DYNAMIC TRANSMISSION, HOST QUALITY, AND POPULATION STRUCTURE IN A MULTIHOST PARASITE OF BUMBLEBEES
Abstract
The evolutionary ecology of multihost parasites is predicted to depend upon patterns of host quality and the dynamics of transmission networks. Depending upon the differences in host quality and transmission asymmetries, as well as the balance between intra- and interspecific transmission, the evolution of specialist or generalist strategies is predicted. Using a trypanosome parasite of bumblebees, we ask how host quality and transmission networks relate to parasite population structure across host species, and thus the potential for the evolution of specialist strains adapted to different host species. Host species differed in quality, with parasite growth varying across host species. Highly asymmetric transmission networks, together with differences in host quality, likely explain local population structure of the parasite across host species. However, parasite population structure across years was highly dynamic, with parasite populations varying significantly from one year to the next within individual species at a given site. This suggests that, while host quality and transmission may provide the opportunity for short-term host specialization by the parasite, repeated bottlenecking of the parasite, in combination with its own reproductive biology, overrides these smaller scale effects, resulting in the evolution of a generalist parasite.
Heterogeneity at different biological scales is responsible for the evolution of both host and parasite traits and, thus, shapes host–parasite interactions (Schmid-Hempel and Koella 1994; Thompson 1999, 2005). The majority of parasites exploit multiple host species, either sequentially or concurrently (Cleaveland et al. 2001; Taylor et al. 2001; Pedersen et al. 2005), and recent work even suggests that multihost parasites can evolve from host specialists (Johnson et al. 2009), in contrast to popular wisdom (Woolhouse et al. 2001). Consequently, an important source of variability in multihost parasite systems is precisely the presence of different host species, where each host species is qualitatively or quantitatively different compared to the others.
Theoretical predictions for the ecology and evolution of such multihost parasites are limited (Rigaud et al. 2010). At an ecological level, Dobson (2004) suggested that when interspecific transmission is less frequent than intraspecific transmission the system may favor parasite persistence, and stronger interspecific transmission may lead to the extinction of one of the host species, although experimental work suggests the opposite (Hellgren et al. 2009). Gandon (2004) predicted that variation in host quantity may affect the transmission rate of the parasite while heterogeneity in host quality (e.g., differences in the immune response) may result in different levels of host exploitation, parasite virulence, and transmission. Thus, the study of evolutionary and coevolutionary outcomes in these complex multihost parasite systems must take into account both relative host quantity and quality and the dynamics of transmission.
Despite the growing interest in parasites that use multiple concurrent hosts, few studies exist of natural systems (Rigaud et al. 2010). Bumblebees (Bombus spp.) coexist across their range as assemblages of different species (Williams 1998) and provide a model system for studies of multihost parasites. Many parasites of bumblebees appear, at first glance, to be generalists, parasitizing multiple species within the genus (Schmid-Hempel 1998; Tay et al. 2005). One such parasite is the trypanosomatid Crithidia bombi (Gorbunov 1987; Lipa and Triggiani 1988). The parasite life cycle is simple. When a bumblebee ingests C. bombi cells, the parasite resides in the gut, actively divides, and releases transmission stages through host feces (Schmid-Hempel 2001). The probability of infection is density dependent, increasing with dose (Ruiz-González and Brown 2006a). Once infected, individuals are likely to remain so, although under highly artificial conditions clearance is possible (Imhoof and Schmid-Hempel 1998a). The parasite is transmitted among individuals of the same colony through contact with feces and infected animals within the nest (Schmid-Hempel 2001; Otterstatter and Thomson 2007). Transmission among individuals from different colonies (and across bumblebee species) occurs via flowers (Durrer and Schmid-Hempel 1994; Ruiz-González and Brown 2006b), when parasite cells from an infected forager that have been left on the flower are picked up by another foraging bee. The risk of infection via these shared resources decreases with increased complexity of the inflorescence (Durrer and Schmid-Hempel 1994) and, possibly, with increased flower complexity as well. The probability of transmission among species will thus depend in large part upon the frequency with which those species forage at the same resources. Bumblebee species differ in their foraging preferences (Pyke 1982; Goulson and Darvill 2004; Williams 2005) and thus the degree of overlap in resource use should vary among different species. Crithidia bombi exhibits a seasonal increase in prevalence (Imhoof and Schmid-Hempel 1999), infecting up to 80% of the foraging workers (Shykoff and Schmid-Hempel 1991a), indicating a change in the transmission dynamics and epidemiology across the seasons. In addition, genotype–genotype interactions between parasite strains and host colonies affect parasite transmission (Shykoff and Schmid-Hempel 1991b,c), prevalence and intensity of infections (Baer and Schmid-Hempel 1999, 2001), and infection success (Schmid-Hempel et al. 1999; Baer and Schmid-Hempel 2003; Mallon and Schmid-Hempel 2004; Schmid-Hempel and Reber-Funk 2004; Yourth and Schmid-Hempel 2006). Recent genetic studies have isolated quantitative trait loci (QTLs) behind these genotype–genotype interactions (Wilfert et al. 2007). These interactions are presumably driven by the heavy fitness impact of the parasite on bumblebee colonies, which occurs through reductions in learning and foraging efficiency (Gegear et al. 2005, 2006) and colony growth rates (Shykoff and Schmid-Hempel 1991b), increases in the mortality rate of food-stressed workers (Brown et al. 2000) and reductions in queen fitness by 40% (Brown et al. 2003a; Yourth et al. 2008). Allopatric infections are sometimes more virulent than sympatric infections, indicating local adaptation (Imhoof and Schmid-Hempel 1998b). Furthermore, the parasite elicits an immune response in its host (Brown et al. 2003b; Rydell et al. 2009; Schlüns et al. 2010), providing a potential mechanism behind the genotype–genotype interactions.
