DNA repair profiles of disease-associated isolates of Neisseria meningitidis
Editor: Johannes Kusters
Abstract
Neisseria meningitidis, or the meningococcus, is the source of significant morbidity and mortality in humans worldwide. Even though mutability has been linked to the occurrence of outbreaks of epidemic disease, meningococcal DNA repair pathways are poorly delineated. For the first time, a collection of meningococcal disease-associated isolates has been demonstrated to express constitutively the DNA glycosylases MutY and Fpg in vivo. DNA sequence analysis showed considerable variability in the deduced amino acid sequences of MutS and Fpg, while MutY and RecA were highly conserved. Interestingly, multi-locus sequence typing demonstrated a putative link between the pattern of amino acid substitutions and levels of spontaneous mutagenicity in meningococcal strains. These results provide a basis for further studies aimed at resolving the genotype/phenotype relationships of meningococcal genome variability and mutator activity.
Introduction
Neisseria meningitidis, or the meningococcus (Mc), colonizes the mucosal surfaces of the human upper respiratory tract. Although it usually resides as part of the commensal flora, Mc may traverse from this nonpathogenic carrier state and disseminate into the blood stream. Despite advances in preventive and therapeutic medicine, meningococcal disease (meningitis and/or septicaemia) is the cause of significant morbidity and mortality in industrialized and developing countries worldwide (Caugant et al., 1986). Mc has humans as its exclusive host and must be uniquely adapted to this environmental niche. This habitat offers challenges such as nutrient deprivation, competition from other microorganisms, high oxygen concentrations, and the immune responses of the host. Mc fitness for survival will be directly linked to its capability of generating change and adaptation to its environment, and strains conferring a selective advantage will excel (Bjedov et al., 2003).
Mutator strains exhibit increased spontaneous mutation rates compared with those commonly found in the corresponding wild-type species (Giraud et al., 2001). Hypermutable strains in bacterial populations might have potential advantages or disadvantages, depending on the nature, rate and magnitude of environmental change (Giraud et al., 2001; Notley-McRobb et al., 2002; Bjedov et al., 2003), and have been described in various human pathogens (LeClerc et al., 1996; Oliver et al., 2000). Mutators have generally been associated with defects in the postreplicative mismatch repair (MMR) pathway responsible for removing insertion/deletion loops (Lahue et al., 1989), suggesting that frameshift mutations and mispairing of nucleotide runs are major pathways for generating variability in pathogenic species, including Mc (LeClerc et al., 1996; Oliver et al., 2000; Richardson & Stojiljkovic, 2001; Richardson et al., 2002). Conflicting evidence exists on the association of Dam methylase variants causing hypermutable neisserial strains with enhanced phase-variable capsule switching (Bucci et al., 1999; Jolley et al., 2004). Moreover, only 40% of the Mc strains investigated by Stojiljkovic and coworkers could be complemented with wild-type alleles of MMR components MutS or MutL (Richardson et al., 2002). This means that more than half of Mc mutators are genetically uncharacterized, suggesting that DNA repair pathways yet to be identified contribute to the pool of Mc mutator phenotypes – a notion supported by findings in other pathogens such as Helicobacter pylori and Streptococcus pneumoniae (Bjorkholm et al., 2001; Morosini et al., 2003).
A number of DNA repair pathways and adaptive responses other than MMR have been related to mutagenicity in laboratory strains of Escherichia coli (Horst et al., 1999). Endogenous DNA damages, including one of the most frequent oxidative lesions, 7,8-dihydro-8oxo-2′-deoxyguanosine (8oxoG), are primarily processed through the base excision repair (BER) pathway (Seeberg et al., 1995). In E. coli, BER is initiated by DNA glycosylases that nick the N-glycosylic bond and remove the damaged base (Lindahl, 1979). The DNA glycosylases MutY and MutM (formamidopyrimidine DNA glycosylase, Fpg) have been reported to reduce mutation rates in bacteria by removing adenine mispaired with 8-oxoG, or removing 8-oxoG when mispaired with cytosine, respectively (Cabrera et al., 1988; Nghiem et al., 1988; Michaels et al., 1992). Other major E. coli DNA repair pathways involved in correcting DNA lesions are the nucleotide excision repair (NER), recombinational repair, and translesion synthesis (TLS). NER generally repairs bulky lesions from exogenous sources interfering with normal basepairing and impairing transcription and replication (Pettijohn & Hanawalt, 1964; Seeberg, 1978). RecA, the major component of recombinational repair, is involved in the repair of double-strand breaks in DNA and control of the inducible SOS response in E. coli (Little & Mount, 1982). TLS polymerases conduct translesion synthesis past blocking lesions, allowing the cell to overcome such obstacles. However, these polymerases might generate mutations at high frequencies (Napolitano et al., 2000).
