone in the kidney has been shown in a tolerant patient with a chronically rejecting renal allograft suggesting that this may allow monitoring of tolerant clones (15).
TcR diversity is seen predominantly in the CDR3 region. The diversity of the CDR3 region is generated by rearrangement of V, (D) and J genes and by junctional N-diversity (16,17). The CDR3 region is critical for the recognition of the peptide in the context of the MHC (18). Therefore, our analysis of the TcR usage is based on the study of the CDR3 length, distribution and sequence. Diversity in the TcR is estimated to be 2.5 × 107 different TcRs in humans. Where the beta chain is fixed, there is still the generation of 4 × 105 functional TcRs using only alpha chain diversity. There are multiple TcRs that can react with a single antigen. In the allotransplant setting this diversity is increased markedly with 5 × 106 likely alloreactive TcRs being used. Molecular screening of CDR3 sequences is used to identify the pathological sequence. In unrelated samples the random chance of selecting a matched TcR by sequencing is highly improbable. So that in a random selection of a single TRBV family TcR cDNA, the chance of finding a matched sequence after sequencing 10 clones would be extremely remote. The appearance of expanded TcR clones is reflected by the matched restriction in TcR length and is confirmed by sequencing (19).
Although the renal biopsy is still considered as an irreplaceable tool in assessing the diagnosis of rejection and guiding the treatment of many renal diseases, the development of noninvasive diagnostic tests to monitor transplant rejection may also have considerable value. Historically, the presence of lymphocytes in the urine has been described as part of the acute rejection process (20,21). Improved cytology and staining has allowed better identification of cytotoxic T cells in the urine (22). More recently, molecular methods have been used to identify rejecting tissue. Analysis of gene-expression patterns by using DNA microarrays in biopsy samples from patients at the time of rejection demonstrates differences in gene expression associated with the clinical course (23). The measurement of mRNA-encoding cytotoxic proteins granzyme and perforin in urinary cells has been studied as a noninvasive means of diagnosing acute rejection of renal allografts. This data showed the levels of mRNA-encoding cytotoxic proteins in urinary cells were significantly higher in the urine of patients at the time of acute rejection than of recipients without acute rejection (24). CD103 mRNA levels in urine has also been shown by the same group to be diagnostic of acute rejection in renal allografts (25). These approaches have been criticized as unable to distinguishing between T cells involved in rejection and other T-cell-associated transplant diseases such as CMV and urinary tract infections (26). Others have shown graft-specific DNA in urine by identifying Y antigen by PCR in the urine of female recipients of male kidneys at the time of rejection (27). Subclinical rejection has also been studied by protocol biopsy or urine spectroscopy as a surrogate marker for the development of chronic rejection (28). However, there have been no studies of TRBV gene usage of urinary T cells in transplant rejection.
The identification of cytotoxic T-cell products in urine suggested that it was possible to identify the T-cell repertoire of the graft-infiltrating T-cell population by analysis of TRBV usage. The aim of this study was to identify and match specific alloreactive T cells involved in acute rejection by analysis of their TRBV gene usage, CDR3 spectratypes and sequencing from the renal biopsy and urine at the time of rejection in pediatric renal transplant patients.
Materials and Methods
Patients
We studied four pediatric renal transplant patients with acute cellular rejection. Their clinical features are shown in Table 1. Two patients received living-related donor (LRD) grafts, while another two received cadaveric grafts. Peripheral blood (PB) and renal biopsies were collected at the time of rejection from all patients. Urine samples of patients were collected during episodes of rejection and various times after the rejection had resolved. Control urine samples from eight patients were collected from transplant patients who had not experienced rejection including one with a urinary tract infection. The patients all received standard triple immunosuppression with FK506, MMF and prednisolone and induction with basiliximab and experienced graft rejection treated with high-dose steroids. A clinical pathologist confirmed all rejection episodes as acute cellular rejection by biopsy and histological assessment.
| Patient | Donor | Date of transplant | Sex Da/Rb | Initial disease | Number of kidney grafts | HLA patient | HLA donor | Time of acute rejection after transplant |
|---|---|---|---|---|---|---|---|---|
| 1 | LRDc Father | 21 Jan. 2001 | Me/M | Dysplasia | 1 | A,1,11, B,44,–, DR,04,07 | A,1,2, B,44,–, DR,07,13, | 12 months |
| 2 | LRD Father | 28 Sept.01 | M/M | FSGSg | 1 | A,11,24, B,39,60, DR,4,8 | A,24,−, B,39,55, DR,8,9 | 2 months |
| 3 | CADd | 11 Dec. 2000 | Ff/M | JNOh | 1 | A,24,–, B,51,52, DR,11,15 | A,3,24, B,7,51, DR,11,15 | 13 months |
| 4 | CAD | 12 Dec. 1998 | F/M | HUSi | 1 | A,24,29, B,7,8, DR,10,17 | A,24,25, B39,44, DR,4,7 | 3 years |
- aDonor.
