Association of disease-predisposition polymorphisms of the melatonin receptors and sunshine duration in the global human populations
Abstract
Abstract: Melatonin is predominantly involved in signaling circadian and seasonal rhythms, and its synthesis is regulated by the environmental light/dark cycle. The selection pressure by geographically different environmental light/dark cycles, which is predominantly determined by sunshine duration, on the global distribution of genetic polymorphisms in the melatonin pathway is not well understood. Recent genetic association studies identified various disease-predisposition polymorphisms in this pathway. We investigated the correlations between the prevalence of these clinically important single nucleotide polymorphisms (SNPs) and sunshine duration among worldwide human populations from twelve regions in the CEPH-HGDP database rs4753426, a recently reported predisposition SNP for type 2 diabetes in the promoter of the MT2 melatonin receptor gene (MTNR1B), which was not included in the CEPH-HGDP genotyping array, was additionally genotyped. This SNP showed a marginally significant correlation in 760 CEPH-HGDP DNA samples (r = −0.5346, P = 0.0733), and it showed the most prominent association among the candidate melatonin pathway SNPs examined. To control for population structure, which may lead to a false positive correlation, we genotyped this SNP in a replication set of 1792 subjects from China. The correlation was confirmed among Chinese populations (r = −0.8694, P = 0.0002), and was also statistically significant after correction of other climatic and geographical covariants in multiple regression analysis (β = −0.907, P = 1.94 × 10−5). Taken together, it suggests that the human melatonin signaling pathway, particularly MT2 melatonin receptor may have undergone a selective pressure in response to global variation in sunshine duration.
Introduction
Sunlight has diverse biological effects on both animals and humans [1–5]. Notably, daytime duration plays a dominant role in entrainment of the circadian and seasonal rhythms for most animals on earth [6]. In animals, multiple physiological and developmental processes of fundamental importance, including sleep, feeding behavior, metabolism, memory, mood, hibernation, and seasonal reproduction are under the regulation of these biological rhythms [7–14]. Also, a wide variety of animals from various taxa assess and use the day length, which is mainly determined by sunshine duration, as an anticipatory cue to time seasonal events in their life cycles [15]. Thus, sunshine duration, together with food availability, temperature, and other environmental factors are of crucial importance for the survival of animals including human beings [16].
Melatonin has been suggested to represent one of the most ancient biological molecules which appeared in living organisms on earth [17]. It exists ubiquitously in nature and has been identified in all major taxa of organisms, including plants, invertebrate, and vertebrate species [18–22]. In animals and humans, the melatonin signaling pathway has been shown to extensively interact with various physiological pathways [5, 23–25]. It plays an important role in the entrainment of circadian and seasonal rhythms through encoding the time-of-day and length-of-day information to the brain and peripheral organs [13]. As such, the melatonin pathway is involved in major physiological processes including sleep regulation, pubertal development, and seasonal adaptation [13, 26, 27].
In mammals and human beings, two melatonin receptors have been cloned and characterized, which are MT1 melatonin receptor (MTNR1A) and MT2 melatonin receptor (MTNR1B) [28, 29]. GPR50, also called melatonin-related receptor, has also been cloned and classified as a member of the melatonin receptor superfamily in different species including humans [30, 31]. A third binding site, initially described as MT3, has been subsequently characterized as the enzyme quinine reductase 2 (NQO2) [32]. Melatonin also binds with relevant nuclear receptors of the retinoic acid receptor family, including RORα1, RORα2, and RORβ (RORA, RORB) [33, 34]. Therefore, melatonin serves as a mediator to transduce the external day-night cycle through these receptors to entrain the circadian rhythm and other internal biological rhythms [3, 35].
Therefore, the signaling pathway involving the phylogenetically ancient molecule, melatonin, is likely to have undergone a selective pressure during the animal or human evolution. From this perspective, the evaluation of the correlation between the environmental sunshine duration and population frequencies of genetic polymorphisms related to the melatonin pathway could shed light on this hypothesis. In this study, we first surveyed disease-predisposition single nucleotide polymorphisms (SNPs) among these genes and then examined the association between disease-predisposition polymorphisms of the melatonin pathway and sunshine duration in global human populations. Simultaneously, the associations with temperature and geographic coordinates were also controlled as covariants. A SNP rs4753426, in the promoter of MTNR1B gene, showed the most prominent correlation with sunshine duration in the melatonin pathway among the worldwide CEPH-HGDP populations (n = 760, r = −0.5346, P = 0.0733). Further study of this SNP confirmed a significant association with sunshine duration in a replication sample of 1792 subjects recruited in China.
