Effect of wine phenolics on cytokine-induced C-reactive protein expression
Abstract
Summary. Background: Elevation of C-reactive protein (CRP) levels in blood was recognized as one of the cardiac disease risk factors. Consumption of wine is shown to reduce the risk from heart disease and improve longevity. Objectives: In the present study, we evaluated the effect of various wine polyphenolic compounds and several active synthetic derivatives of resveratrol on the inflammatory cytokines (IL-1ß + IL-6)-induced CRP expression in Hep3B cells. Results: Among the wine phenolics tested, quercetin and resveratrol, in a dose-dependent manner, suppressed cytokine-induced CRP expression. Two of the synthetic derivatives of resveratrol, R3 and 7b, elicited a fiftyfold higher suppressive effect compared with resveratrol. The inhibitory effects of resveratrol and its derivatives on CRP expression were at the level of mRNA production. Investigation of signaling pathways showed that the cytokines induced the phosphorylation of p38 and p44/42 MAP kinases. Inhibitors of p38 and p44/42 mitogen-activated protein kinase (MAPK) activation inhibited CRP expression, implicating the involvement of both pathways in cytokine-induced CRP expression. These data revealed a previously unrecognized role of the p44/42 MAPK signaling pathway in CRP expression. Wine polyphenolics or the synthetic compounds of resveratrol did not affect cytokine-activated phosphorylation of these MAPKs. Conclusions: Wine phenolics inhibit CRP expression; however, to do so, they do not utilize the MAPK pathways.
Introduction
C-reactive protein (CRP) is an acute phase protein whose concentration also increases under chronic inflammatory conditions [1]. Epidemiological studies suggest that CRP is both a marker of and a causal agent in the development of atherosclerosis [2–5]. Although it is generally accepted that CRP is a well-proven clinical marker of increased cardiovascular risk, still it is debated whether CRP is a causal agent in atherogenesis [6,7]. Nonetheless, an accumulating evidence from in vitro and in vivo studies strongly suggest that CRP acts as a proatherogenic factor and promotes atherothrombosis [8,9]. CRP is shown to promote endothelial cell activation and dysfunction [10,11], affect vascular smooth muscle cell migration and proliferation [7,12,13], induce changes in matrix biology [14], and promote coagulation [15]. If CRP plays a role in pathogenesis of atherosclerosis, then the blockade of CRP synthesis or its actions would be beneficial in inhibiting the development of atherosclerosis.
Overwhelming epidemiological evidence suggests that moderate consumption of alcoholic beverages, particularly red wine, lowers mortality rates from coronary heart diseases [16–21]. Cardiovascular benefits associated with moderate wine consumption have been thought to stem, at least partly, from antioxidant [22–24], anti-inflammatory [25–27], antiplatelet [28–30] and anticoagulant [31,32] activities of wine phenolics, particularly resveratrol. Resveratrol is shown to mimic calorie restriction by stimulating Sir2 (sirtuin 2, a histonedeacetylase), increasing DNA stability and extending lifespan of yeast by 70% [33]. Recent studies showed that resveratrol improves health and survival of mice in a high calorie diet by producing changes associated with longer lifespan, such as increased insulin sensitivity, reduced insulin-like growth factor-1 levels, and increased mitochondrial numbers [34].
Recent epidemiological studies found an inverse/U-shaped relation between alcoholic beverage consumption and plasma concentration of CRP expression [35]. In age-adjusted analyses, wine consumption appears to be more effective in reducing CRP levels compared with other alcoholic beverages. However, this difference disappeared when BMI was taken into account. In the present study, we investigated the effect of wine phenolics on cytokine-induced CRP expression in a cell model system. Further, we also tested whether chemically-modified derivatives of resveratrol could inhibit cytokine-induced CRP expression much more effectively than resveratrol. CRP is produced primarily in liver [36] and can be experimentally induced in human hepatoma Hep3B cells by treatment with proinflammatory cytokines [37]. Therefore, cytokine-induced CRP expression in Hep3B cells was chosen as a model system to investigate the effect of wine phenolics.