Until recently, this work has been conducted almost exclusively in a single host species, B. terrestris. However, C. bombi infects multiple host species in both Europe and North America (Shykoff and Schmid-Hempel 1991a; Colla et al. 2006; Gillespie 2010; Schmid-Hempel and Tognazzo 2010; Kissinger et al. 2011). A recent study suggested that the parasite's population structure across host species might be related to resource sharing (Salathé and Schmid-Hempel 2011). Here, using experimental, observational, and molecular techniques, we ask how host heterogeneity and transmission patterns structure C. bombi populations across their multiple host species (Dobson 2004). Specifically, we (1) use a cross-infection experiment to determine both whether hosts differ in quality with respect to parasite epidemiology and if parasite strains are adapted to specific host species, (2) examine host immune defense across species to determine whether it underlies differences in host quality, (3) use observational and sampling approaches to determine the relative intra- and interspecific transmission potential of the parasite, and use microsatellite analyses to both, (4) determine whether the dynamics of transmission are reflected in parasite population structure across host species, and (5) ask whether this structure is maintained across sites and years, with the latter being a prerequisite for long-term parasite specialization across host species.
Materials and Methods
HOST QUALITY AND PARASITE SPECIALIZATION—A CROSS-INFECTION EXPERIMENT
We reared colonies of B. terrestris and B. lucorum from queens collected during March to May of 2005 from sites in Co. Dublin (Table S1). We chose these species because they can be easily reared in laboratory conditions and produce sufficient worker bees for experiments. They were also two of the most common species in our study area (M. J. F. Brown, unpubl. data). Colony rearing followed Rutrecht and Brown (2009) and is described in Supporting information.
For the cross-infection experiment, after the second batch of workers emerged each colony was checked twice daily to remove callows (newly hatched workers), enabling us to conduct the experiment with age-controlled animals. In this study a parasite strain was defined as the parasites within a queen; queens filter the parasite population (Ulrich et al. 2011) and thus our null hypothesis is that the parasites within a given queen are adapted to that host species. To control for variation among strains from the same host species, three queens infected with C. bombi from each different host species, B. lucorum and B. terrestris, were randomly selected and labeled as the source of parasite strains to conduct the cross-infection experiment within the two host species, B. lucorum and B. terrestris. In addition, one extra parasite strain from a queen of B. pascuorum was used to infect B. terrestris workers and confirm the parasite's ability to infect multiple host species. Sample sizes and experimental design are shown in Table 1.
| Parasite strain from | Strain | B. terrestris colonies | B. lucorum colonies | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | ||
| B. lucorum | A | 6 | 6 | 5 | - | - | 5 | 4 | 4 | - |
| B | 6 | 6 | 5 | - | - | 6 | 4 | 4 | - | |
| C | 6 | 6 | 6 | - | - | 5 | 3 | 3 | - | |
| B. terrestris | D | - | - | 6 | 6 | 4 | - | - | 5 | 4 |
| E | - | - | 6 | 6 | 5 | - | - | 5 | 4 | |
| F | - | - | 6 | 6 | 4 | - | - | 5 | 6 | |
| B. pascuorum | G | 5 | - | 5 (1) | 5 (2) | - | - | - | - | - |
All callows were randomly assigned to one of each experimental infection group. For each individual inoculation, a final parasite concentration of 5000 cells/μl was obtained. To prepare the inocula, feces were collected from infected queens of either B. lucorum, B. pascuorum, or B. terrestris (see Supporting information for inoculation protocol). On each of the following 10 days postinoculation, a single sample of feces was taken from each animal in parallel, and at the same time on each day, and all parasite cells present in each sample of feces were counted under 400× magnification using an improved Neubauer slide. Seven animals that did not develop the infection, all of which were inoculated with the B. pascuorum-sourced strain, were excluded from further analyses.
Because life-history timing varied between the bumblebee species, it was not possible to complete a fully crossed design (see Table 1). Consequently, we analyzed our data using the following two-way ANOVAs, and the number of parasite cells as the dependent variable. First, we combined strains within species origin and colonies within host species to examine the effect of host species, parasite origin (at the species level), and their interaction on the internal epidemiology (see below) of C. bombi. Second, we examined whether parasite strain (irrespective of host-species origin), host species, or their interaction had an impact on internal epidemiology. Third, we examined whether parasite origin (at the species level), host colony (irrespective of species), or their interaction affected internal epidemiology. Fourth, we asked whether parasite strain (irrespective of host-species origin), host colony (irrespective of species), or their interaction had an effect on the development of the infection inside the host. We had three measures of development—the total number of parasite cells shed over 10 days, the first day parasite cells appeared in the host species, and the peak day for the production of parasite cells; consequently we ran 12 analyses.
SPECIES DIFFERENCES IN IMMUNE BACKGROUND AND RESPONSE TO INFECTION BY C. BOMBI
We used 87 workers of three common species (B. lapidarius, B. pascuorum, and B. terrestris), that were either infected with C. bombi or uninfected, collected from the Botanical Gardens in Co. Dublin, Ireland on 21 July 2005. By collecting workers from the field, we were able to mimic the transmission/infection challenge faced by C. bombi in the field. Workers were kept individually in plastic vials and chilled until their return to the laboratory, where each was maintained in standard conditions (e.g., Ruiz-González et al. 2009). Uninfected workers were randomly allocated into two categories; uninfected or experimentally infected workers. The experimentally infected workers were inoculated with 2000 cells of C. bombi (Supporting information), which is around the expected concentration of parasite cells in the feces of an infected bee, to simulate natural exposure to the parasite (Schmid-Hempel and Schmid-Hempel 1993; Ruiz-González and Brown 2006a; Koch and Schmid-Hempel 2011).
After 5 days postinfection all animals were checked to confirm the first diagnosis and the success of the inoculations. Immune measurements were then taken. The constitutive immune response was studied by analyzing both the levels of phenoloxidase enzyme activity (PO) and its proenzyme activity, pro-phenoloxidase (proPO), following Barnes and Siva-Jothy (2000) and Ruiz-González et al. (2009). The induced immune response was measured as the antibacterial activity in haemolymph following Moret and Schmid-Hempel (2000). After measuring the enzyme activity and while saving the output, the Windows operative system generated an unexpected error and the PO and proPO values for more than 34% of the samples (30 and 31 out of 87 samples, respectively) were lost.