- bReceipt.
- cLiving-related donor.
- dCadaveric donor.
- eMale.
- fFemale.
- gFocal segmental glomerulosclerosis.
- hJuvenile nephronopthisis.
- iAtypical hemolytic uraemic syndrome.
Collection of PB, renal biopsy and urine samples
Peripheral blood lymphocytes (PBLs) were isolated by Ficoll-paque™ PLUS (Pharmacia Biotech) density gradient centrifugation from recipient PB. Samples of diagnostic renal biopsies were taken during routine care of patients with suspected acute rejection. Renal biopsy tissues were preserved in either OCT (Miles Laboratories) or placed in RNAlater (Ambion) for RNA extraction. About 40 mL of urine was collected and put in an Rnase-free container at 4°C. Urine was centrifuged at 500 × g for 5 min at 4°C. The supernatant was removed and 1 mL TRIzol reagent (Invitrogen) was added for RNA extraction. The yield of RNA from 40 mL of urine ranged from 550 ng to 7490 ng. From 780 ng to 2500 ng was used for RT-PCR for TRBV repertoire (23 TRBV families) of urine at the time of rejection based on the TRBC ratio to GAPDH.
Real-time PCR
Real-time PCR were performed in triplicate on urine samples from transplanted patients at the time of rejection and posttransplantation and control samples from transplant patients without rejection on an ABI 7700 Sequence Detection System. Primers and probes were designed for human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and TRBC forward and reverse primers were designed using Primer Express software version 1.5 (Applied Biosystems, CA) (Table 2). Reaction conditions involved the amplification of 1–2 μL of cDNA in a 25 μL reaction consisting of 2.5 μL 10 × PCR buffer containing 1.5 mM Magnesium Chloride (Roche), 2 μL 2.5 mM dNTPs (final concentration 0.2 mM), 1.5 μL 5 mM each GAPDH and TRBC forward and reverse primer (final concentration 0.3 mM), 0.125 μL 5 U/mL Taq DNA Polymerase (Roche) and 0.25 μL 5 mM dual-labeled fluorescent probe (final concentration 0.05 mM). Reactions were made up to final volume with sterile water. GAPDH and TRBC measurements were performed separately.
| IMGT name | Sequence from 5′ to 3′ |
|---|---|
| TRBC forward | TCCAGTTCTACGGGCTCTCG |
| TRBC reverse | AGGATGGTGGCAGACAGGAC |
| TRBC probe | 6-FAM ACGAGTGGACCCAGGATAGGGCCAA NFQ |
| GAPDH forward | TGCACCACCAACTGCTTAGC |
| GAPDH reverse | GGAAGGCCATGCCAGTGA |
| GAPDH probe | VIC CCTGGCCAAGGTCATCCATGACAACTT TAMRA |
RT-PCR for TRBV repertoire
Total RNA was extracted from PBLs, renal biopsies and urine samples using the guanidinium thiocyanate–phenol–chloroform extraction method (29). TRIzol reagent was added into samples. RNA was then extracted according to the manufacturer's instructions. RNA sample was stored at −80°C. First-Stand complementary DNA was synthesized using the SUPERSCRIPT™ II RNase H−reverse transcriptase (Invitrogen). Human TRBV families were named according to the international ImMunoGeneTics (IMGT) database. For TRBV families, TcR cDNAs were amplified with 5′ TRBV primers and a 3′ TRBC universal (TRBCuni) primer. Oligonucleotide primers for human TRBCuni, and TRBV2, 4, 9, 15, 18, 19, 20, 24, 25, 27, 28, 29 and 30 are from Current Protocols in Immunology by Coligan et al. (30). Oligonucleotide primers of human TRBC forward and reverse, which amplify all TRB, are also from Current Protocols in Immunology by Coligan et al. (30). Oligonucleotide primers for human TRBV11 were designed by Maslanka et al. (31). Oligonucleotide primers used for human GAPD, human TRBC probe and TRBV3, 5, 6, 7, 10, 12, 13, 14 and 16 were designed using Primer3 software (BioNavigator). Table 3 shows all primers and relationship between the IMGT name of TRBV and the previous TcR Vβ name.