Despite the profound influence of genome instability on the occurrence of hypervirulent Mc lineages (Richardson & Stojiljkovic, 2001; Richardson et al., 2002), Mc DNA repair pathways have so far not been delineated in detail (Davidsen et al., 2005; Davidsen & Tonjum, 2006). The aim of this study was to characterize key DNA repair components in Mc clinical isolates to assess whether obvious defects, other than those in MMR, were associated with mutator phenotypes. The spontaneous mutation frequencies were determined in a collection of Mc disease-associated isolates. Cell lysates from these strains were tested for their in vivo DNA glycosylase activity. Moreover, Mc strains were assessed for survival under oxidative stress, and the mutation spectrum of rifampicin-resistant strains was determined. Selected Mc DNA repair genes with the potential to be part of the mutator-inducing pool were subjected to DNA sequence analysis. Multi-locus sequence typing (MLST) data were used as a reference for sequence polymorphisms and epidemiologic links of the Mc strains under study. On this basis, the sequence conservation and variation of DNA repair components in Mc wild-type and mutator isolates were compared.
Materials and methods
Strains and growth conditions
The Mc strains included in the study are listed in Table 1. Mc strains were selected based on their being the source of major outbreaks worldwide and on mutagenicity studies previously performed by other workers (Richardson & Stojiljkovic, 2001; Richardson et al., 2002). The strains represent a broad variety of serogroups and sequence types (ST-). Mc strains Z1099, Z1227, Z1269, Z1466, Z4756, Z5826, Z1318, Z3515 and Z5037 were kindly provided by D.A. Caugant (National Institute of Public Health, Oslo, Norway). Mc strains were propagated on 5% blood agar plates, GC plates or GC plates containing rifampicin to a final concentration of 3 μg mL−1 when appropriate. All incubations were performed in 5% CO2 at 34°C.
| Mc Z number and strain | Relevant characteristics (serogroup/ST designation/MLEE designation/origin) | Source |
|---|---|---|
| Z1099 139M | Serogroup A, ST1, subgroup I, isolated in Philippines 1968 | CDC, US* |
| Z1227 S4355 | Serogroup A, ST5, subgroup III, isolated in Denmark 1974 | CDC, US* |
| Z1269 10 | Serogroup A, ST4, subgroup IV-I, isolated in Burkina Faso 1963 | CDC, US* |
| Z1466 S5611 | Serogroup A, ST1, subgroup I, isolated in Australia 1977 | CDC, US* |
| Z4756 371 | Serogroup A, ST1, subgroup I, isolated in India 1980 | CDC, US* |
| Z5826 92001 | Serogroup A, ST7, subgroup III, isolated in China 1992 | CDC, US* |
| Z6244 Z2491 | Serogroup A, ST4, subgroup IV-1, isolated in Gambia 1983 | Parkhill et al. (2000) |
| –† M1080 | Serogroup B, ST44, isolated in the USA 1970 | Frasch & Gotschlich (1974) |
| Z3842 H44/76 | Serogroup B, ST32, ET-5 complex, isolated in Norway 1976 | Holten (1979) |
| Z7176 MC58 | Serogroup B, ST74, ET-5 complex, isolated in UK 1983 | McGuinness et al. (1991) |
| Z4259 FAM18 | Serogroup C, ST11, ET-37 complex, isolated in USA 1983 | Aho et al. (1991) |
| Z1318 255 | Serogroup A, ST4, subgroup IV-I, isolated in Burkina Faso 1966 | CDC, US* |
| Z3515‡ F4698 | Serogroup A, ST5, subgroup III, isolated in Saudi Arabia 1987 | CDC, US* |
| Z3524 F6124 | Serogroup A, ST5, subgroup III, isolated in Chad 1988 | Klee et al. (2000) |
| Z5005 106 | Serogroup A, ST1, subgroup I, isolated in Morocco 1967 | Klee et al. (2000) |
| Z5037 322/85 | Serogroup A, ST2, subgroup IV, isolated in DDR 1985 | CDC, US* |
| Z6466 890592 | Serogroup A, ST60, subgroup IX, isolated in Netherlands 1989 | Klee et al. (2000) |
| Z6835† 585/88 | Serogroup A, ST2, isolated in Russia 1988 | Klee et al. (2000) |
| Z4662 BZ 10 | Serogroup B, ST8, cluster A4, isolated in Netherlands 1967 | Klee et al. (2000) |