To test whether species differed in their background immune levels and whether natural or experimental inoculation with C. bombi had an impact on immunity, we used a two-way MANOVA with host species and infection status (naturally uninfected, naturally infected, laboratory infected) as fixed factors and ZI, PO, and PPO values as the dependent variables. Overall results are reported as the Pillai's trace statistic, which is the most conservative F-statistic within MANOVA analyses. The data were analyzed with SPSS 16 for MacOSX and IBM SPSS Statistics 19.
MEASURING THE RELATIVE POTENTIAL RATES OF INTRA- AND INTERSPECIFIC PARASITE TRANSMISSION IN THE FIELD
To determine the potential for transmission in the field, we conducted an observational study at Irishtown Nature Reserve (see Table S1), a florally rich habitat. Observations were made between 5 and 15 July, and 27 July to 12 August 2004. The foraging behavior of different bumblebee species on all available resources in the park was studied. We undertook direct observations of potential resources to determine which resources were being used by which bumblebee species. A total of 58 1-h intervals were timed and the landing time and species identity of each bumblebee species was recorded for each resource species. Once it was determined which resources were being used, we took a further 57 h of direct observations (see Table S2). In these observations, we studied the levels of visitation of the same flower by two different animals to obtain a measure for potential parasite transmission. To do so, for each resource we observed until a bumblebee foraged (first visitor) and, then, we observed for up to 1 h. If a second bumblebee (second visitor) landed on the same flower the elapsed time and the species were recorded (see Table S3). Observations were terminated after the second bumblebee or the end of the hour. One-hour intervals were chosen because transmission and infection success are expected to be higher the sooner the parasite infects a new host after deposition in the transmission arena, before the parasite suffers desiccation or UV radiation (Schmid-Hempel et al. 1999). At the end of the foraging observations, we sampled animals for a 1-h period and we determined the proportion of them infected by C. bombi in the laboratory (see above).
We calculated the relative abundance of each flowering plant species used by bumblebees as explained in Supporting information.
Data on second visitors (from the whole 115 h of observation) for each species pair at each resource, the relative rates of flower visitation of the different host species (again, from the whole 115 h), parasite prevalence within species, and relative resource availability were combined to calculate the probability of intra- and interspecies transmission for all the host species found in the study area (detailed in Supporting information). Essentially, we calculated for each species the cumulative probability of it foraging at any resource after that resource had been visited either by an infected conspecific or by an infected animal from another species. These species-pair interactions were combined to produce a potential transmission matrix for each observational period.
DOES C. BOMBI EXHIBIT LOCAL POPULATION GENETIC STRUCTURE ACROSS HOST SPECIES?
To address this question, the bumblebees collected after the first and second bouts of observation in the field (see above) were examined for infection by C. bombi (see above). The guts from all the animals infected with C. bombi were dissected out and total DNA was extracted from the guts using the Chelex® extraction method (Walsh et al. 1991). The genetic structure of the parasite strains was analyzed with four microsatellite primers (Cri16, CriP4, Cri2F10, and Cri1B6) following the protocols described in Schmid-Hempel and Reber Funk (2004) (see Supporting information). To test whether the sampling size per species was sufficient to find the majority of all alleles present in that population, a set of rarefaction curves was generated in a simulation model (Ruiz-González 2007; see Supporting information). After the simulations, all populations with less than eight infected individuals and those individuals where amplification of three out of the four loci had failed were removed from the analysis.
Our microsatellite analyses revealed the parasite alleles present in infected animals, but not the strains to which these alleles belonged. However, we can infer possible strains for each bee and then bootstrap FST values based on these inferred strains. We weight the inferred strains according to the number of alleles found in the bee and the number of homozygous loci in the strain. Because the parameters for this weighting are hard to estimate, we made a best guess and then used biologically extreme and conservative parameters to predict a range of FST values (see Supporting information).
DOES THE PARASITE EXHIBIT POPULATION STRUCTURE ACROSS SPECIES, SPACE, AND TIME AT THE START OF THE ANNUAL EPIDEMIC?
We collected queens in the spring of 2004 (March–April) and 2005 (March–May) from sites around Co. Dublin (see above). Some of these queens were used in the cross-infection experiment (see above). We checked each queen to determine whether it was infected by C. bombi (see above). Parasite strains were isolated as above, and population structure of the parasite across host species was analyzed for queens in the same way as for workers.
Results
VARIATION IN HOST QUALITY BUT LITTLE EVIDENCE FOR PARASITE SPECIALIZATION
We experimentally inoculated workers of two host species, B. lucorum and B. terrestris (see Table 1). All parasite strains isolated from these two species were capable of infecting workers from both host species, and a strain isolated from B. pascuorum was capable of infecting B. terrestris workers. Whereas all inoculations with strains from B. lucorum and B. terrestris were successful, only eight of the 15 B. terrestris (53.3%) workers inoculated with parasite cells from B. pascuorum became infected. Grouping data across B. terrestris colonies, and across strains within species origin, there were significant differences in the ability of parasite strains from B. terrestris, B. lucorum, and B. pascuorum to infect B. terrestris workers (G= 32.149, df = 2, P < 0.001), which are clearly due to the lower success of the B. pascuorum-derived strain. Because there were insufficient workers from B. lucorum to test the B. pascuorum isolate, and because there was no replication in isolates from B. pascuorum, only data from inoculations with strains from B. lucorum and B. terrestris were used for further analyses. Crithidia bombi infections released cells (transmission stages) into the feces of their host more quickly if they were growing in B. lucorum colonies than if they were in B. terrestris colonies (Fig. 1A). There were significant effects of either host species or host colony in all four ANOVAs (Table 2). Interestingly, strains originating from B. terrestris produced transmission stages more quickly (Fig. 1B), irrespective of which host species they were growing in (Table 2). There were no significant interaction effects for strain origin × infected host species (Table 2).