| IMGT name | Old TCR name | Sequence from 5′ to 3′ |
|---|---|---|
| TRBV2 | Vβ22 | ATGAAATCTCAGAGAAGTCT |
| TRBV3 | Vβ9 | CACCTAAATC TCCAGACAAAGCT |
| TRBV4 | Vβ7 | CCTGAATGCCCCAACAGCTCTC |
| TRBV5 | Vβ5 | CTGATCAAAA CGAGAGGACA GCA |
| TRBV6 | Vβ13 | CTCTCCTGTG GGCARGTC |
| TRBV7 | Vβ6 | TCAGGTGTGA TCCAATTTC |
| TRBV9 | Vβ1 | GCACAACAGTTCCCTGACTTGCAC |
| TRBV10 | Vβ12 | GRCATGGGCT GAGGCTGAT |
| TRBV11 | Vβ21 | GGCTCAAAGG AGTAGACTCC |
| TRBV12 | Vβ8 | GGTGACAGAG ATGGGACAAG A |
| TRBV13 | Vβ23 | GATCAAAGAA AAGAGGGAAA C |
| TRBV14 | Vβ16 | AAAGAGTCTAAACAGGATGAGTCC |
| TRBV15 | Vβ 24 | TACCCAGTTTGGAAAGC |
| TRBV16 | Vβ 25 | CAGGTATGCC CAAGGAAAGA |
| TRBV18 | Vβ18 | AGCCCAATGAAAGGACACAGTCAT |
| TRBV19 | Vβ17 | TTTCAGAAAGGAGATATAGCT |
| TRBV20 | Vβ2 | TCATCAACCATGCAAGCCTGACCT |
| TRBV24 | Vβ15 | AGTGTCTCTCGACAGGCACAG |
| TRBV25 | Vβ11 | TGTTCTCAAACCATGGGCCATGAC |
| TRBV27 | Vβ14 | ACCCAAGATACCTCATCACAG |
| TRBV28 | Vβ3 | GTCTCTAGAGAGAAGAAGGAGCGC |
| TRBV29 | Vβ4 | ACGATCCAGTGTCAAGTCGAT |
| TRBV30 | Vβ20 | CTCTGAGGTGCCCCAGAA |
| TRBCuni | Cβuni | TTCTGATGGCTCAAACAC |
| TRBCuni -biotin | Cβuni -biotin | TTCTGATGGCTCAAACAC |
| TRBC probe | Cβ probe | GTGTTCCCACC CGAGGTCGCT |
| TRBC forward | C-Cβ forward | GAGGACCTGAA(C/A)AA(G/C)GTG |
| TRBC reverse | C-Cβ reverse | CATTCACCCACCAGCTCAGCT |
| TRBC reverse-biotin | C-Cβ reverse-biotin | CATTCACCCACCAGCTCAGCT |
| GAPDH forward | TGCACCACCAACTGCTTAGC | |
| GAPDH reverse | GGAAGGCCATGCCAGTGA |
- The table also shows relationship between the IMGT name and the previous TcR Vβ name.
PCR amplification was performed using a Perkin Elmer 9600 thermal cycler (PE Biosystems) with an individual program for each set of primers. For 23 families of human TRBV expression, the PCR profile was as follows: denaturation at 95°C for 1 min, annealing at 54°C for 1 min and extension at 72°C for 1 min for 34 cycles. For the GAPDH, TRBC forward and reverse, 32 cycles were used. Standard curves of PCR amplifications for these primers were established to avoid amplification plateau. The specificity of PCR products was confirmed using the QPCR System 5000 (Applied Biosystems, CA).
Individual TRBV gene usage expression was expressed as a percentage of total TcR signal. TRBV repertoires were assessed for PBLs, renal biopsies and urine. In order to determine overexpanded families occurring in both allograft and urine, ratios of individual TRBV for urine and biopsy relative to PB were analyzed using the percentage of each TRBV family in urine and biopsy relative to the percentage of the same TRBV family in PBLs. In LRD patients, prerejection and rejection PBL samples were used. In cadaveric patients, PBL at the time of rejection were used.