| Z4667 BZ 147 | Serogroup B, ST48, lineage 3, isolated in Netherlands 1963 | Klee et al. (2000) |
| Z4673 BZ 198 | Serogroup B, ST41, lineage 3, isolated in Netherlands 1986 | Klee et al. (2000) |
| Z4683† NG 4/88 | Serogroup B, ST30, isolated in Norway 1988 | Klee et al. (2000) |
| Z4690†,‡ NG G40 | Serogroup B, ST25, isolated in Norway 1988 | Klee et al. (2000) |
| Z4707†,‡ 297-0 | Serogroup B, ST49, isolated in Chile 1987 | Klee et al. (2000) |
| Z6904 ROU | Serogroup W135, ST11, ET-37 complex, isolated in France 1995 | Klee et al. (2000) |
| – Mc 5 | Serogroup W135, ET-34 complex, isolated in Norway 1984 | Allunans & Bovre (1996) |
| – Mc 7 | Serogroup C, ET-35 complex, isolated in Norway 1984 | Allunans & Bovre (1996) |
| – Mc 17 | Serogroup C, ET-37 complex, isolated in Norway 1984 | Allunans & Bovre (1996) |
| – Mc 40 | Serogroup C, ET-40 complex, isolated in Norway 1984 | Allunans & Bovre (1996) |
- Strains listed above the line were assessed for spontaneous mutation rates, survival under oxidative stress and in vivo DNA glycosylase activity, as well as being subjected to sequence analysis of rpoB and selected DNA repair and housekeeping genes. Strains listed below the line were analysed for spontaneous mutation rates and in vivo MutY DNA glycosylase activity only.
- * Mc isolates selected from a collection at the Meningitis and Special Pathogens Branch, Centers for Disease Control and Prevention, Atlanta, GA.
- † † Multi-locus enzyme electrophoresis (MLEE) designation not available.
- ‡ ‡ Carrier isolate.
Assessment of Mc spontaneous mutation frequency
Selected Mc disease-associated isolates (Table 1) were propagated and assessed for spontaneous mutation rates by rifampicin resistance selection as previously described (Davidsen et al., 2005). In short, Mc cells were grown in 5% CO2 at 34°C and inoculated on plain GC plates and GC plates containing rifampicin to a final concentration of 3 μg mL−1. The ratio of rifampicin-resistant cells to the total number of cells yielded the mutation rate. The assay was repeated ten times for each strain. Differences in spontaneous mutation rates between strains were monitored by comparing the mutant cells giving rise to colonies using the Wilcoxon test with continuity-adjusted P-values. Corrected P-values were calculated by the Steel−Dwass method. Additional Mc strains (Table 1) were classified as exhibiting low, intermediate or high mutation rates by screening for colonies on rifampicin plates.
Determination of mutations in rpoB conferring rifampicin resistance
Rifampicin-resistant single colonies were propagated overnight in 5% CO2 at 34°C. Three individual rifampicin-resistant colonies as well as one wild-type rifampicin-sensitive colony from each selected clinical isolate (Table 1) were analysed. The 230-bp region of the Mc rpoB gene harbouring nucleotides prone to changes conferring rifampicin resistance (Nolte et al., 2003) was PCR-amplified and sequenced using the primers listed in Table S1 (Supplementary Material).
Assessment of Mc survival rate under oxidative stress
Nonimpregnated paper disks manufactured by Becton Dickinson Microbiology Systems (Cockeysville, MD) were saturated with 10, 20 or 30 mM hydrogen peroxide (H2O2) (Sigma, UK). H2O2 generates hydroxyl radicals in the presence of iron (Imlay & Linn, 1988) and thereby induces oxidative stress conditions. Selected Mc strains (Table 1) were inoculated on GC plates, H2O2-saturated discs were placed on top of the inoculate, and the cultures were preincubated for 10 min at room temperature and then incubated in 5% CO2 at 34°C for 20 h. Survival of Mc cells was monitored by measurement of the diameter of the inhibition zone around the H2O2-saturated disks. The assay was repeated twice for each strain.