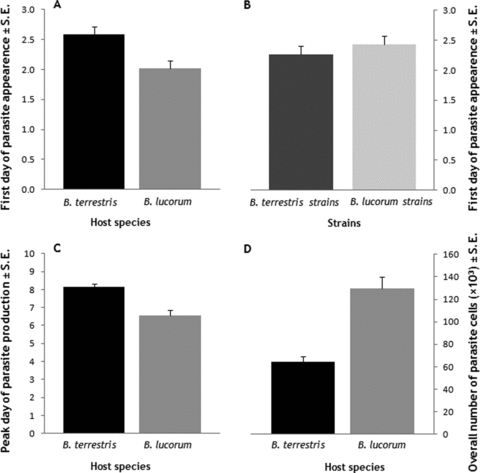
Effects of parasite origin and current host on within-host growth of Crithidia bombi. (A) Parasite strains, independently of the host species from where they were isolated, grow more rapid in Bombus lucorum hosts. (B) Parasite strains isolated from B. terrestris hosts grow more quickly than those from B. lucorum, independently of the new host species. (C) Parasite productivity peaks earlier in B. lucorum hosts. (D) Bombus lucorum supports higher parasite reproduction than B. terrestris (see text and Table 2 for statistics).
| First day of parasite cells in host feces | Peak day of parasite cell emission | Total number of parasite cells shed in 10 days | |||||||
|---|---|---|---|---|---|---|---|---|---|
| F | df | P | F | df | P | F | df | P | |
| Host | |||||||||
| Host species×origin species | 9.418 | 1164 | 0.003 | 24.394 | 1164 | <0.001 | 21.237 | 1164 | <0.001 |
| Host species×parasite strain | 10.751 | 1164 | 0.001 | 24.912 | 1156 | <0.001 | 22.124 | 1156 | <0.001 |
| Host colony×origin species | 4.752 | 1164 | <0.001 | 4.038 | 8157 | <0.001 | 5.562 | 8157 | <0.001 |
| Host colony×parasite strain | 4.938 | 1164 | <0.001 | 3.965 | 8135 | <0.001 | 5.507 | 8135 | <0.001 |
| Parasite | |||||||||
| Host species×origin species | 1.609 | 1164 | 0.206 | 0.001 | 1164 | 0.981 | 2.299 | 1164 | 0.131 |
| Host species×parasite strain | 4.248 | 5156 | 0.001 | 1.172 | 5156 | 0.325 | 1.117 | 5156 | 0.354 |
| Host colony×origin species | 4.237 | 1157 | 0.041 | 1.624 | 1157 | 0.204 | 0.925 | 1157 | 0.338 |
| Host colony×parasite strain | 6.203 | 5135 | <0.001 | 1.404 | 5135 | 0.227 | 1.218 | 5135 | 0.304 |
| Host×parasite | |||||||||
| Host species×origin species | 2.274 | 1164 | 0.134 | 1.207 | 1164 | 0.273 | 0.333 | 1164 | 0.565 |
| Host species×parasite strain | 0.940 | 5156 | 0.457 | 0.806 | 5156 | 0.547 | 1.213 | 5156 | 0.306 |
| Host colony×origin species | 0.067 | 1157 | 0.795 | 0.324 | 1157 | 0.570 | 1.141 | 1157 | 0.287 |
| Host colony×parasite strain | 1.548 | 19,135 | 0.079 | 0.787 | 19,135 | 0.719 | 1.111 | 19,135 | 0.347 |
Overall, parasites peaked earlier in B. lucorum than in B. terrestris (Fig. 1C). This was significant for both host species and host colony analyses (Table 2). There were no significant effects of strain origin, strain, or the interaction between any measure of parasite strain and host identity (Table 2). Parasites in B. lucorum in general produced more transmission cells overall than parasites in B. terrestris (Fig. 1D). There were significant effects of either host species or host colony in all four analyses (Table 2). Again, there were no effects of strain origin, strain, or the interaction between any measure of parasite strain and host identity (Table 2).
SPECIES DIFFER IN IMMUNE BACKGROUND BUT INFECTION DOES NOT ELICIT IMMUNE CHANGES
There were significant differences in natural immune levels across species of wild caught bees (F6,92= 3.268, P= 0.006). Overall, host species only had a significant impact on phenoloxidase activity (ZI: F2,55= 2.339, P= 0.108; PO: F2,55= 4.633, P= 0.015; PPO: F2,55= 2.213, P= 0.121; Fig. 2A–C). Scheffé post hoc pairwise comparisons detected higher PO activity in B. terrestris than in B. pascuorum (P= 0.006). In contrast, there were no overall (F6,92= 0.253, P= 0.957) or individual effects of infection status on any of the immune parameters (ZI: F2,55= 0.048, P= 0.953; PO: F2,55= 0.338, P= 0. 715; PPO: F2,55= 0.496, P= 0.612; Fig. 2A–C). Similarly, there were no overall or individual effects of the interaction between host species and infection status (F12,141= 1.410, P= 0.168; ZI: F4,55= 1.405, P= 0.247; PO: F4,55= 0.757, P= 0.558; PPO: F4,55= 2.094, P= 0.097; Fig. 2A–C).

Immune function across host species with and without parasites. Species differ in (A) antimicrobial defenses, (B) phenoloxidase activity, and (C) total phenoloxidase, but neither natural nor controlled infection with C. bombi has an effect on immunity.
POTENTIAL FOR INTRA- AND INTERSPECIFIC TRANSMISSION
Six different species of bumblebees were found foraging on 19 food-plant species during the 1-h observations at Irishtown Nature Reserve (Tables S2 and S3). Both bumblebee and resource species varied in relative abundance. Because B. terrestris and B. lucorum workers are impossible to distinguish in the field both species were grouped together (B. terr/luc). Only one B. pratorum observation was obtained and this species was therefore not included in the final analysis. Bombus pascuorum was found to be the most generalist species foraging on 12 different species of plants while B. muscorum was the most specialist visiting four resources; B. lapidarius and B. terrestris/lucorum foraged at eight different resources each. There was an overall density of approximately 10 flowering plants (independently of species) per square meter.
