Detailed Structure of Pneumocystis carinii Chromosome Ends
Pneumocystis carinii has three gene families, MSG, MSR and PRT1, the members of which tend to be grouped together in clusters located at chromosome ends (reviewed in reference 6). Each of these gene families encodes a family of surface proteins, which are known as Major Surface Glycoprotein (MSG), MSG-Related (MSR), and Protease (PRT1). Determining the size and composition of these gene arrays is important for understanding their evolution and function. A question of particular interest is the possibility of co-expression of a specific MSG gene and a specific PRT1, because MSG proteins may be processed by proteases encoded by PRT1 genes [3, 7]. Prior to the work described here, segments of the genome that contain members of these gene families had been characterized, but an entire cluster had not yet been isolated.
As part of the Pneumocystis Genome Project, the Sanger Center (http://www.sanger.ac.uk) determined the sequence of gene arrays contained within two cosmids, 3G5 and 1B2. Both cloned gene clusters contained members of all three gene families, but the order of the genes in the two clones was different. In addition to full size MSG and MSR genes, both gene clusters contained what appear to be fragments of MSG or MSR genes.
The library contained additional cosmids that shared with 1B2, and with each other, a core region of about 15 kilobases at the telomere-distal end of the sequence. Surprisingly, two of these 1B2-related cosmids mapped to one chromosome, and another mapped to a different chromosome. These linkage data show that the same gene array can be at the ends of two different chromosomes.
MATERIALS AND METHODS
A cosmid library was made from genomic DNA from a population of P. carinii from a single infected rat [5].
Methods for the following procedures were as described in reference 4. Cosmids that contained MSG genes and telomeres were identified by hybridization of bacterial colonies with DNA probes. Thirty cosmids were analyzed by digestion with restriction endonuclease EcoRI. Two cosmids that were clearly different were selected for sequence analysis. The DNA in the cosmid was fragmented by shearing and fragments were inserted into a plasmid vector. The inserts in one thousand plasmids were sequenced. The small sequences were assembled into one contiguous sequence by methods created by the Sanger Centre. The sequence of each cosmid was queried for restriction enzyme sites and fragments sizes and numbers were predicted. Cosmid DNA was tested for the presence of the predicted restriction enzyme cleavage sites by Southern blot analysis using a battery of radioactive DNA probes specific for MSG, MSR, PRT1 and telomeres. Some regions of each cosmid were resequenced to verify the sequence assembled from random fragments. DNA for resequencing was produced by the PCR using primers based on the assembled random sequence. Expression of putative genes was tested by Northern blot analysis. The accession numbers for the cosmid sequences are: AL592382 and AL592263 for cosmids 3G5 and 1B2, respectively. Orthologues of genes were initially identified by the Sanger Centre and then subjected to BLASTX searches using the NCBI web site (http://www.ncbi.nlm.nih.gov). Genes were mapped to P. carinii chromosomes by hybridization to Southern blots carrying electrophoretically separated chromosomes prepared as described previously [1, 2].
RESULTS AND DISCUSSION
Figure 1 shows maps derived from the two sequenced cosmid inserts. The map of 3G5 was produced by analysis of the sequence. The map labeled 1B2+11H12 was produced by analysis of the complete 1B2 sequence and by partial sequencing of the insert in cosmid 11H12, which overlapped the 1B2 insert, and served to link it to a chromosome via the chitin biosynthesis gene. While the maps shown were generated solely from sequence data, these maps were consistent with the results of analysis of the cosmids by Southern blotting of DNA fragments produced by digestion with restriction endonucleases, by amplification of segments of each cosmid, and by sequencing of these amplified segments (data not shown). Therefore, we conclude that the two complete sequences are accurate, and that none of the features shown in the Figure were artifacts generated by erroneous assembly, which was a concern given the repetitive nature of much of the DNA under analysis.