Assessment of in vivo DNA glycosylase activity in Mc clinical isolates
Mc whole-cell extracts were prepared as previously described (Davidsen et al., 2005) by a combination of plasmolysis and lysozyme treatment (Seeberg, 1978). Duplex DNA substrates containing either a single A:8oxoG, C:8oxoG or G:5-hydroxy-cytosine (G:5OHC) basepair were generated by 32P 5′ end labelling of oligonucleotides 5′-GATGGGCCTC-A-GGGTCATGCCGCC-3′, 5′-GGCGGCATGACCC 8oxoG-GAGGCCCATC-3′ and 5′-GCATGCCTGCACGG[5OH-dc]-CATGGCCAGATCCCCGGGTACCGAG-3′ hybridized with 5′-GGCGGCATGACCC-8oxoG-GAGGCCCATC-3′, 5′-GATGGGCCTC-C-GGGTCATGCCGCC-3′ and 5′-CTCGGTACCCGGGGATCTGGCCATG-G-CCGTGCAGGCATGC-3′, respectively (Eide et al., 1996). DNA glycosylase reactions were performed by mixing whole-cell extracts with radioactive DNA substrates in reaction buffer and incubating at 37°C for 30 min (Eide et al., 1996; Davidsen et al., 2005). Recombinant Mc M1080 MutY (Davidsen et al., 2005) and E. coli Fpg (New England Biolabs, MA) were included as positive controls. The products of the reactions were analysed by 20% denaturing DNA sequencing gel and visualized by phosphor imaging.
DNA sequence analysis of DNA repair genes
The Mc homologues of mutY, fpg, nth, mutS, dinB and recA genes were PCR-amplified from selected strains listed in Table 1 using the primers specified (Table S1). DNA sequencing was performed using an Applied BioSystems 3730 genetic analyzer system (Applied Biosystems, NC) with an ABI BigDye Terminator v. 3.1 DNA sequencing kit (Applied Biosystems, N.C.) using primers listed in Table S1. DNA sequence assembly was generated using the software sequencher v. 2. The nucleotide sequences are available at GenBank, accession numbers DQ465615–DQ465670. The deduced amino acid sequences for all genes analysed were predicted on http://www.expasy.org and aligned by genedoc v. 2.6.002 (http://www.psc.edu/biomed/genedoc/). Putative catalytic residues and signature motifs were identified in the DNA repair components. Both nucleotide sequences and deduced amino acid sequences were subjected to dendrogram analysis in treeview v. 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/rod.html). Available MLST sequences were retrieved from the Neisseria MLST homepage (http://pubmlst.org/neisseria) to obtain a reference to the general sequence variation/conservation in the isolates examined (Maiden et al., 1998). In the MLST analysis, the nucleotide sequences of c. 500 bp of seven housekeeping genes were assessed, revealing the variation at each locus.
Results and discussion
The majority of hypermutable Mc isolates examined to date are genetically uncharacterized (Richardson et al., 2002). We have investigated a collection of disease-related Mc strains and employed functional DNA repair assays and DNA sequence analysis to define why some of these isolates display high mutation frequencies.
The spontaneous mutation frequencies of the Mc clinical isolates listed in Table 1 ranged from <0.1 to 9.9 per 108 CFU (Table 2). Five isolates, Z1227, Z1269, Z3515, Z6244 and M1080, had spontaneous mutation frequencies that were significantly higher (1.3–9.9 × 10−8 CFU) (P<0.05) than those of the other isolates (<0.1 × 10−8 CFU) (Table 2). Mc strains Z1466 and Z5826 both had spontaneous mutation frequencies of 0.2 × 10−8 CFU, but could not be clearly distinguished from the isolates that exhibited ‘low’ spontaneous mutation frequencies (P>0.05) (Table 2). In addition, three strains were classified as having intermediate mutation frequencies by screening for rifampicin-resistant colonies only (Table 2).