Materials and methods
Reagents
Wine phenolics (resveratrol, quercetin, rutin, catechin and epicatechin) and 2,2’-azino-bis(3-ethyl) benzthiazoline-6-sulfonic acid were obtained from Sigma (St Louis, MO, USA). Resveratrol derivatives were synthesized as described earlier [38,39] and were a kind gift from Dr M. Roberti, Università di Bologna, Italy, and Dr F. Raul, Laboratory of Nutritional Cancer Prevention, Strasbourg, France. They were stored as lyophilized powders in dark glass vials wrapped with aluminum foil until they were reconstituted on the day of the experiment. IL-1ß and IL-6 were obtained from R&D Systems (Minneapolis, MN, USA) and chemiluminescence reagent was from PerkinElmer Life Sciences Inc. (Boston, MA, USA). Cell culture media and reagents were obtained from Invitrogen (Carlsbad, CA, USA). Phosphospecific antibodies and relevant control antibodies were from Cell Signaling Technology (Beverly, MA, USA). Polyclonal rabbit antihuman CRP antibodies were obtained from Sigma and monoclonal anti-CRP (HD 2.4) was from ATCC (Rockville, MD, USA).
Cell culture
Hep3B cells were obtained from ATCC (Rockville, MD, USA) and cultured to confluence at 37 °C under 5% CO2 in DMEM supplemented with 10% heat-inactivated FBS, 1% penicillin/streptomycin and glutamine.
CRP induction and treatment with phenolic compounds
Monolayers of Hep3B cells were serum-starved overnight by replacing the serum-containing medium with serum-free DMEM. The medium was replaced with fresh serum-free medium and stabilized the cells for 2 h before starting experimental treatments. Hep3B cells were pretreated with wine phenolics, resveratrol derivatives or control vehicle for 1 h before the cells were stimulated with IL-1ß (10 ng mL−1) and IL-6 (20 ng mL−1) for 24 h to induce CRP expression. All wine phenolics and resveratrol derivatives were dissolved in DMSO before they were diluted in serum-free medium to treat the cells. An equal amount of DMSO was added to serum-free medium to serve as a control vehicle. The final concentration of the solvent (DMSO) in control and experimental groups was 0.1%.
Measurement of CRP levels in ELISA
Secreted CRP levels in conditioned media were quantified using a sandwich ELISA. Briefly, a 96-well plate was coated with 100 μl of polyclonal rabbit antihuman CRP antibody (1:1000) in TBS (10 mm Tris–HCl, 150 mm NaCl, pH 7.2) overnight at 4 °C, and free binding sites were blocked with G-block (10 mm Tris–HCl, 150 mm NaCl, pH 7.2 and 0.5% gelatin) for 3 h, and the plates were washed thrice with TBS. Cell supernatants (100 μL, concentrated 10 times or more prior to the assay by the ultrafiltration centrifugation method using 10 000 Da mol. wt. cut-off membrane filter assembly) were added to the well. After 4 h of incubation, the unbound material was removed, the wells were washed twice with G-ELISA buffer (10 mm Tris–HCl, 150 mm NaCl, pH 7.2, 0.1% gelatin and 0.02% Tween-20) and then incubated with anti-CRP monoclonal antibody (100 μL of 0.5 μg mL−1) for 1 h, followed by HRP-labeled goat antimouse IgG (diluted 1:000 in G-ELISA buffer, 100 μL) for 1 h. After removing the unbound material and washing the wells, HRP substrate 2,2’-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (15 mg mL−1 in 0.1 m citrate buffer, pH 4.0 containing 0.001% v/v of 30% H2O2) was added to the wells and the color developed after 30 min was measured at 405 nm using a microplate reader (Molecular Devices, Sunnyvale, CA, USA). Known quantities of recombinant CRP were employed in the assay to construct a standard curve, from which absorbance readings were converted to ng mL−1 CRP.