To measure the proportion of infected animals in the field, 106 bumblebees in Period 1 and 83 bees in Period 2 were sampled. The parasite was found in all host species with a prevalence per host species as shown in Table S3. In Period 2, B. muscorum was absent due to species-specific phenology.
As can be seen in Figure 3, we found that the potential for interspecific transmission between pairs of host species was variable and asymmetric. For example, B. lapidarius was seven times more likely to be a source of the parasite to B. pascuorum than it was to be a recipient of parasites from this species in Period 1. Species also varied in their potential for intraspecific transmission, with this being highest in B. lapidarius and lowest in B. muscorum. Overall, the proportion of potential self-infection was highest in B. lapidarius (Period 1 = 0.72, Period 2 = 0.86), much lower in B. pascuorum (Period 1 = 0.2, Period 2 = 0.071), B. muscorum (Period 1 = 0.125), and then lowest in the B. terr/luc aggregate (Period 1 = 0.1, Period 2 = 0.063). These results suggest that intraspecific transmission may dominate in B. lapidarius and B. pascuorum in Period 1, and in B. lapidarius only in Period 2. Bombus lapidarius was consistently the dominant source of interspecific transmission for all the other species in both periods.
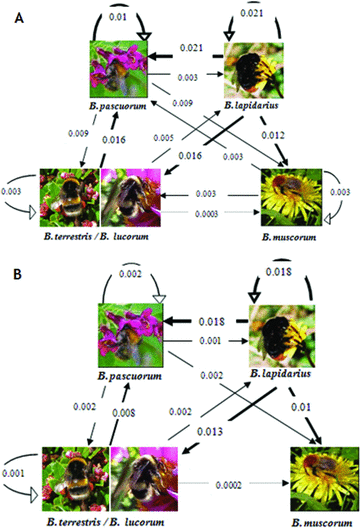
Field transmission matrices. The thickness of the connector represents the probability of a second visitor becoming infected after foraging on a previously visited resource. The calculi are described in the text. The head of the arrows point to the second visitor and line thickness increases with the likelihood of transmission. (A) Period of sampling 1; (B) Period of sampling 2.
LOCAL POPULATION GENETIC STRUCTURE OF C. BOMBI ACROSS HOST SPECIES
A total of 106 bumblebees were collected in Period 1 and 86 in Period 2. Of these, 59 were infected with C. bombi in Period 1 and 23 in Period 2 (Table S4). The amplification of the four microsatellite markers was successful for nearly all the 82 infected animals: three animals only amplified the Cri16 locus, three animals amplified three of the four microsatellite markers and one bee did not amplify loci CriF10 and Cri1B6. Crithidia bombi is diploid and all samples amplified one or more bands, and thus denoted homozygote and heterozygote strains, and multiply infected hosts (more than two bands in seven out of the 59 animals from Period 1, and three out of the 23 animals from Period 2). Five different alleles were found in loci Cri16 and CriP4, six alleles in locus Cri2F10, and 14 alleles in locus Cri1B6. Some alleles appeared only in one of the two periods or in one of the species (see Table S4). Because of low sample sizes in Period 2, we conducted two analyses. First, we analyzed the population genetic structure of the parasite in Period 1 across B. lapidarius, B. pascuorum, and B. terr/luc. Second, we asked whether the population of the parasite in B. lapidarius varied from Period 1 to Period 2.
There was significant population structuring across the parasite population in the three host species in Period 1 (Table 3). The parasite subpopulation in B. terr/luc was significantly different from that in B. lapidarius and in B. pascuorum (Table 3). The parasite subpopulation in B. lapidarius, however, was not different from that in B. pascuorum (Table 3). Finally, there was significant differentiation between the C. bombi populations hosted by B. lapidarius in Period 1 versus Period 2 (Table 3).
| Best FST±SD | Best P-value | Minimum FST±SD | Minimum P-value | Maximum FST±SD | Maximum P-value | |
|---|---|---|---|---|---|---|
| Overall | 0.107±0.002 | <0.001 | 0.243±0.002 | <0.001 | 0.071±0.002 | <0.001 |
| B. lapidarius versus B. pascuorum | 0.015±0.006 | 0.005 | 0.002±0.006 | 0.359 | 0.151±0.004 | <0.001 |
| B. lapidarius versus B. terr/luc | 0.154±0.003 | <0.001 | 0.105±0.003 | <0.001 | 0.302±0.002 | <0.001 |
| B. pascuorum versus B. terr/luc | 0.130±0.004 | <0.001 | 0.088±0.004 | <0.001 | 0.182±0.004 | <0.001 |
| B. lapidarius 1 versus B. lapidarius 2 | 0.039±0.004 | <0.001 | 0.010±0.004 | 0.013 | 0.214±0.003 | <0.001 |
HOST SPECIES, SPATIAL, AND TEMPORAL POPULATION GENETIC STRUCTURE OF C. BOMBI AT THE START OF THE ANNUAL EPIDEMIC
A total of 148 queens were collected in 2004, and 359 in 2005, and their infection status was checked. Crithidia bombi was found in six host species, in 72 queens (49%) in 2004 and 109 queens (30%) in 2005 (Table 4). Parasite prevalence was higher in the more abundant species (blanket collection took place at all sites, so numbers collected serve as a proxy for abundance; Table 4).The amplification of the four microsatellite markers was successful for nearly all of the 181 parasitized queens: four animals only amplified the Cri4 locus, two animals only amplified the Cri16 and Cri2F10 loci, six animals did not amplify the Cri1B6 locus, and two animals failed to amplify locus Cri2F10 and only one animal failed to amplify locus Cri16. The alleles found in parasites from founding queens are listed in Tables S5 and S6.