| Mc Z numberand strain | RifR mutants per 108 CFU | Previous data | RifR screening | |
|---|---|---|---|---|
| Median | (Q1/Q3)* | |||
| Z1099 139M | <0.1 | (<0.1/0.2) | 1.5 Richardson et al. (2002) | − |
| Z1227 S4355 | 4.9 | (3.3/7.2) | 4.0†Richardson et al. (2002) | ++ |
| Z1269 10 | 9.9 | (7.0/12.2) | 5.8 Richardson et al. (2002) | ++ |
| Z1466 S5611 | 0.2 | (<0.1/0.8) | 1.0 Richardson et al. (2002) | (+) |
| Z4756 371 | < 0.1 | (<0.1/0.2) | 0.9 Richardson et al. (2002) | − |
| Z5826 ‡ 92001 | 0.2 | (<0.1/0.6) | 3.0*Richardson et al. (2002) | (+) |
| Z6244 ‡ Z2491 | 1.3 | (0.5/2.0) | <0.5§Richardson & Stojiljkovic (2001) | ++ |
| – M1080 | 7.1 | (3.5/8.9) | ND | ++ |
| Z3842 H44/76 | < 0.1 | (0.0/0.3) | ND | − |
| Z7176 MC58 | < 0.1 | (<0.1/<0.1) | 0.3 Martin et al. (2004) | − |
| Z4259 FAM18 | < 0.1 | (<0.1/<0.1) | ND | − |
| Z1318 255 | ND | ND | NA | + |
| Z3515 F4698 | ND | ND | NA | ++ |
| Z3524 F6124 | ND | ND | NA | + |
| Z4662 BZ 10 | ND | ND | NA | − |
| Z4667 BZ 147 | ND | ND | NA | − |
| Z5005 106 | ND | ND | NA | − |
| Z5037 322/85 | ND | ND | NA | + |
| Z6466 890592 | ND | ND | NA | − |
| Z6835 585/88 | ND | ND | NA | − |
| Z4673 BZ 198 | ND | ND | NA | − |
| Z4683 NG 4/88 | ND | ND | NA | − |
| Z4690 NG G40 | ND | ND | NA | − |
| Z4707 297–0 | ND | ND | NA | − |
| Z6904 ROU | ND | ND | NA | − |
| – MC 5 | ND | ND | NA | − |
| – MC 7 | ND | ND | NA | − |
| – MC 17 | ND | ND | NA | − |
| – MC 40 | ND | ND | NA | − |
- The results are given as the median of ten independent measurements or as rifampicin resistance screening by comparison of the number of spontaneous rifampicin-resistant colonies. ++ denotes high; + denotes intermediate; and − denotes a low mutation frequency. When possible, the data are compared with findings of previous studies on the same isolate.
- * Q: quartile.
- † Complemented by mutL.
- ‡ ‡ The discrepancy between the mutagenicity found in strains Z2491 and Z5826 in different laboratories might be explained by strain variation, the experimental design and culture media employed, or by other complex factors.
- § Evidence for functional mutL copy.
- ND, not done; NA, not applicable.
To search for any uniformity in the pattern of mutation spectrum conferring rifampicin resistance, such as the GC→TA transversions one previously detected for Mc MutY (Davidsen et al., 2005), part of the gene encoding RpoB in selected Mc disease-associated isolates in the ‘high’ and ‘low’ mutation frequency groups were subjected to DNA sequencing (Table 3). All rifampicin-resistant strains, except Z1269 and Z4259, consistently exhibited CG→TA transitions; indeed, isolates Z1227, Z1466, M1080, Z3842 and Z7176 exclusively harboured this mutation (Table 3). Furthermore, Z1099, Z1269, Z5826, Z6244 and Z4259 harboured one CG→AT, AT→TA, AT→GC, TA→CG or GC→AT transversion/transition, while strain Z4756 harboured a triplet expansion (Table 3). The results indicate that Mc strains Z1099, Z1269, Z4756, Z5826 Z6244 and Z4259 exhibit a broader rpoB mutation spectrum than Z1227, Z1466, M1080, Z3842 and Z7176, suggesting the presence of more than one mutagenicity pathway in the six former strains. However, no relation between rpoB mutation pattern and high/low mutation rate could be observed.