Materials and methods
Selection of SNPs in the melatonin pathway in the HGDP database
Several SNPs in the melatonin pathway have been identified to predispose to clinical diseases. We conducted an extensive literature search for these SNPs in the melatonin pathway covering the following genes: genes encoding for melatonin receptors and co-receptors (MTNR1A, MTNR1B, GPR50, NQO2, RORA, and RORB). A PubMed search using the gene name and ‘polymorphism’ was carried out. Thirty-seven SNPs were found for the six genes, among which 21 SNPs were significantly associated with clinical phenotypes (Table S1). We retrieved the data for the positively associated polymorphisms from the CEPH-HGDP dataset, using the linkage disequilibrium (LD) based tagging SNP selection strategy. Finally, nine SNPs, six in the MTNR1B gene, two in the NQO2 gene, and one in the GPR50 gene, which have been associated with disease-predisposition, were included from PubMed (Table 1). Subsequently, we retrieved the polymorphic data of the candidate SNPs from the HGDP database except rs4753426, which was not genotyped and did not locate in a same LD bin with other genotyped SNPs, and thus we performed an independent genotyping in the same set of DNA samples of HGDP subjects.
| Gene | SNP | Disease/phenotype (Reference) |
|---|---|---|
| GPR50 | rs2072621 | Schizophrenia in females [59] |
| MTNR1B | rs1387153 | Fasting plasma glucose (FPG) [53, 60] |
| rs1447352 | Glucose metabolism [52] | |
| rs1597023a | Type 2 diabetes (rs7121092, r2 > 0.8) [52] | |
| rs2166706a | Higher fasting plasma glucose concentrations and reduced OGTT- and IVGTT-induced insulin release (rs10830962, r2 = 0.911) [51] | |
| rs4753073a | Type 2 diabetes (rs7121092, r2 = 1.0) [52] | |
| Rheumatoid factor in rheumatoid arthritis patients (rs1562444, r2 = 0.979) [61] | ||
| rs4753426 | Adolescent idiopathic scoliosis [36] | |
| Fasting plasma glucose (FPG) [51] | ||
| NQO2 | rs1143684a | NQO2 activity [62] |
| Clozapine-induced agranulocytosis (rs2071003, r2 > 0.8) [63] | ||
| rs4149370 | NQO2 activity (rs1143684, r2 = 0.801) [64] |
- a Indicates the SNPs are chosen as surrogates for the disease-predisposition polymorphisms within the same LD bin (r2 > 0.8; MAF > 0.05).
Climatological and geographic information
For HGDP-CEPH populations, the data of average daily sunshine duration, temperature were extracted from the World Meteorological Organization publication ‘1961–1990 Global Climate Normals’ compiled by National Climatic Data Center of the United States. They are 30-yr average values for the period 1961–1990. The data was also available from Hong Kong Observatory website (http://gb.weather.gov.hk). Geographic coordinates of these populations were included by CEPH-HGDP dataset.
Climatological information for Chinese populations was collected from China Meterological Data Sharing Service System (http://cdc.cma.gov.cn). The data was mainly extracted from ‘The Annual Surface Climate Normals of International Exchanging Stations of China (1971–2000)’. They are 30-yr average values for the period 1971–2000. Geographic coordinate information was also obtained from this source.
For all populations, if the specific information was not available, information for the most closely neighboring site was used as a substitute. In addition, an average would be taken if samples were collected from more than one nearby location.
DNA samples of CEPH-HGDP panel (n = 952) for genotyping of rs4753426
For rs4753426 worldwide screening, we used DNA samples of CEPH-HGDP panel, which included 952 unrelated individuals from 52 populations from diverse geographic areas, including Sub-Saharan Africa, North Africa, Europe, the Middle East, South/Central Asia, East Asia, Oceania, and the America.