| Year | Host species | N | Prevalence |
|---|---|---|---|
| 2004 | B. hortorum | 1 | 100.00 |
| B. jonellus | 11 | 9.09 | |
| B. lapidarius | 4 | 50.00 | |
| B. lucorum | 54 | 55.56 | |
| B. pratorum | 40 | 32.50 | |
| B. terrestris | 38 | 65.79 | |
| TOTAL | 148 | 48.65 | |
| 2005 | B. hortorum | 2 | 0.00 |
| B. jonellus | 8 | 12.50 | |
| B. lapidarius | 1 | 0.00 | |
| B. lucorum | 68 | 20.59 | |
| B. muscorum | 2 | 0.00 | |
| B. pascuorum | 45 | 37.78 | |
| B. pratorum | 59 | 25.42 | |
| B. terrestris | 174 | 32.76 | |
| TOTAL | 359 | 28.97 |
Due to unbalanced and variable sample sizes across species, sites, and years, we ran the following analyses. First, prior to lumping data from all species together across sample sites, we determined whether there was parasite population structure within a species across sites within a year. Second, because these analyses indicated significant spatial population structure (see below), we asked whether there was parasite population structure across host species within individual sites. Third, we asked whether parasite population structure differed between years for a single host species at a single site.
Crithidia bombi exhibited significant population structure (i.e., significant values of FST) in its host B. lucorum in 2004 across four sample sites (Glenasmole, Glenveagh, Archbishop Ryan's Park and Howth: Table 5). This structure was driven by differences between all pairs of populations (Table 5). In 2005, the parasite exhibited significant population structure in both B. lucorum and B. terrestris across two sites (Archbishop Ryan's Park and Howth: Table 5).
| Comparison | Best FST±SD | Best P-value | Minimum FST±SD | Minimum P-value | Maximum FST±SD | Maximum P-value |
|---|---|---|---|---|---|---|
| B. lucorum across sites 2004 | 0.099±0.002 | <0.001 | 0.182±0.002 | <0.001 | 0.064±0.002 | <0.001 |
| B. lucorum 2004: GLM versus GLV | 0.077±0.003 | <0.001 | 0.048±0.003 | <0.001 | 0.101±0.004 | <0.001 |
| B. lucorum 2004: GLM versus HOW | 0.091±0.003 | <0.001 | 0.051±0.003 | <0.001 | 0.196±0.002 | <0.001 |
| B. lucorum 2004: GLM versus ARP | 0.101±0.004 | <0.001 | 0.048±0.005 | <0.001 | 0.157±0.004 | <0.001 |
| B. lucorum 2004: GLV versus HOW | 0.106±0.004 | <0.001 | 0.046±0.004 | <0.001 | 0.277±0.003 | <0.001 |
| B. lucorum 2004: GLV versus ARP | 0.192±0.006 | <0.001 | 0.124±0.005 | <0.001 | 0.275±0.011 | <0.001 |
| B. lucorum 2004: HOW versus ARP | 0.091±0.005 | <0.001 | 0.055±0.004 | <0.001 | 0.160±0.005 | <0.001 |
| B. lucorum 2005: HOW versus ARP | 0.089±0.012 | <0.001 | 0.043±0.010 | <0.001 | 0.284±0.010 | <0.001 |
| B. terrestris 2005: HOW versus ARP | 0.027±0.010 | 0.006 | 0.006±0.008 | 0.022 | 0.136±0.016 | <0.001 |
| HOW 2004: B. jonellus versus B. lucorum | 0.063±0.014 | <0.001 | 0.039±0.013 | <0.001 | 0.109±0.017 | <0.001 |
| ARP 2005: B. lucorum versus B. terrestris | 0.043±0.008 | <0.001 | 0.028±0.008 | <0.001 | 0.223±0.009 | <0.001 |
| Across all species in HOW 2005 | 0.093±0.003 | <0.001 | 0.289±0.002 | <0.001 | 0.060±0.003 | <0.001 |
| HOW 2005: B. jonellus versus B. lucorum | 0.113±0.005 | <0.001 | 0.076±0.005 | <0.001 | 0.154±0.009 | <0.001 |
| HOW 2005: B. jonellus versus B. pascuorum | 0.136±0.007 | <0.001 | 0.074±0.006 | <0.001 | 0.285±0.010 | <0.001 |
| HOW 2005: B. jonellus versus B. pratorum | 0.139±0.005 | <0.001 | 0.068±0.006 | <0.001 | 0.399±0.003 | <0.001 |
| HOW 2005: B. jonellus versus B. terrestris | 0.215±0.007 | <0.001 | 0.145±0.006 | <0.001 | 0.345±0.010 | <0.001 |
| HOW 2005: B. lucorum versus B. pascuorum | 0.072±0.005 | <0.001 | 0.045±0.005 | <0.001 | 0.122±0.007 | <0.001 |
| HOW 2005: B. lucorum versus B. pratorum | 0.064±0.004 | <0.001 | 0.042±0.005 | <0.001 | 0.288±0.003 | <0.001 |
| HOW 2005: B. lucorum versus B. terrestris | 0.068±0.005 | <0.001 | 0.043±0.005 | <0.001 | 0.138±0.007 | <0.001 |
| HOW 2005: B. pascuorum versus B. pratorum | 0.086±0.005 | <0.001 | 0.054±0.004 | <0.001 | 0.311±0.003 | <0.001 |
| HOW 2005: B. pascuorum versus B. terrestris | 0.383±0.006 | <0.001 | 0.052±0.006 | <0.001 | 0.147±0.010 | <0.001 |
| HOW 2005: B. pratorum versus B. terrestris | 0.098±0.005 | <0.001 | 0.061±0.005 | <0.001 | 0.323±0.003 | <0.001 |
| HOW B. jonellus: 2004 versus 2005 | 0.222±0.025 | <0.001 | 0.127±0.020 | <0.001 | 0.323±0.050 | <0.001 |
| HOW B. lucorum: 2004 versus 2005 | 0.078±0.011 | <0.001 | 0.047±0.014 | <0.001 | 0.211±0.008 | <0.001 |
| ARP B. lucorum: 2004 versus 2005 | 0.110±0.014 | <0.001 | 0.048±0.011 | <0.001 | 0.286±0.012 | <0.001 |
Within a site, and within a year, there was significant parasite population structure across pairs of host species for B. jonellus and B. lucorum from Howth in 2004, B. lucorum and B. terrestris from Archbishop Ryan's Park in 2005, and for all pairwise comparisons of B. jonellus, B. lucorum, B. pascuorum, B. pratorum, and B. terrestris from Howth in 2005 (Table 5).