| Mc Z numberand strain | Transition/transversion† | Tripletexpansion† | |||||
|---|---|---|---|---|---|---|---|
| CG→AT | CG→TA | AT→TA | AT→GC | TA→CG | GC→AT | ||
| Z1099 139M | 103 × 2 | 103 × 1 | |||||
| Z1227‡ S4355 | 92 × 2 | ||||||
| Z1269 10 | 104 × 1 | 128 × 2 | |||||
| Z1466 S5611 | 103 × 3 | ||||||
| Z4756 371 | 103 × 2 | 120–122 × 1 | |||||
| Z5826 92001 | 92 × 1122 × 1 | 74 × 1 | |||||
| Z6244 Z2491 | 221 × 1 | 128 × 1 | 130 × 1 | ||||
| – M1080 | 103 × 3 | ||||||
| Z3842 H44/76 | 103 × 3 | ||||||
| Z7176‡ MC58 | 122 × 2 | ||||||
| Z4259 FAM18 | 74 × 1 | 103 × 2 | |||||
- * Three independent rifampicin-resistant strain of the meningococcal strain examined were analysed for nucleotide changes conferring rifampicin resistance.
- † The number in front of ‘x’ indicates the nucleotide within rpoB that has been changed. The first nucleotide in primer TD170 is set as number 1. The number following ‘x’ indicates the number of Rif-resistant derivatives with the given transition/transversion or triplet expansion among the three derivatives tested.
- ‡ ‡ One Rif-resistant derivative of strains Z1227 and Z7176 exhibited a nucleotide change conferring rifampicin resistance outside the sequenced area.
In addition, the Mc clinical isolates were subjected to oxidative stress by exposure to H2O2. The Mc isolates exhibited differences in H2O2 sensitivity, as observed in the close relative Neisseria gonorrhoeae (Alcorn et al., 1994), and no clear correlation could be detected between high or low mutation rates and hyper-resistance or hyper-sensitivity to H2O2 (Table 4). In this context, it should be pointed out that Mc, in addition to catalase and a number of other scavenger enzymes (Seib et al., 2004), hosts no fewer than three DsbA homologues (Sinha et al., 2004), rendering this species particularly fit for handling oxidative stress.
| Mc Z numberand strain | Sensitivity to hydrogen peroxide (inhibition zonein mm) | ||
|---|---|---|---|
| 10 | 20 | 30 | |
| Z1099 139M | 6.3 | 8.0 | 9.7 |
| Z1227 S4355 | 6.8 | 10.4 | 11.9 |
| Z1269 10 | 7.2 | 9.9 | 11.2 |
| Z1466 S5611 | 6.0 | 9.3 | 10.3 |
| Z4756 371 | 6.6 | 8.7 | 10.4 |
| Z5826 92001 | 7.2 | 9.3 | 11.0 |
| Z6244 Z2491 | 6.8 | 8.8 | 11.3 |
| – M1080 | 7.2 | 9.5 | 10.8 |
| Z3842 H44/76 | 7.9 | 10.6 | 12.0 |
| Z7176 MC58 | 7.0 | 9.7 | 10.6 |
| Z4259 FAM18 | 6.9 | 9.3 | 10.7 |
- Survival was monitored by measurement of the diameter of the inhibition zone (in mm) around the H2O2-containing disks. The results are reported as the average of two experiments.
The DNA glycosylase MutY of the BER pathway has previously been implicated in mutator activity in pathogenic species – four Pseudomonas aeruginosa mutator isolates from cystic fibrosis patients were found to have defects in MutY (Oliver et al., 2000). Moreover, we have recently demonstrated a striking increase in the spontaneous mutation frequency in MutY-defective Mc strains (Davidsen et al., 2005). Furthermore, Mc mutS and fpg mutants have been shown to give rise to increased mutation frequencies, but these were far below the rate induced by the mutY mutant (Davidsen & Tonjum, 2006; Davidsen et al., unpublished results). Fpg is, together with MutY, important in the protection against mutations caused by 8oxoG (Michaels et al., 1992; Davidsen et al., 2005). Defects in MutY and Fpg might thus contribute to the naturally occurring Mc mutator pool. However, MutY and Fpg were constitutively expressed in vivo in all of the Mc clinical isolates tested (Table 1), even in the strong mutator strains, as detected by their excision of A:8oxoG, 8oxoG:C and 5-hydroxy-C:G DNA lesions (Fig. 1). This is the first demonstration of in vivo expression of MutY and Fpg in Mc isolates. The findings might indicate that the excessive mutator phenotype seen in the mutY mutant (Davidsen et al., 2005) is infrequent in Mc natural populations. Such defects might be selected against, be very transient, or be rapidly repaired by transforming DNA (Davidsen et al., 2004). If the mutY gene is damaged, its uptake seems to be facilitated, because it exhibits the highest density of DNA uptake sequences (DUS) required for transformation of all Mc DNA repair genes (Davidsen et al., 2004).