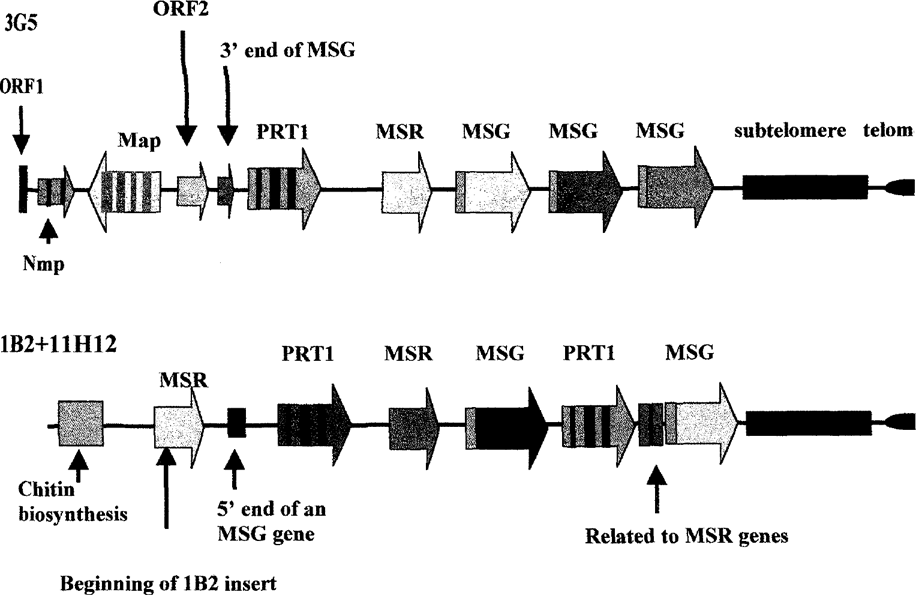
Maps of two complete gene clusters.
The DNA in 3G5 begins with four presumptive genes (ORF1, Nmp, Map, ORF2) that appeared to be unique because they each hybridized to the same chromosome which was approximately 315 kb in size (data not shown). Two of these genes, Nmp and Map, are orthologous to genes of known function. Nmp encodes a protein related to one involved in nuclear migration in mycelia. Map encodes a protein related to one associated with microtubules. The other two unique sequences, ORF1 and ORF2, encode peptides with no known relatives. However, the protein encoded by ORF2 has a 65 amino acid tract related to one found in MSG proteins (33% identity, 66% similarity). Following ORF2 is a 1 kb region that matches the 3′ends of MSG genes. This juxtapositioning suggested that ORF2 and the 1 kb MSG-like region might be transcribed together, but Northern blot experiments showed that this was not the case. ORF2 hybridized to a 1.2 kb band on a Northern blot, but this transcript did not hybridize to a probe that detects the 3′ends of MSG transcripts, showing that the fragmentary MSG gene downstream of ORF2 is not co-transcribed with it.
The 1 kb MSG-like region marks the beginning of the repeated region of the 3G5 insert. After this comes a cluster of genes containing members of the three repeated gene families. The end of the gene cluster is marked by subtelomeric DNA which in turn is followed by a short stretch of telomere. The gene cluster was not interrupted, and all of the genes in the cluster were oriented in the same direction. These two features of the cluster are consistent with it being formed by unequal reciprocal recombination events. The three MSG genes are also linked directly one to another without interruption, suggesting that this group of genes was also formed by unequal reciprocal recombination. However, the three MSG genes are not more closely related to each other than they are to other known MSG genes. Therefore, this triplication appears to not be a recent event.
Figure 1 also shows the structure of the genome segment carried by cosmids 11H12 and 1B2, which overlapped. This cluster of repeated genes begins with a gene orthologous to a gene that in other fungi encodes an enzyme used to synthesize chitin. The repeated gene cluster begins with an MSR gene, followed by a PRT1 gene, which is followed by an MSR and an MSG, but then there is a second PRT1 gene. Thus the arrangement of repeated genes in the segment of the genome covered by the inserts in cosmids 1B2 and 11H12 is quite different from that in cosmid 3G5. In addition, the arrangement in 11H12+1B2 is the first example of an array that contains more than one PRT1 gene. After the second PRT1 gene is an aberrant MSR gene (see below). The cluster ended with an MSG gene, but the reading frame of this gene is shifted by one base at a point about 20% in from the 3′end.
It seemed possible that the frameshift mutation in the sequence of the last MSG gene in cosmid 1B2 could have been caused by a sequencing error. To examine this possibility, the region in question was amplified from cosmid 1B2 DNA by the PCR, and both strands of the amplicon were scqucnced. This experiment showed that the frameshift was present in cosmid 1B2. However, sequence analysis of the same region from two other cosmids that carried the same segment of the genome as 1B2, such as 11H12, showed that the frameshift mutation was unique to cosmid 1B2. Comparison of the 1B2 sequence to that of the other two cosmids showed that the frameshift was caused by deletion of one basepair from the 1B2 sequence. This deletion could have occurred during cloning, or it may have existed in some of the P. carinii in the population that was used to make the cosmid library.