Replication samples of Chinese subjects from China (n = 1792)
To better control for underlying population structure, the results obtained from CEPH-HGDP populations were validated in a replication sample set of 1305 Han Chinese and 487 Chinese ethnic minorities obtained from China. The replication sample set of 1305 Han individuals were sampled randomly from seven different provinces of China: 130 from Yunnan, 196 from Guangdong, 69 from Guizhou, 176 from Wuhan, 239 from Sichuan, 304 from Liaoning, 164 from Shandong, and 27 from Xinjiang. In addition, other four Chinese ethnic minorities (total n = 487) were also genotyped in the present study: Miao (n = 155) collected from Guizhou and Hunan Provinces; Dai (n = 68) collected from Yunnan Province; Zhuang (n = 223) was from Yunnan and Guangxi Provinces; and Russian (n = 41) collected from inner Mongolia Autonomous Region. The geographic origin of each individual was ascertained by both self-reported geographic precoding tenancy of the three generations and absence of family history of migration.
SNP genotyping
Genomic DNA was extracted from the whole blood of the replication Chinese samples by standard phenol/chloroform methods. The rs4753426 polymorphism was analyzed in both CEPH-HGDP panel DNA and replication Chinese samples by PCR-RFLP method as reported previously [36]. The 127bp PCR products with the ancestral allele (T) was not cut while mutant allele (C) was cut by HaeIII (Takara) into two (23bp, 104 bp) fragments (Fig. S1). Representative sample of each genotype called by PCR-RFLP and samples with suspicious genotypes by PCR-RFLP method were confirmed by sequencing. Sequencing reaction was performed by using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). Electrophoresis of purified sequencing product was performed on a 3730 DNA Analyzer (Applied Biosystems). Sequencing data were edited and aligned by use of DNASTAR software (DNAStar Inc., Madison, WI, USA) (Fig. S2).
Data analyses
Among the nine included disease-predisposition SNPs in the melatonin pathway, the frequencies of genotypes and alleles were calculated for all the populations. Hardy–Weinberg equilibrium (HWE) was tested by the software PEDSTATS V0.6.8 (http://www.sph.umich.edu/csg/abecasis/). Statistical analysis of alleles and genotype frequencies between genders and among different age groups was performed by chi-squared test (SPSS for Windows, 13.0; SPSS Inc., Chicago, IL, USA).
Correlation analysis between the derived allele frequency and daily sunshine duration was carried out using Pearson bivariate correlation analysis (SPSS for Windows; 13.0), respectively. The effect of other environmental factors, namely, temperature, longitude, and latitude was evaluated using multiple linear regression analysis weighted by sample size (SPSS for Windows; 13.0). The data of longitude and latitude for world populations were transformed into the form of natural logarithm to insure normality. In a similar manner, correlation analysis and multiple linear regression analysis were carried out in the replication study using Han Chinese populations for SNP rs4753426.
Results
The CEPH-HGDP enabled us to evaluate the worldwide distributions of the prevalence (frequencies) of polymorphisms for these melatonin pathway SNPs among global populations originating from different parts of the world [37]. In this study, the SNPs in the melatonin pathway which have been previously shown to predispose to clinical diseases were identified (Table S1). We extracted the genotype frequencies data of worldwide populations for these disease-predisposition melatonin pathway SNPs from the CEPH-HGDP database containing 760 subjects of the worldwide populations. SNPs that were not genotyped in this database were represented by other SNPs within the same LD bin. Ultimately, nine SNPs were included in the following analysis (Table 1). The genotype data for rs2072621, rs2166706, rs1447352, rs1387153, rs4753073, rs1597023, s4149370, and rs1143684 (as they had been genotyped in CEPH-HGDP) were retrieved from this dataset. The data for the remaining rs4753426, located in the MTNR1B gene, were not represented by any SNPs in the CEPH-HGDP dataset and thus were supplemented through new genotyping experiment here (see Materials and methods for details). Therefore, there were nine SNPs in total in the melatonin pathway which were analyzed in details.