Detection of RT-PCR products by QPCR System 5000 and densitometry
For establishing a standard curve, the specificity of each PCR product was verified by separate hybridization with a tris (2,2-bipyridine) ruthenium (II) chelate (TBR)-labeled sequence-specific oligonucleotide probe directed at segments internal to the amplified PCR segments as previously described (31). The electrochemiluminescent signal of the hybridized probe was detected with a QPCR 50000 system (Perkin Elmer) according to the manufacturer's recommendations.
We also used densitometry to detect RT-PCR products. PCR products mixed with gel-loading buffer were analyzed on 2% agarose gels. The PCR products were photographed using Gel Doc100 (Bio-Rad). The relative quantity of each of 23 TRBV families was detected using the densitometry method in the Gel Doc100 program (32).
CDR3 spectratyping of PCR products for screening clonal expansion
CDR3 spectratyping was used to analyze the TRBV families to screen for clonal expansion of T cells at rejection. A second round of PCR, in which 1 μL PCR product from each TRBV family was used as cDNA, was performed. Primers were as before with the addition a Fam-labeled TRBCuni reverse primer. PCR product (1 μL) from this reaction was mixed with Hi Di Formamide and GeneScan-500 size standard (Applied Biosystems). The sample was denatured and electrophoresed on the ABI Prism 310 Genetic Analyzer (Applied Biosystems). Genescan and Genotyper software (Applied Biosystems) were used to analyze the running results (32).
Cloning and sequencing of PCR products for matched spectratypes of TRBV families
CDR3 sequencing was performed on restricted, well-represented TRBV families with matched spectratypes from renal biopsies and urine in four patients at the time of acute rejection. PCR products were purified using a Concert™ Rapid PCR purification system (Life Technologies) and cloned into pGEM-T Easy vector system I (Promega). Products of the ligation reactions were transformed into Escherichia coli JM 109 competent cells. Sequencing reaction was performed using BigDye™ Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems). The samples were electrophoresed on the ABI Prism 310 Genetic Analyzer. The data were analyzed using Sequencing Analysis software (Applied Biosystems).
Statistical analysis
Paired t-tests were used in comparisons of matched groups to detect significance.
Results
Expression level of TRB in urine at the time of acute rejection
Urinary expression of TRB RNA was greatest in patients with acute rejection. It fell following treatment and was lower again in urine from patients with transplants who had not experienced clinical rejection in either urine less than or greater than 12 months posttransplantation. TRB levels were measured by TRBC by real-time PCR adjusted for GAPDH (Figure 1).
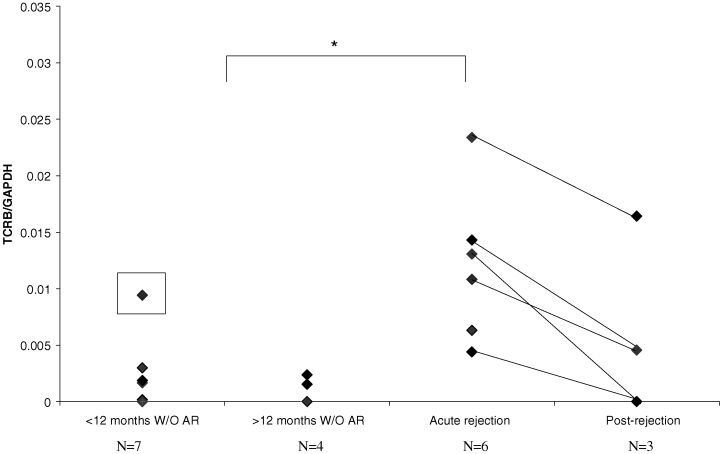
The expression level of TRB in urine samples of the patients with acute rejection (six urine samples from four patients) and postrejection (three urine samples from three patients, one patient lost to follow-up), and without acute rejection (W/O AR) (seven urine samples less than 12 months after transplantation and four urine samples more than 12 months from seven patients, and one urine from a patient less than 12 months posttransplant with a urinary infection which is marked with a black square). The level of TRBC relative to GAPDH PCR product as measured by real-time PCR is shown. The level of TRBC in urine was significantly higher in the patients with acute rejection than without rejection (p = 0.000708).