Like 3G5, which carried one incomplete MSG gene, 1B2 carried two segments of DNA that appear to be segments of either MSR or MSG genes. Following the first MSR gene, is a sequence resembling the 5′end of an MSG gene. This sequence includes what appears to be a degenerate copy of the CRJE, which is a 25 basepair element that is identical in every MSG gene. The second aberrant sequence in 1B2 resides between the second PRT1 gene and the last MSG gene. To determine if cloning or sequencing error generated these aberrant sequences, the regions of interest were subjected to the PCR. Both regions were amplified and sequencing showed them to exactly the same as the 1B2 sequence. Furthermore, the segment resembling an MSG 5′end was also present in cosmid 11H12. We conclude that cloning or sequencing errors did not generated these sequences.
In both cosmids 3G5 and 1B2, the genes and gene fragments in the repeat arrays are all oriented in the same direction. This structure would make it possible for all of the genes in the cluster to be controlled in unison by a single transcriptional promoter upstream of the first gene in the cluster. However, it is known that MSG genes are not expressed unless they reside at a specific locus, called the UCS locus. Therefore, coordinate expression of all the genes in a cluster would seem to require that the first gene in the cluster be an MSG gene, because only then would the entire cluster be able to move to the UCS locus along with the MSG gene. However, in neither cosmid was the first gene in each cluster an MSG gene. Instead, the first full gene is either a PRT1 or an MSR.
From EcoRI endonuclease mapping, it was shown that, in addition to 11H12, several other cosmids in the library carried an insert highly related to that in 1B2. The mapping data defined a region of about 15 kb that was present in both 1B2 and in all of the other cosmids. One such cosmid, 12D10, contained an insert that, like 11H12, began in unique DNA. However, the unique DNA in 12D10 DNA did not map to the same chromosome as the unique DNA in 11H12 (data not shown). Whereas cosmid 11H12 was linked to chromosome 9 via a chitin biosynthesis gene, cosmid 12D10 was linked to chromosome 4 via a unique sequence encoding a protein related to a nuclear protein (see Figure 2). These linkage data show that the same gene array can be at the ends of two different chromosomes. In addition, the gene arrays in 1B2 and 12D10 were linked to different downstream DNA sequences (see Figure 2). A segment of the genome containing the 15 kb shared region may have been inserted into at least two different locations in the genome by gene conversion events. The shared region begins in the first MSR gene and ends either in or just after the second PRT1 gene. Cosmid 12D10 has not been sequenced yet, so the precise boundaries of the shared region are not yet known.
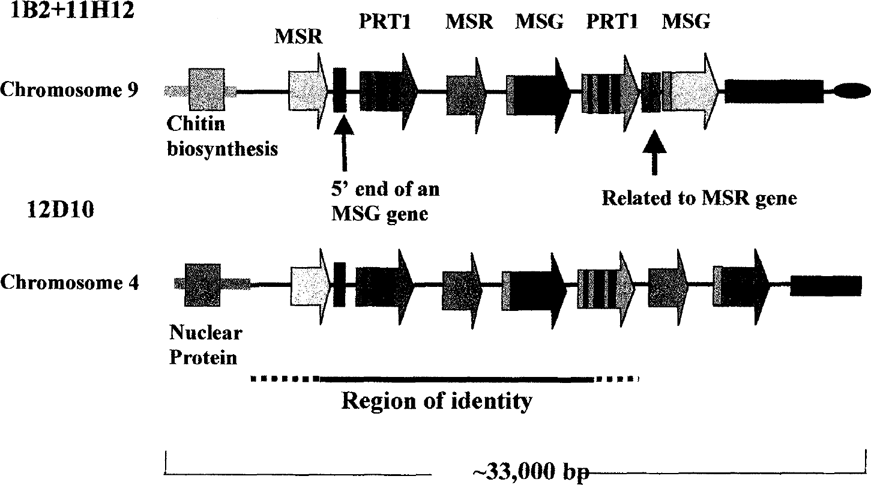
The same gene cluster linked to two different chromosomes.
It is interesting to note that the MSR and MSG genes in the shared region were identical, and most of the second PRT1 gene very similar, to the sequence of a previously described MSG array (GenBank Accession number D31909). This match may reflect a propensity of this gene cluster to be copied into different sites in the genome.
These data establish that repeated gene arrays in P. carinii can be quite different from one another. The gene clusters in cosmids 3G5 and 1B2 each contained members of all three gene families, but the order of the genes was different. In addition to full size MSG and MSR genes, both gene clusters contained what appear to be fragments of MSG or MSR genes. These data also imply that a gene array can move, as has been previously suspected based on the heterogeneous nature of the DNA downstream of the locus that controls transcription of MSG genes.
ACKNOWLEDGEMENTS
Supported by the following grants the NIH: R01AI36701, R01AI44651, and AIDS/FIRCA TW01200–02, and by a grant from The Wellcome Trust.