For the nine polymorphisms, we examined their correlations with the sunshine duration in the worldwide CEPH-HGDP dataset (Table 2). Among them, only SNP rs4753426 showed the best correlation with sunshine duration at a marginally significant level (r = −0.5346, P = 0.0733). There was no significant differences between genders or among different age groups in both genotype and allele frequencies (P > 0.05). So we analyzed the polymorphism of SNP rs4753426 in the unit of populations, without separating sexes or age groups.
| Gene | SNP | Location | Sunshine duration | |
|---|---|---|---|---|
| r | P-value | |||
| GPR50 | rs2072621A | 150,096,517 | 0.1429 | 0.6577 |
| MTNR1B | rs1387153T | 92,313,476 | 0.0627 | 0.8465 |
| rs1447352A | 92,362,409 | −0.0991 | 0.7593 | |
| rs1597023Aa | 92,360,967 | −0.0991 | 0.7593 | |
| rs2166706Ta | 92,331,180 | −0.4304 | 0.2871 | |
| rs4753073Aa | 92,357,123 | −0.2686 | 0.5201 | |
| rs4753426C | 92,341,244 | −0.5346 | 0.0733 | |
| NQO2 | rs1143684C | 2,955,389 | −0.3829 | 0.2193 |
| rs4149370Ta | 2,964,653 | 0.2335 | 0.5779 | |
- ‘r’ represents correlation coefficient.
- a Indicates the SNPs are chosen as surrogates for the disease-predisposition polymorphisms within the same LD bin (r2 > 0.8; MAF > 0.05). The SNP in bold shows the most prominent association.
The distributions of the allele frequencies of rs4753426 for worldwide populations were illustrated in Fig. 1, in which T is the ancestral allele while C is the derived one. The SNP demonstrated high derived allele frequencies in non-African populations. For example, it was monomorphic in populations from Columbia and Brazil, where the C allele had been fixed. No significant deviation from HWE was observed except the sample of Nigeria (two-tailed P = 0.01).
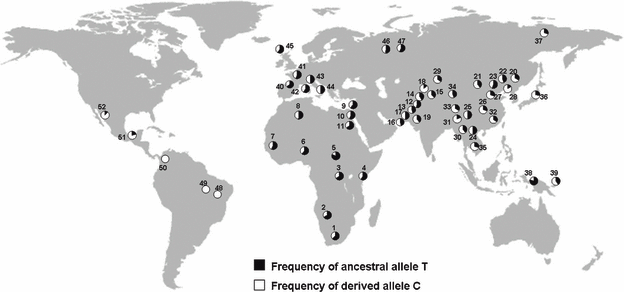
Global frequencies of the alleles of tagging single nucleotide polymorphisms rs4753426 in a panel of 948 individuals. For each population, the country of origin, number of individuals sampled (N), and frequencies of the derived allele C are listed (in parentheses) as follows: 1, Southeastern and Southwestern Bantu (South Africa, 8, 37.50%); 2, San (Namibia, 6, 33.33%); 3, Mbuti Pygmy (Democratic Republic of Congo, 13, 34.62%); 4, Northeastern Bantu (Kenya, 11, 40.91%); 5, Biaka Pygmy (Central Africa Republic, 23, 19.57%); 6, Yoruba (Nigeria, 22, 38.64%); 7, Mandenka (Senegal, 22, 43.18%); 8, Mozabite [Algeria (Mzab region), 29, 48.28%]; 9, Druze [Israel (Carmel region), 42, 38.10%]; 10, Palestinian [Israel (Central), 46, 43.48%]; 11, Bedouin [Israel (Negev region), 46, 35.87%]; 12, Hazara (Pakistan, 22, 47.73%); 13, Balochi (Pakistan, 24, 54.17%); 14, Pathan (Pakistan, 24, 58.33%); 15, Burusho (Pakistan, 25, 62.00%); 16, Makrani (Pakistan, 25, 54.00%); 17, Brahui (Pakistan, 25, 50.00%); 18, Kalash (Pakistan, 23, 86.96%); 19, Sindhi (Pakistan, 24, 60.42%); 20, Hezhen (China, 9, 66.67%); 21, Mongola (China, 10, 60.00%); 22, Daur (China, 10, 55.00%); 23, Orogen (China, 9, 44.44%); 24, Miaozu (China, 10, 50.00%); 25, Yizu (China, 