Alteration in TRBV repertoires in urinary cells and infiltrating kidney cells during acute rejection
Altered TRBV repertoires were observed in both the renal biopsy and urine at the time of acute rejection compared with PBLs of patients (relative expression in Table 4). In both LRD and cadaveric patients, at least 22 of 23 TRBV families were detected in PB samples. In urine and biopsy, the majority of TRBV families (21–23 TRBV families) were also detected in these samples, though some families were overexpressed compared with PB. However, the renal biopsy repertoire in patient 4 showed a limited TRBV repertoire (15 TRBV families).
| Patient | Sample | Ratios of TRBV expression for urine and biopsy to PBLs | ||||
|---|---|---|---|---|---|---|
| 0–1.0 (TRBV) | 1.1–1.2 (TRBV) | 1.3–1.4 (TRBV) | 1.5–1.6 (TRBV) | >1.7 (TRBV) | ||
| 1 | Urine | 7, 9, 12, 14, 15, 18, 19, 20, 25, 28, 29 | 2, 10, 13, 24 | 3, 5, 27, 30 | 4, 6 | 11 |
| Biopsy | 7, 10, 14, 16, 19, 20, 28, 29, 30 | 9, 12, 24 | 3, 11, 18 | 4 , 13, 15, 25 | 2, 5, 6 | |
| 2 | Urine | 2, 3, 4, 5, 6, 7, 9, 10, 11, 12, 13, 29, 30 | 14, 18, 25, 27 | 15, 28 | 20, 24 | 16, 19 |
| 3 | Urine | 2, 3, 5, 6, 12, 14, 15, 18, 20, 30 | 11, 24, 29 | 4, 28 | 9, 10, | 7, 13, 19, 27 |
| Biopsy | 3, 4, 5, 6, 7, 10, 11, 12, 14, 25 | 2, 9, 24, 29 | 13, 15, 27 | 18, 20, 28 | 30, 19 | |
| 4 | Urine | 2, 3, 5, 12, 13, 15, 18, 19, 20, 24, 25 | 4 , 6, 11, 27 | 7 , 10, 14, 28 | 29 | 30, 9 |
| Biopsy | 5, 12 | 2, 10 | 15, 18 | 3, 11, 20, 28 | 4 , 7, 9, 19, 30 | |
- A ratio over 1.1 represents an increased expression of TRBV families in urine and renal biopsy compared with PBLs. In LRD patients prerejection and rejection peripheral blood (PB) samples were used. In cadaveric patients PB at the time of rejection were used. In all samples, multiple TRBV families are overexpressed. Several families are overexpressed in both biopsy and urine samples (shown in bold). Underlined families show matched and restricted TRBV spectratypes in both urine and biopsy at the time of acute rejection.
In order to identify overexpanded families in the TRBV repertoires at that time of rejection, ratios of Vβ expression for urine and biopsy relative to PB were analyzed in four patients (Table 4). A ratio over 1.1 represented an increased expression of TRBV families in urine and the renal biopsy samples at the time of rejection. An increased ratio both in biopsy and urine was found in TRBV2, 3, 4, 5, 6, 11, 13 and 24 in patient 1, TRBV9, 13, 19, 24, 27, 28 and 29 in patient 3 and TRBV4, 7, 9, 10, 11, 28 and 30 in patient 4. In the urine of patient 2 an increased ratio was observed in 10 TRBV families (BV14, 15, 16, 18, 19, 20, 24, 25, 27 and 28) (biopsy was not assessed for repertoire but saved for spectratype and clonal analysis because of limited cDNA in patient 2).
Restricted and matched TRBV CDR3 spectratypes in urine and biopsy at acute rejection
Spectratyping of PCR products showed both matched and restricted TRBV spectratypes in both urine and biopsy at the time of acute rejection in all four patients. TRBV13 in patient 1, TRBV19, 24 and 27 in patient 3 and TRBV4, 7, 9 and 11 in patient 4, had a relatively increased expression (ratio over 1.1 in Table 4) in both biopsy and urine, and displayed a restricted and matched spectratype. In patient 2 increased expression in urine (ratio over 1.1) of TRBV19 and 28 was associated with restricted and matched spectratypes in both urine and biopsy. Of the TRBV that were not overexpressed TRBV14 in patient 1, TRBV2 in patient 3 and TRBV15, 18 and 19 in patient 4 also showed a restricted and matched spectratypes. Two TRBV spectratypes from each patient are shown in Figure 2 and these were chosen based on similarity of the spectratype pattern. Follow-up urines were available from three children. In three TRBV families there was insufficient cDNA expression of those families for spectratype analysis. In the three TRBV spectratypes analyzed, there is loss of oligoclonality in two of three spectratypes but the dominant peak is still visible in one TRBV family.