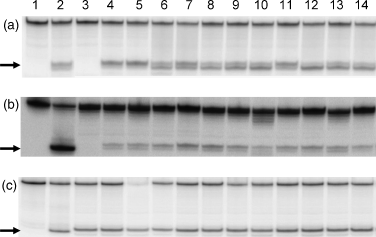
Neisserial DNA glycosylases MutY and Fpg are expressed in vivo. Whole-cell extracts of a collection of Neisseria meningitidis (Mc) disease-associated isolates were tested for activity in a DNA base excision assay using A:8oxoG, 8oxoG:C and 5OHC:G substrates. (a) MutY-specific A:8oxoG substrate, (b) Fpg-specific 8oxoG:C substrate, and (c) Fpg/Nth-specific 5OHC:G substrate*. Lanes 1, substrate; 2, positive control, (a) Mc M1080 purified MutY, (b) and (c): Escherichia coli Fpg; 3: (a) Z3842 (H44/76) mutY-deficient strain, (b) and (c) Z3842 (H44/76) fpg-deficient strain; 4, Z3842 (H44/76); 5, M1080; 6, Z7176 (MC58); 7, Z4259 (FAM18); 8, Z1099 (139M); 9, Z1227 (S4355); 10, Z1269 (10); 11, Z1466 (S5611); 12, Z6244 (Z2491); 13, Z4756 (371); 14, Z5826 (92001). The remaining strains listed in Table 1 were tested for A:8oxoG nicking only. All strains were identified as exhibiting such activity (data not shown). *The H44/76 fpg mutant (negative control) shows base excision of 5OHC opposite guanine, suggesting the presence of an Mc Nth (endonuclase III) activity.
DNA sequence analysis of genes encoding selected components representing major DNA repair pathways was performed. The deduced amino acid sequences of these DNA repair components from the Mc clinical isolates were aligned (Supplementary Material, Figs S1−S7). Interestingly, none of the amino acid changes detected in the DNA repair components occurred in recognized DNA binding sites, catalytic residues or important motifs. Amino acid changes were, however, found in the regions between these motifs, and some of the amino acid substitutions detected might influence protein structure, particularly when they involve a change in amino acid class. The patterns of amino acid substitutions in strain M1080 were distinctly different from those of the other strains with ‘high’ mutation rates, and M1080 might thus host a different mutator-inducing genetic background (see below).
The DNA repair genes were moderately to highly conserved with regard to both nucleotide and amino acid sequences, as were the MLST sequences (Table 5). The mutY gene exhibited the lowest average nucleotide variation (0.9%) among the isolates, while fpg, nth and uvrA exhibited the highest nucleotide variability (>3%) (Table 5). At the deduced amino acid level, however, RecA showed complete sequence conservation (no variability), while the MutY amino acid sequence was the second most conserved (0.5% variation). Despite the relatively high variability of the nth gene at the nucleotide level, the Nth deduced amino acid sequence was remarkably conserved (0.5% variation). The deduced amino acid sequences for MutS and Fpg exhibited the highest numbers of average amino acid substitutions (>1%) (Table 5). The sequence conservation observed in some of the DNA repair enzymes might underline the significance of these components in vivo. No clear relationship between frequencies of amino acid substitutions, functional assays and mutator phenotype was evident in the Mc isolates.