10, 50.00%); 26, Tujia (China, 10, 70.00%); 27, Han (China, 44, 69.32%); 28, Xibo (China, 9, 83.33%); 29, Uygur (China, 10, 35.00%); 30, Dai (China, 10, 60.00%); 31, Lahu (China, 8, 81.25%); 32, She (China, 10, 65.00%); 33, Naxi (China, 9, 66.67%); 34, Tu (China, 10, 55.00%); 35, Cambodian (Cambodia, 10, 70.00%); 36, Japanese (Japan, 25, 72.00%); 37, Yakut [Russia (Siberia region), 25, 70.00%]; 38, Papuan (New Guinea, 17, 20.59%); 39, NAN Melanesian (Bougainville, 11, 59.09%); 40, French Basque (France, 24, 35.42%); 41, French (France, 28, 44.64%); 42, Sardinian (Italy, 28, 41.07%); 43, North Italian [Italy (Bergamo region), 13, 50.00%]; 44, Tuscan (Italy, 8, 50.00%); 45, Orcadian (Orkney Islands, 15, 40.00%); 46, Russian (Russia, 25, 48.00%); 47, Adygei [Russia (Caucasus region), 17, 44.12%]; 48, Karitiana (Brazil, 14, 1); 49, Surui (Brazil, 8, 1); 50, Colombian (Colombia, 7, 1); 51, Pima (Mexico, 14, 78.57%); 52, Maya (Mexico, 21, 88.10%).
A subset of data from twelve countries/regions (China, France, Israel, Italy, Japan, Kenya, Nigeria, Pakistan, Russia, Senegal, Siberia, and South Africa) were included for a multiple regression analysis as detailed climatic information was available. As mentioned above, the frequency of rs4753426C allele was marginally correlated with average daily sunshine duration (r = −0.5346, P = 0.0733; Fig. 2 and Table 3) in the Pearson bivariate correlation analysis. In multiple regression weighted by sample size, the correlation with sunshine duration was further strengthened after other environmental variants (temperature, longitude, and latitude) were controlled (β = −0.424, P = 0.043).
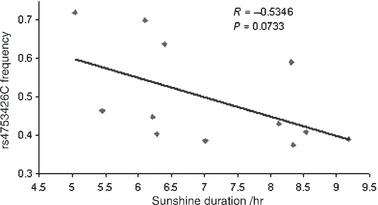
The distribution of the rs4753426C allele frequencies and their correlation with sunshine duration for worldwide human populations from 12 countries/regions. They are China (15 subpopulations including Hezhen, Mongola, Daur, Orogen, Miaozu, Yizu, Tujia, Han, Xibo, Uygur, Dai, Lahu, She, Naxi, and Tu), France (two subpopulations including French Basque and French), Israel (three subpopulations including Druze, Palestinian, and Bedouin), Italy (three subpopulations including Sardinian, North Italian, and Tuscan), Japan, Kenya, Nigeria, Pakistan (eight subpopulations including Hazara, Balochi, Pathan, Burusho, Makrani, Brahui, Kalash, and Sindhi), Russia (two subpopulations including Russian and Adygei), Senegal, Siberia, and South Africa.
| Population | rs4753426C frequency | Sunshine duration (hr) | n | HWE P-value |
|---|---|---|---|---|
| Japan | 0.72 | 5.04 | 25 | 0.1490 |
| Russia | 0.46 | 5.45 | 42 | 0.0820 |
| Siberia | 0.70 | 6.09 | 25 | 1.0000 |
| Italy | 0.45 | 6.22 | 49 | 0.8030 |
| France | 0.40 | 6.28 | 52 | 1.0000 |
| China | 0.64 | 6.39 | 178 | 0.2690 |
| Nigeriaa | 0.39 | 7.01 | 22 | 0.0100 |
| Senegal | 0.43 | 8.13 | 22 | 0.4040 |
| Pakistan | 0.59 | 8.31 | 192 | 0.3170 |
| South Africa | 0.38 | 8.34 | 8 | 0.5220 |
| Kenya | 0.41 | 8.53 | 11 | 0.5580 |
| Israel | 0.39 | 9.18 | 134 | 1.0000 |
- HWE: two-tailed P-value, simulation of 1000.