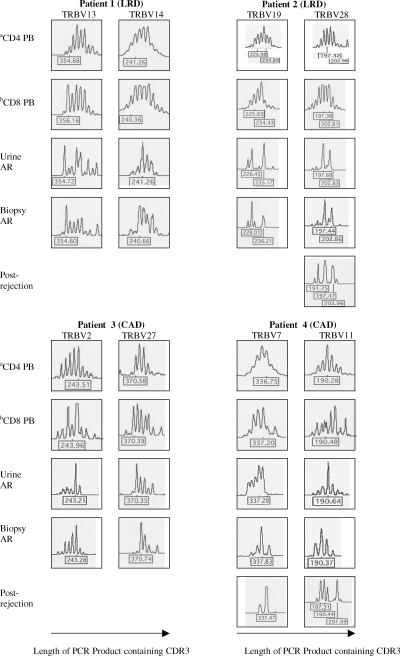
Matched and restricted TRBV spectratypes in both urine and biopsy at acute rejection. aCD4 cells from PBLs at that time of acute rejection. bCD8 cells from PBLs at that time of acute rejection. The figure shows matched and restricted TRBV spectratypes (TRBV13 and 14 in patient 1, TRBV19 and 28 in patient 2, TRBV2 and 27 in patient 3, TRBV7 and 11 in patient 4) were observed in both urine and biopsy at acute rejection compared with their PBLs at the time of acute rejection. Postrejection spectratypes are shown for three TRBV families in two patients. In the other samples there were insufficient cDNA for spectratyping and patient 3 was lost to follow-up. They show a loss of clonality in two of the three spectratypes but still demonstrate the predominant peak seen at the time of rejection in TRBV7 in patient 4.
Identical TRBV CDR3 sequences in urine and renal biopsy at the time of acute rejection
CDR3 sequencing was performed on restricted, well-represented TRBV families with matched spectratypes from renal biopsies and urine in four patients. Identical TRBV CDR3 sequences were observed matching predominant spectratypes sizes in the patient's renal biopsies and urine (Table 5). In patient 1 there is an identical TRBV13 (ratio > 1.1) sequence at the dominant size 355 in both urine and biopsy. The sequence shows clonal expansion with frequencies of 3/12 sequences in the urine. In patient 2 two identical TRBV19 (ratio > 1.7 in urine) sequences were found in urine and biopsy at the predominant sizes 227 and 236. The sequence at size 236 also shows clonal expansion with frequencies of 8/24 sequences in urine and 3/9 in the renal biopsy. We also noted the expression of TRBJ 2.1 in most of the detected clones at size 236. Patient 3 also showed an identical and over-represented TRBV2 sequence at the predominant size 243 in both urine and renal biopsy. In patient 4 an identical and over-represented TRBV7 (ratio > 1.1) sequence was observed at the dominant size 337.