| Cellular role andfunction of protein | Gene | Nucleotidesubstitutions (in %) | Amino acidsubstitutions (in %) |
|---|---|---|---|
| Base excision repair (BER): A/G–specific adenine glycosylase | mutY | 0.9 | 0.5 |
| Base excision repair (BER): Formamidopyrimidine glycosylase | fpg | 3.1 | 1.3 |
| Base excision repair (BER): Endonuclease III, Nth | nth | 4.0 | 0.5 |
| Mismatch repair (MMR): DNA mismatch repair protein MutS | mutS | 1.9 | 1.3 |
| Nucleotide excision repair (NER): Exinuclease ABC, subunit A | uvrA | 4.8 | 1.0 |
| Recombinational repair: RecA protein | recA | 1.2 | 0.0 |
| Translesion synthesis (TLS): DNA damage inducible protein B (DNA polymerase) | dinB | 1.3 | 1.0 |
| Transport and binding protein: ABC transporter, ATP-binding protein | abcZ | 4.6 | 0.6 |
| Nucleotide and nucleoside interconversions: Adenylate kinase | adK | 1.6 | 0.6 |
| Amino acid biosynthesis: Shikimate 5-dehydrogenase | aroE | 5.4 | 4.5 |
| Energy metabolism: Fumarate hydratase, class II, aerobic | fumC | 3.0 | 0.6 |
| Amino acid biosynthesis: Glutamate dehydrogenase | gdh | 2.3 | 0.1 |
| Energy metabolism: Pyruvate dehydrogenase, E1 | pdhC | 4.2 | 1.5 |
| Cell envelope and energy metabolism: Phosphoglucomutase | pgm | 3.2 | 3.5 |
Interestingly, a putative correlation between the deduced amino acid sequence patterns and spontaneous mutation frequencies was found. Mc strains Z1099, Z1466, Z4746, Z5005, Z5826, Z7176 and Z3842 all exhibited ‘low’ mutation frequencies, while Z1318, Z3524 and Z5037 were classified as having intermediate mutation frequencies, and Z1227, Z1269, Z3515, Z6244 and M1080 were defined as mutators. Although the Mc strains were isolated from various geographical regions worldwide and at different times (Table 1), dendrograms of MLST data and DNA repair component sequences both indicated that strains Z1099/Z1466/Z4756/Z5005, Z7176/Z3842, Z1227/Z3515/Z3524 and Z1269/Z6244/Z1318 were linked (Fig. 2; Supplementary Material, Figs S8 and S9). Despite the small sample size, an association between sequence patterns and mutation phenotypes may exist. How much of this is the result of clonal expansion of hypermutagenic serogroup A lineages or selective pressures ensuring that only certain lineages of mutator strains will survive is unknown.
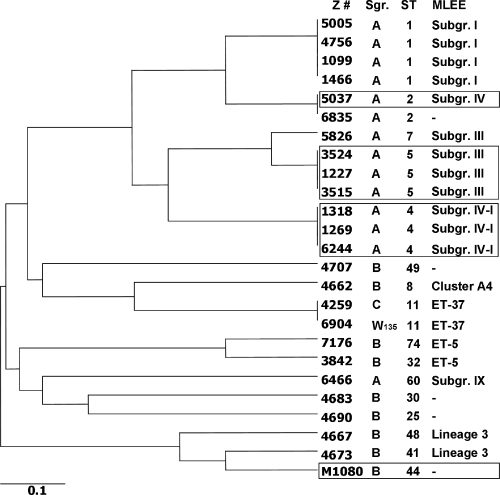
Relationships of Neisseria meningitidis (Mc) disease-associated isolates by MLST. The sequences of alleles abcZ, adk, aroE, fumC, gdh, pdhC and pgm were subjected to MLST/UPGMA analysis to display links between the various Mc strains (http://pubmlst.org/neisseria). The strains are specified by their Z number (Z #). Serogroup (Sgr), sequence type (ST) identified by MLST and subgroup/complex identified by multi-locus enzyme electrophoresis are depicted. Strains exhibiting intermediate and high spontaneous mutation rates are boxed.
The evolution and adaptation of microorganisms are influenced by two opposing forces: the maintenance of genetic information and the generation of a suitable level of genetic variation upon which selection can act. Although the analysis of the potential role of Mc mutY, fpg, nth, mutS, uvrA, recA and dinB in the generation of mutator phenotypes has been initiated, a number of other putative mutator target genes remain (Horst et al., 1999). As a starting point, these findings represent a basis for understanding the mechanisms configuring the genetic profiles of Mc natural populations. In turn, this information could lead to models that elucidate how mutator phenotypes are linked to microbial fitness and hypervirulence.
Acknowledgements
This work was supported by grants from The Research Council of Norway. We thank D.A. Caugant for providing N. meningitidis strains. H.K. Tuven and L. Davidsen are gratefully acknowledged for their technical assistance.