- China includes 15 CEPH-HGDP subpopulations (Han, Tujia, Yizu, Miaozu, Oroqen, Daur, Mongola, Hezhen, Xibo, Uygur, Dai, Lahu, She, Naxi, and Tu); France includes two subpopulations (French and French Basque); Israel includes three subpopulations (Negev, Carmel, and Central); Italy includes three subpopulations (Sardinian, Bergamo, and Tuscan); Pakistan includes eight subpopulations (Brahui, Balochi, Hazara, Makrani, Sindhi, Pathan, Kalash, and Burusho); Russia includes two populations (Russian Caucasus and Russia).
- aThe genotype frequencies of rs4753426 in the samples of Nigeria did not follow HWE.
To better understand the association and exclude the confounding effect of ethnic population structure, we did further genotyping in a replication sample set of Chinese populations (n = 1792), in which the ethnic background is better defined. The distribution of the allele frequencies of rs4753426 among Chinese populations was illustrated in Fig. 3. No significant deviation from HWE was observed (P > 0.05). For the subjects from 12 populations within China (see Materials and methods), the rs4753426C allele was also associated with decreased daily sunshine duration (r = −0.8694, P = 0.0002; Fig. 4 and Table 4) in the Pearson bivariate correlation analysis. In weighted multiple regression analysis, the correlation with this SNP was also statistically significant (β = −0.907, P = 1.94 × 10−5). Furthermore, the significant association between sunshine duration and the polymorphism of rs4753426 was also evident in the Han Chinese replication samples after exclusion of ethnic minorities (r = −0.8083, P = 0.015).
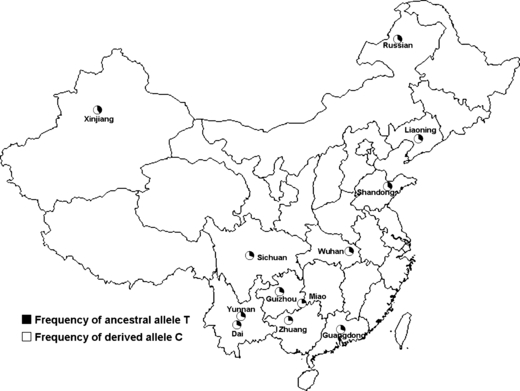
Frequencies of the alleles of tagging single nucleotide polymorphisms rs4753426 in 12 Chinese populations. For each population, the ethnic origin, number of individuals sampled, and frequencies of the derived allele C are given (in parentheses) as follows: Guangdong (Han, 196, 70.41%); Yunnan (Han, 130, 70.77%); Guizhou (Han, 69, 70.29%); Sichuan (Han, 239, 69.25%); Wuhan (Han, 176, 67.33%); Shandong (Han, 164, 64.33%); Liaoning (Han, 304, 66.78%); Xinjiang (Han, 27, 59.26%); Dai (Dai, minority, 68, 69.85%); Zhuang (Zhuang, minority, 223, 73.77%); Miao (Miao, minority, 155, 77.74%); Russian (Russian, minority, 41, 64.63%).
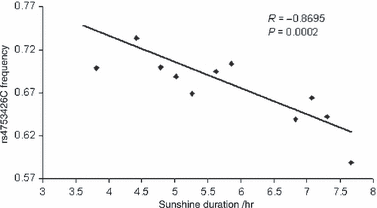
The distribution of the rs4753426C allele frequencies and their correlation with sunshine duration for the 12 Chinese populations.
| Population | rs4753426C frequency | Sunshine duration (hr) | n | HWE P-value |
|---|---|---|---|---|
| Miaoa | 0.78 | 3.60 | 155 | 0.4780 |
| Guizhou Han | 0.70 | 3.84 | 69 | 0.7960 |
| Zhuanga | 0.74 | 4.44 | 223 | 0.5650 |
| Guangdong Han | 0.70 | 4.80 | 196 | 0.2530 |
| Sichuan Han | 0.69 | 5.04 | 239 | 1.0000 |
| Hubei Han | 0.67 | 5.28 | 176 | 0.1860 |
| Daia | 0.70 | 5.64 | 68 | 1.0000 |
| Yunnan Han | 0.71 | 5.88 | 130 | 1.0000 |
| Shandong Han | 0.64 | 6.84 | 164 | 0.4540 |
| Liaoning Han | 0.67 | 7.08 | 304 | 0.7160 |
| Russiana | 0.65 | 7.32 | 41 | 1.0000 |
| Xinjiang Han | 0.59 | 7.68 | 27 | 0.1730 |
- HWE: two-tailed P-value, and the number of iteration for simulation is 1000.