| Patient | Sample | Sequence | BV | BJ | Frequency | Size in spectratypes |
|---|---|---|---|---|---|---|
| 1 | Urine | CASGQGSREQY | 13 | 2.7 | 3/12 | 355 (354.72) |
| CASSLRGKPQH | 1.5 | 1/12 | 355 (354.72) | |||
| CASSLRLGLTSPW | 2.7 | 1/12 | 361 | |||
| CASSLGGQLYGYT | 1.2 | 1/12 | 361 | |||
| CASRNLKDGNQPQH | 1.5 | 1/12 | 364 | |||
| CASSTRRDLGNEQY | 2.7 | 1/12 | 364 | |||
| CASRNGGVRAKNIQY | 2.4 | 1/12 | 367 | |||
| CASSFRSAGYTYEQY | 2.7 | 1/12 | 367 | |||
| CASRPRRLAGDYYNEQF | 2.7 | 1/12 | 373 | |||
| CASSKFRDWGATGFWQY | 2.1 | 1/12 | 373 | |||
| Biopsy | CASGQGSREQY | 13 | 2.7 | 1/5 | 355 (354.60) | |
| CASSLDGFEAF | 1.1 | 1/5 | 355 (354.60) | |||
| CASSFPQGTDTQY | 2.3 | 1/5 | 361 | |||
| CASSPGTSPTDTQY | 2.3 | 1/5 | 364 | |||
| CASSQPGTSGRTLGMDEQF | 2.1 | 1/5 | 379 | |||
| 2 | Urine | CASSLQRGPLH | 19 | 1.6 | 1/24 | 224 |
| CASSIVTGYGYT(1) | 1.2 | 1/24 | 227 (226.45) | |||
| CASSGGSNQPQH | 1.5 | 2/24 | 227 | |||
| CASTYMNTGELF | 2.2 | 2/24 | 227 | |||
| CASSIRTGLRDYGYT | 1.2 | 1/24 | 236 (235.17) | |||
| CASSLRTEFTNYGYT | 1.2 | 1/24 | 236 (235.17) | |||
| CASSIPLGGRGVEQF | 2.1 | 4/24 | 236 (235.17) | |||
| CASSIRASGSWDEQF | 2.1 | 2/24 | 236 (235.17) | |||
| CASSTRTSGRLDEQF(2) | 2.1 | 8/24 | 236 (235.17) | |||
| CASSIGQGQTFQETQY | 2.5 | 1/24 | 239 | |||
| Biopsy | CASEGGSGELF | 19 | 2.2 | 1/9 | 224 | |
| CASSIVTGYGYT(1) | 1.2 | 1/9 | 227 (228.01) | |||
| CASTYMNTGELF | 2.2 | 1/9 | 227 | |||
| CASSNGGSYNEQF | 2.1 | 1/9 | 230 | |||
| CASSIRDYHETQY | 2.5 | 1/9 | 230 | |||
| CASSTRTSGRLDEQF(2) | 2.1 | 3/9 | 236 (236.21) | |||
| CASSIEPGLAGETQY | 2.5 | 1/9 | 236 (236.21) | |||
| 3 | Urine | CASSERLNYGYT | 2 | 1.2 | 1/11 | 237 |
| CASSDMGAYVLT | 2.6 | 1/11 | 237 | |||
| CASKGFASDNEQF | 2.1 | 1/11 | 240 | |||
| CASSDGRVNTGELF | 2.2 | 1/11 | 243 | |||
| CASSELVASTDTQY | 2.3 | 1/11 | 243 | |||
| CASSEGWLSSYEQY | 2.7 | 5/11 | 243 | |||
| CASSDLAGGRTGELF | 2.2 | 1/11 | 246 | |||
| Biopsy | CASSLRETQY | 2 | 2.3 | 1/11 | 231 | |
| CASSASRATEAF | 1.1 | 2/11 | 237 | |||
| CASSEDRELEQY | 2.7 | 1/11 | 237 | |||
| CASSGTGSNNEQF | 2.1 | 1/11 | 240 | |||
| CASNGLAGKDTQY | 2.3 | 2/11 | 240 | |||
| CASSEGRPWDTQY | 2.3 | 1/11 | 240 | |||
| CASSVKGMPYNEQF | 2.1 | 1/11 | 243 | |||
| CASSEGWLSSYEQY | 2.7 | 2/11 | 243 | |||
| 4 | Urine | CASSLAQINTQY | 7 | 2.3 | 1/4 | 325 |
| CASSLSGGPNTEAF | 1.1 | 1/4 | 331 | |||
| CASSALGGGEGEQF | 2.1 | 1/4 | 331 | |||
| CASSLAGGVTSTDTQY | 2.3 | 1/4 | 337 | |||
| Biopsy | CASSPTGGEETQY | 7 | 2.5 | 2/5 | 328 | |
| CASSFYTTGERNQPQH | 1.5 | 1/5 | 337 | |||
| CASSLAGGVTSTDTQY | 2.3 | 2/5 | 337 | |||
- The sequences of selected TRBV families with matched and restricted CDR3 spectratypes in the urine and renal biopsies at that time of acute rejection. The sequences TRBV13 of patient 1, TRBV19 of patient 2, TRBV2 of patient 3 and TRBV7 of patient 4 in urine and renal biopsy cDNA are shown in the table. Identical sequences in both urine and renal biopsies are shown in bold. There are two identical TRBV19 sequences present in the urine and biopsy of patient 2.