Immunoblotting
For detecting CRP, conditioned media were concentrated tenfold using ultrafiltration with 10 000 mol. wt. cut-off filter membrane before they were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE). For others, cell extracts were prepared by solubilizing monolayers of Hep3B cells cultured in six-well plates in 70 μL of cell lysis buffer (50 mm Tris–HCl, pH 7.4; 150 mm NaCl; 1 mm EDTA; 1% Triton-X 100; 1 mm Na3VO4; 2.5 mm NaPPi; 2 mg mL−1 leupeptin and aprotinin; 1 mm PMSF and 15% glycerol). Twenty μl of samples were subjected to SDS–PAGE (12% polyacrylamide gels), followed by immunoblot analysis with appropriate antibodies.
Northern blot analysis
Total RNA was extracted with Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Total RNA (30 μg) was subjected to gel electrophoresis in 1.1% agarose/6% formaldehyde gels and transferred onto the nitrocellulose membrane by a capillary blot method. Northern blots were prehybridized at 42 °C with a solution containing 50% formamide in 5 × SSC (50 mm Tris–HCl, pH 7.5, 0.1% sodium pyrophosphate, 1% SDS, 1% polyvinylpyrrolidone, 1% Ficoll, 25 mm EDTA, 100 μg mL−1 denatured salmon sperm DNA, and 1% BSA), followed by hybridization with 32P-labeled CRP cDNA probe. The hybridized membrane was exposed to X-ray film. To quantify the hybridization signal intensities, the blots were exposed to phosphor screens, and the screens were read in a phosphor imager (Molecular Imager, Bio-Rad, Richmond, CA, USA).
Transient transfection and luciferase assay
Hep3B cells seeded in 12-well plates (50 000 cells well−1) were cultured overnight in growth medium, and then transferred to serum-free DMEM (500 μL). Cells were transfected with –157/+9 CRP-Luc promoter construct [40], and 6 h after the transfection an additional 500 μl of serum-free medium was added. The transfected cells were subjected to pretreatment with experimental compounds for 1 h, followed by cytokine treatment for 24 h. The cells were washed once with PBS and lyzed by adding reporter lysis buffer (50 μL) and subjecting them to two cycles of freeze and thawing. Luciferase activity in cell lysates was measured by taking 20 μL of cell lysate and adding 100 μL of luciferase reagent, and measuring the light intensity in a luminometer. The values were normalized to the protein content of cell lysates.
Results
Earlier studies showed that a combination of IL-1ß and IL-6 treatment was necessary to induce CRP maximally in Hep3B cells [37]. Consistent with these data, we found that the combination of IL-1ß and IL-6 treatment increased CRP expression in Hep3B cells by 5- to thirtyfold over unstimulated cells (a much wider difference in fold CRP induction among various experiments reflects variations in very low basal levels of CRP expression in unstimulated cells).
Effect of wine phenolics on cytokine-induced CRP expression
Catechin, epicatechin, rutin, quercetin and resveratrol are major and active components of wine phenolics in red wines [41]. Therefore, we examined the effect of these compounds on IL-1ß and IL-6-induced CRP expression in Hep3B cells. As shown in Fig. 1A, when tested at a concentration of 50 μm, resveratrol and quercetin, and not other wine phenolics, inhibited cytokine-induced CRP expression. Additional studies showed that quercetin and resveratrol lowered CRP expression levels in Hep3B cells in a dose-dependent manner. Both the compounds appeared to have a similar potency in suppressing the cytokine-induced CRP expression (Fig. 1B,C). For 50% inhibition of CRP expression, 25–50 μm of quercetin or resveratrol was required.
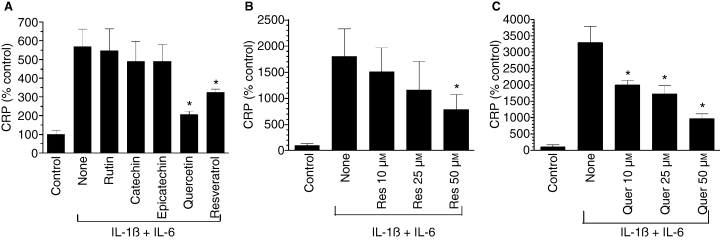
Effect of some major wine compounds on cytokine (IL-1ß + IL-6)-induced C-reactive protein (CRP) expression. Hep3B cells were pretreated for 1 h with various wine polyphenolics (50 μm) (panel A), varying doses of resveratrol (panel B) or quercetin (panel C), and then stimulated for 24 h with a combination of IL-1ß (10 ng mL−1) and IL-6 (10 ng mL−1). At the end of 24 h, the conditioned media were collected, concentrated and CRP levels were measured in ELISA. *Denotes that the CRP level in cells treated with the experimental compound was significantly lower compared with CRP levels in stimulated cells that were exposed to a control vehicle.