- aEthnic minority.
Discussion
It has been speculated that sunlight, as a dominant feature in the environment, plays an important role in the evolution of life forms [1, 2, 4]. The intensity of sunlight has been recognized as a selective pressure on the evolution of human skin pigmentation [38]. Furthermore, animals from multiple taxa including human mainly use the length of sunlight to time the daily and seasonal events during their life histories [39]. The role of day length is especially important for the adaptation in various types of seasonal environments and evolution of seasonal behavior [15, 40]. Therefore, the selective pressure caused by sunshine duration has been increasingly recognized and studied [15, 39, 41].
The melatonin signaling pathway is predominantly responsible for entraining of the circadian and seasonal rhythms in animals and humans [5]. The melatonin secretion is suppressed by visual exposure to light [3, 42]. Thus, this pathway, which mediates the biological effect of light exposure, is of great importance to the investigation of the selective role of sunshine duration upon human genome diversity. Meanwhile, genetic variation in photoperiodic response has been suggested to be due to the effects of melatonin on target organs downstream from the circadian clock in Peromyscus leucopus and Peromyscus maniculatus [41, 43, 44]. Thus, the melatonin signaling pathway is a potential candidate under the selection of circadian and/or seasonal rhythms. From this perspective, the evaluation of the correlation of the environmental sunshine duration and population frequencies of genetic polymorphisms related to the melatonin pathway could shed some light on the selective pressure due to sunshine duration upon human genome diversity.
In this study, only the SNP rs4753426 showed significant correlation with sunshine duration among the nine disease-predisposition SNPs in the melatonin pathway. This polymorphism of the MT2 melatonin receptor gene (MTNR1B) was differentially distributed in different ethnic populations, originating from different parts of the world. Moreover, the SNP demonstrated high derived allele frequencies in human populations out of Africa, suggesting an underlying positive selection. Next, in the exploration of the association of this polymorphism with possible environmental factors, namely, sunshine duration and temperature in our study, we found this polymorphism was the one showing strongest correlation with sunshine duration among the nine candidate disease-predisposition SNPs among the worldwide human populations. More importantly, we replicated the significant association between the polymorphism and sunshine duration in global Chinese populations, which included four ethnic minorities as well as the only Han Chinese populations sampled from different parts of China. Collectively, these data support the notion that this locus is under natural selection of environmental sunshine duration.
The melatonin signaling pathway has a number of important biological roles, ranging from acting as a major circadian rhythm transducer, to a potent free radical scavenger as well as modulator of gene transcription [45, 46]. The MT2 melatonin receptor regulates the phase shift of circadian rhythms [35]. In agreement with the function of the melatonin signaling pathway and the MT2 receptor in biological rhythm entrainment, the polymorphism of rs4753426 was significantly associated with sunshine duration, and this association was found to be specific. By contrast, there was no significant association between this polymorphism and temperature. These results indicate that selection of the derived rs4753426C allele could provide a selective advantage to populations adapting to climates with different sunshine durations, through a much more efficient temporal adaptation. As mentioned above, the improved photoperiodic response has been suggested to be due to the effects of melatonin pathway in P. leucopus and P. maniculatus [41, 43, 44].
A daytime-related signal to the immune system by melatonin has been previously suggested [47]. Immune functions in some specific mammals follow daily and seasonal rhythms, showing an enhancement during short days, which correlates well with the duration of melatonin secretion [48]. This immunomodulatory effect is predominantly through MT2 receptor [35]. It is interesting to note that MTNR1B encoding the MT2 receptor had an extensive and replicated association with glycemic trait and diabetes, where multiple SNPs of this genes were identified by the latest genome-wide association study as disease-predisposition SNPs [49–53]. For example, the rs4753426C allele was suggested in association with a higher fasting plasma glucose concentration and reduced OGTT- and IVGTT-induced insulin release [51]. In addition, considering its location in the promoter of MTNR1B gene (Fig. 5), this polymorphism presumably affects the gene expression by altering transcription factor binding sites [51]. As the regulation of immune reaction is of great importance for the survival of our ancestors during the evolution [54], it is also conceivable that the rs4753426C allele with increased immunostimulatory effect of melatonin can provide the carriers with a selective advantage. Probably, its association with type 2 diabetes is the result of changeable environment during the migration of humans that archaic advantage has become disadvantageous recently.