Finally, within a site and for individual species, there was significant parasite population structure across years for B. lucorum in Archbishop Ryan's Park and Howth and B. jonellus in Howth (Table 5).
Discussion
Variation in host quality and asymmetries between intra- and interspecific transmission are expected to drive population structure in multihost parasites (Gandon 2004). In turn, such population structure provides the background from which host generalism or specialism can evolve. Significant differences across bumblebee host species in their quality, measured by parasite growth rate, and their potential as inter- and intraspecific transmitters may indeed generate population structure in the multihost parasite C. bombi. However, this structure is highly dynamic across species, space, and time, and thus is unlikely to facilitate the evolution of long-term host specialization in this system.
Numerous lines of evidence show that C. bombi is a true multihost parasite across Bombus spp. (e.g., results from our cross-infection experiments, phylogenetic [Schmid-Hempel and Tognazzo 2010] and population genetic analyses [Salathé and Schmid-Hempel 2011]). However, from a parasite perspective, these host species are not equal. Hosts differ in their quality with respect to the rate at which parasite populations can grow, as well as the number of transmission stages produced by these populations. Consequently, host species will vary in their value to the parasite, with some species disseminating the parasite earlier and at a higher level than others. An underlying mechanism for these differences may be variation across species in immune function. In particular, we found that the phenoloxidase branch of the immune system varied across host species, and previous studies have found phenoloxidase to be related to infections of C. bombi in B. terrestris (Brown et al. 2003b). Surprisingly, in contrast to previous studies (Brown et al. 2003b; Otterstatter and Thomson 2006; Rydell et al. 2009; Schlüns et al. 2010), we found no impact of experimental infection on immune function in wild-caught animals. One explanation for this may be immune exhaustion, with wild-caught animals likely to have a history of repeated immune stimulation (Plaistow et al. 2003) leaving them less able to respond actively to novel threats (but see Allander and Schmid-Hempel 2000).
Host species also differ in their quality as transmitters. Our potential transmission matrices demonstrated that host species differ in their contribution to inter- versus intraspecific transmission, as well as their relative importance as sources or sinks in the web of transmission. For example, B. lapidarius is likely to host a parasite pool dominated by intraspecific transmission, while acting as an important transmission route to other host species. These differences are mediated by overlap in, and species-specific utilization of foraging resources, which is a general feature of bumblebee assemblages (e.g., Goulson and Darvill 2004). Interestingly, our potential transmission matrices varied across time, due to changes in host assemblages (the presence/absence and relative abundance of different host species) and available resources. For example, the relative importance of intraspecific transmission in B. pascuorum decreased by an order of magnitude from Period 1 to Period 2, while the contribution of interspecific transmission to B. pascuorum from B. terr/luc decreased by half. As both bumblebee assemblages (e.g., Alford 1975) and floral resources vary over space and time, the key feature of transmission matrices in this system should be that they are spatially and temporally dynamic.
While we believe that our potential transmission matrices are likely to be broadly accurate, incorporating, as they do, host behavior, floral abundance, parasite prevalence, and the dynamics of parasite transmission stages, at least three other factors may modify them. First, results from our cross-infection experiments, and previous studies of host–parasite genotype–genotype interaction (e.g., Shykoff and Schmid-Hempel, 1991b,c) suggest that not all host colony–parasite strain combinations are equally likely to result in infection. However, there is no a priori reason to expect such interactions to be distributed asymmetrically across host species, and thus they are unlikely to alter the relative force of intra- and interspecific infection identified in our matrices. Second, inflorescence architecture has been shown to affect the likelihood of picking up C. bombi from a contaminated flower (Durrer and Schmid-Hempel 1994). In our study, some complex resources, such as Dipsacus fullonum or Trifolium pratense, were only visited by B. pascuorum, the species with the longest tongue (Goulson and Darvill 2004). Rubus fruticosus, with the simplest flower morphology, was the most visited resource by five bee species. Bombus lapidarius and B. pascuorum visited flowers or inflorescences of all levels of complexity while B. terrestris visited R. fruticosus most frequently. We did not explore the impact of flower or inflorescence complexity on the potential parasite transmission matrices. However, inflorescence complexity and, possibly, also flower complexity, reduce the probability of infection (Durrer and Schmid-Hempel 1994) and long-tongued bumblebees forage on more complex flowers (see above). Consequently, the contribution of intraspecific interactions to transmission is likely to be reduced in long-tongued bumblebee species (Ruiz-González and Brown 2006b). Finally, our matrices do not include within-nest transmission. This is undoubtedly high (Otterstatter and Thomson 2007) and thus is likely to enhance the intraspecific transmission component for all host species.
Differences in host quality and asymmetric patterns of transmission may generate parasite population structure. However, while the parasite population indeed exhibited significant genetic structure across foraging workers of different species, it bore no simple relationship to the asymmetries observed in the potential transmission matrices. Two parasite subpopulations were found, one shared by the host species B. lapidarius and B. pascuorum and one in B. terr/luc. While the transmission matrix predicted the potential for a parasite subpopulation in B. lapidarius, it did not predict that this subpopulation would also span another host species (i.e., B. pascuorum). This divergence between observations and population genetics may result from differences in host quality with respect to infection and growth by the parasite, a time lag in the impact of transmission on parasite population structure, the absence of intracolony transmission from our matrix, strain filtering by individual colonies (Schmid-Hempel et al. 1999) or as yet undetermined factors. Nevertheless, the presence of population structure in the parasite indicates the potential for the evolution of specialization. Interestingly, similar results were found in a recent study by Salathé and Schmid-Hempel (2011), despite using a much more conservative estimate of population structure. Parasite transmission via flowers is analogous to vectored transmission, with flower preferences of bumblebees being equivalent to feeding preferences by vectors. Simpson et al. (2011) showed that feeding preferences by mosquitoes predict parasite epidemiology in the West Nile Virus system. Together with our results, these studies suggest that asymmetries in transmission in multihost parasites may play a general role in structuring parasite populations and driving epidemiology.