Discussion
In pediatric patients with kidney allograft rejection we have identified T lymphocytes in the urine by using RT-PCR. Analysis of the TRBV gene usage and CDR3 spectratyping of each family in both urine and renal biopsies showed restricted and matched spectratyping in a number of highly expressed TRBV families. Further analysis of restricted, well-represented TRBV families with matched spectratyping showed identical TRBV CDR3 sequences in both the urine and biopsy at the time of rejection. These results are consistent with the previous data showing TRBV repertoire alterations in acute rejection and the presence of cytotoxic T-cell products in the urine. The presence of matching sequences in the urine suggests that urinary TcRs may be used to look for pathogenic clones in the urine.
The frequency of the lymphocyte population involved in alloresponses is between 1% and 10% (7,33,34). This suggests that the alloreactive T-cell population will have a very diverse T-cell repertoire. However, in many diseases involving T-cell activation (cancer immunology, autoimmunity and infection) there is a selection for expansion of higher affinity T cells leading to ongoing restriction of the T-cell repertoire to those of highest affinity (35–37). High affinity T cells outcompete T cells of lesser affinity. This leads to expansion of high affinity T-cell clones in the T-cell repertoire (38). All our patients have major HLA mismatches as shown in Table 1. Two patients who have received LRDs have a one haplotype mismatch, one patient is mismatched at five antigens and the final patient is mismatched for two Class I antigens. All these mismatches are enough to elicit a very broad T-cell response. The CDR3 region of the TcR is critical for the recognition of the peptide and MHC complex. In renal transplantation, analyzing the CDR3 sequence of the infiltrating T-cell clones of renal allografts assists in identification of the T cell involved in acute rejection and possibly tolerance. The majority of studies of TcR gene usage in rejection focus on TRBV family repertoire or on alloreactive clones in vitro. (10–12,39–41). In chronic rejection a restricted usage of TRBV has been shown by spectratyping and ‘immunoscope’ analysis (13). Our study demonstrates that restricted TRBV usage can be found in the urine and that it matches the restriction seen in the renal biopsy at that time of acute rejection. Of note is the greater number of matching clones seen in the urine than the biopsy suggesting the repertoire in the urine may more accurately reflect the alloreactive cells selected by crossing the tubule. In the biopsy the repertoire may be diluted by less specific T cells in the graft or by contamination by blood in the biopsy.
Cellular rejection is in part defined by the presence of tubulitis involving T-cell infiltration of tubules (Banff criteria) (42). This in turn leads to identification of T cells in the urine. Thus T cells found in the urine are most likely to be effector cells having crossed the tubule into the urine. Identification of effector T-cell markers has clinical predictive significance in acute rejection (24,25). Noninvasive methods of identifying acute rejection avoid the morbidity associated with renal allograft biopsy though at present there is no method that can satisfactorily match a renal biopsy in identifying both rejection and risk of a poor outcome (26). However, development of further noninvasive indications of rejection will be clinically useful.
However, there have been no studies of TRBV gene usage of urinary T cells in transplant rejection. This has the advantage of better characterizing the T-cell population in the urine and may in the future allow discrimination between infection and rejection, if alloreactive T cells can be defined at a CDR3 level (as can be seen in Figure 1, one patient with a urinary infection has elevated levels of TRBC). It is therefore likely that specific alloreactive T cells involved in acute rejection can be identified both in renal biopsies and urine at that time of acute rejection by analysis of their TRBV gene usages. Our data suggests that αβ T cells are detectable in both renal biopsy and urine during the process of acute rejection. Altered TRBV repertoires were observed in both renal biopsy and urine in all four patients at the time of rejection. Increased expression of multiple TRBV families was observed in both urine and biopsy. Restricted and very similar TRBV spectratypes were observed both in urine and renal biopsies in all four patients at the time of acute rejection. Furthermore, identical and clonally expanded sequences, were identified in restricted, well-represented TRBV families with similar spectratypes, and were observed both in urine and biopsy at the time of acute rejection. The high variability of the TcR suggests that matching clones are highly unlikely to be found by chance but reflect clones that are highly expanded in both the graft and the urine. It is however not possible by our techniques to confirm this alloreactivity by functional assays.
Current clinical trials in transplantation using immunosuppression or tolerance regimes increasingly require improved methods of monitoring immune responses. Our results suggest that it is possible to identify T-cell clones that may be involved in acute rejection and follow them after transplantation. It also raises the possibility of urinary T-cell studies that can allow for more specific and sensitive monitoring of TcR gene usages in addition to monitoring of functional cytotoxic molecules.