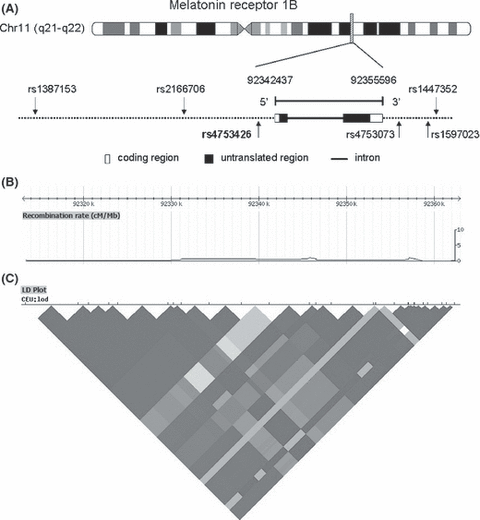
Positions of single nucleotide polymorphisms (SNP) examined in this study and linkage disequilibrium (LD) structure of MTNR1B gene. (A) Schematic representation of the melatonin receptor 1B (MTNR1B) gene with the location of the six disease-predisposition SNPs. The figure shows a schematic representation of the MTNR1B gene structure, with arrow indicating the location of rs4753426 polymorphism. Coding regions are depicted as black rectangles, and the open rectangles represent untranslated regions. (B) The recombination rate across the MTNR1B gene. (C) LD plot across the MTNR1B locus in Caucasians. LD plot is based on the measure D’. Each diamond indicates the pairwise magnitude of LD, with dark gray indicating strong LD (D’ > 0.8) and a logarithm of odds score of greater than 2.0.
The rs4753426C allele might also play a role in developmental process of skeletal vertebrate. It is well known that melatonin during development can communicate information about photoperiod and thereby insure that vital functions especially bone formation occur in an appropriate and precise temporal sequence and in accordance with cyclic environmental changes [25, 55]. The rs4753426 has been associated with the occurrence of adolescent idiopathic scoliosis, an abnormality of skeletal development during puberty [36]. Therefore, it is also possible that selection of rs4753426C allele could be attributed to a developmental advantage in the adaptation to diverse sunshine durations.
Current archeological and genetic evidence is generally considered to support the spread of anatomically modern humans within Africa across the rest of the globe within the past ∼100 kyr [56, 57]. Migration out of Africa and subsequent colonization throughout the world entailed many novel challenges that favored the selections of enhanced survival probability [58]. Therefore, it is not surprising that the different sunshine duration, from long in Africa to short in non-Africa regions, would have placed a selective pressure on the melatonin signaling pathway. The selection of rs4753426C allele would insure that carriers adapted to local daily and seasonal rhythms more quickly and efficiently. Perhaps, this allele could also enhance immunomodulatory effects as well as other developmental advantage to increase survival and reproduction successes.
Collectively, our data suggest that sunshine duration has exerted a selective force on the melatonin signaling pathway. More importantly, our results also confirm the idea that climate has been an important selective pressure upon candidate genes for common metabolic disorders and developmental disorders, which is of great clinical implications for the pathogenesis of these disorders.
Acknowledgements
This work was supported by grant from the National Natural Science Foundation of China (30621092), Bureau of Science and Technology of Yunnan Province. We thank Wang ling for providing help in data analysis. We are also grateful to Gou Shi-Kang, Wu Shi-Fang and Zhu Chun-ling for technical assistance.
Author contributions
Ya-ping Zhang and Nelson L. S. Tang conceived and designed the experiments. Lin-dan Ji performed the experiments. Lin-dan Ji, Dong-dong Wu, and Jin Xu analyzed the data. Lin-dan Ji, Jin Xu, Nelson L. S. Tang and Si-da Xie wrote the paper.