The evolution of host specialization is most likely if parasite population structure across host species is stable over time. To test this, we examined population structure across 2 years in spring queens. These queens have just emerged from hibernation and, if infected, should be carrying strains picked up from their maternal colonies (Ulrich et al. 2011). The parasite exhibited significant population structure across host species in both years. This structure likely reflects the maintenance of species-specific parasite populations in their mother colonies via transmission and host quality asymmetries (see above; Schmid-Hempel and Reber-Funk 2004). However, this parasite population structure is not stable across time, with significant differences in parasite population structure within a given host species from one year to the next. These differences presumably result from differential colony founding success by queens from different maternal lineages (Gerloff and Schmid-Hempel 2005), differential transmission potential due to changes in floral community and bumblebee assemblages from one year to the next, filtering of strains, and thus bottlenecking of the parasite populations by queens (Ulrich et al. 2011), and the combination of clonal and sexual reproduction in this parasite (Schmid-Hempel et al. 2011). Similar patterns of population structure have been found in a parasitic mite of bats, with transmission and winter bottlenecking again appearing to be the major determinants of parasite population structure (Bruyndonckx et al. 2009). The presence of interannual population structure suggests that, while short-term patterns of transmission may structure the parasite population, over annual time scales such structure is rapidly broken down, which is likely to negate the potential for long-term parasite adaptation to particular host species. This would explain the general lack of parasite origin effects in our cross-infection experiments.
We also found significant spatial structure within host species in the parasite population. The scale of this spatial structure matches that of queen dispersal in the spring (Lepais et al. 2010), suggesting that it is being driven by host philopatry. Such spatial structure may help to explain the presence of local adaptation in this host–parasite relationship, where allopatric infections exhibit higher virulence (Imhoof and Schmid-Hempel 1998b), although we note that the scale at which we identified this structure is smaller than that at which local adaptation has been detected.
While most of our results indicate that C. bombi has evolved a generalist strategy, and despite the absence of consistent parasite lineages across time in different host species, we nevertheless found some evidence for at least short-term host specialization. Parasite origin had a small effect on epidemiology within hosts in our cross-infection experiments, and the relative failure of one strain from B. pascuorum is also suggestive of host specialization. An alternative explanation for these results is the close genotype–genotype associations seen in this system (Schmid-Hempel and Reber Funk 2004), with apparent specialization being due to simple host–parasite genetic mismatches (e.g., Schmid-Hempel et al. 1999). Further work is required to distinguish between these two explanations. Nevertheless, the possibility remains that at least short-term adaptation to individual host species exists under natural conditions. One mechanism for this may be strain filtering, which has been shown to reduce allocolonial infectivity by C. bombi after five passages in a laboratory study (Yourth and Schmid-Hempel 2006).
To conclude, theoretical models of multihost parasites suggest that transmission, host quality, and trade-offs in the exploitation of different host species are likely to drive the evolution of parasite specialization and virulence (Regoes et al. 2000; Woolhouse et al. 2001; Gandon 2004). Empirical evidence for these predictions, however, is lacking (Rigaud et al. 2010). Furthermore, using an experimental system, Bedhomme et al. (2011) showed that the specialist-generalist trade-off assumed by the models above may be absent, resulting in cost-free maintenance of a generalist strategy. Thus, more broadly, the maintenance of generalist multihost-parasite populations might be favored if adaptation to specific hosts is costly. Our results suggest that asymmetry in transmission and host quality can indeed drive parasite population structure, at least over short spatiotemporal scales, which may in turn enable the short-term evolution of specialist strains. However, the parasite population structure is highly dynamic, perhaps due to the idiosyncrasies of both host and parasite biology, rendering long-term evolution of specialist strains unlikely in this system, irrespective of costs. The reproductive biology of C. bombi (Schmid-Hempel et al. 2011) combined with host filtering of parasite strains (Schmid-Hempel et al. 1999; Ulrich et al. 2011) is likely to be important in structuring the parasite population across annual scales, although this remains to be confirmed. While we did not examine the virulence of C. bombi across host species, our results suggest that species-specific virulence driven by parasite adaptation is unlikely to emerge in this system. Further studies are required if we are to understand how having multiple host species impacts on parasite epidemiology and virulence.
Associate Editor: A. Read
ACKNOWLEDGMENTS
MJFB conceived the study. MXRG, YM, PSH and MJFB designed the experiments. MXRG conducted the experimental work, with additional immune work by YM. CRF & PSH aided the interpretation of the molecular data. JB designed and implemented the algorithm for the analysis of the molecular data. MXRG and MJFB led the writing of the paper, with significant contributions from all authors. We thank R. M. Falcao, A. M. Flanagan, and C. Strevens for assistance in the field and in the laboratory, S. Cornet and P. Stafford for technical support, S. T. Rutrecht for providing some C. bombi infected queens, and E. van Leeuwen, K. Dexter, and R. Schmid-Hempel for valuable comments on the microsatellite data analysis and its interpretation. This study was supported by a Basic Research from Enterprise Ireland SC/2002/209 to MJFB and a Ulysses grant to MJFB and YM MJFB and JB were supported by a grant funded jointly by the Biotechnology and Biological Sciences Research Council [grant nr. BB/I000151/1]; the Department for Environment, Food and Rural Affairs; the Natural Environment Research Council; the Scottish Government; and the Wellcome Trust: under the Insect Pollinators Initiative (to MJFB). YM was supported by the Centre National de la Recherche Scientifique (CNRS). PSH and CRF were supported by the Swiss NSF (grant nr.31003A-116057 to PSH). This work complied with the laws governing animal research in Ireland. The authors have no competing financial interests. The manuscript was significantly improved thanks to comments by two anonymous reviewers.