Synthetic derivatives of resveratrol are potent inhibitors of CRP expression
A number of cis- and trans- stilbene-based resveratrol derivatives were synthesized with the aim of improving the effectiveness of resveratrol in modulating various cellular activities [38,39,42]. Among them, a cis- form of methyl derivative of resveratrol (R3) [38] and resveratrol analogs bearing the 3,5-dimethoxy motif at the A phenyl ring with amino, methoxy and hydroxyl moieties at the 3′- and 4′-positions (7b, 8b, 11b and 12b) [39] were found to be potent analogs of resveratrol. Therefore, we examined the effect of these compounds on cytokine-induced CRP expression. R3 and 7b compounds appeared to be approximately fiftyfold more effective than resveratrol in suppressing the CRP expression (i.e. 1 μm of these compounds was sufficient to inhibit CRP expression by 50%) (Fig. 2A,B). Other derivatives were 2- to 5-fold more effective than resveratrol in suppressing CRP expression (Fig. 2C–E).
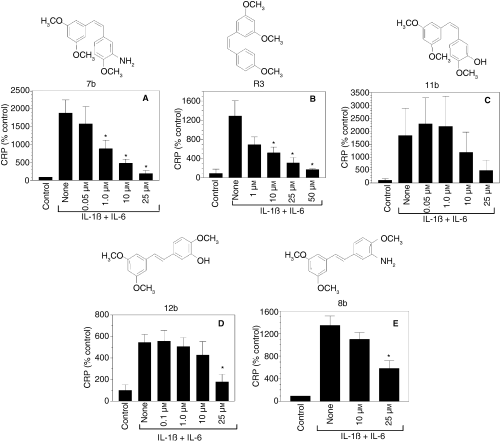
Effect of various chemically synthesized resveratrol derivatives on C-reactive protein (CRP) expression. Hep3B cells were pretreated for 1 h with varying concentrations of resveratrol derivatives and then stimulated for 24 h with a combination of IL-1ß and IL-6. At the end of 24 h, the conditioned media were collected, concentrated and CRP levels were measured in ELISA. *Denotes that the CRP level in cells treated with the experimental compound was significantly lower compared with CRP levels in stimulated cells that were exposed to a control vehicle.
Quercetin, resveratrol and resveratrol derivatives suppress CRP mRNA expression
To determine whether suppression of CRP expression in cytokine-stimulated cells by wine phenolics is at the transcriptional level, we investigated the effect of these compounds on CRP mRNA expression. Earlier studies showed that IL-6 + IL-1ß increased CRP mRNA accumulation in Hep3B cells maximally at 24 h and the inclusion of dexamethasone (1 μm) to the culture medium increased the transcription of endogenous CRP gene, which may be essential in detecting the cytokine-induced CRP mRNA accumulation by Northern blot analysis [43]. Therefore, Hep3B cells were stimulated with IL-1ß and IL-6 for 24 h in the presence of dexamethasone to induce CRP mRNA. The cells were pretreated with a control vehicle, quercetin, resveratrol or its derivatives before they were subjected to cytokine treatment. CRP mRNA was undetectable in unstimulated cells. Cytokine treatment resulted in readily detectable CRP mRNA levels (Fig. 3A). Resveratrol and quercetin (50 μm) completely suppressed the cytokine-induced CRP mRNA accumulation. R3 and 7b compounds, at much lower concentrations, inhibited cytokine-induced CRP mRNA accumulation. Consistent with the observations of Northern blots and CRP ELISA, immunoblot analysis also showed a marked decrease in CRP protein secreted from cells treated with quercetin, resveratrol and its derivatives prior to stimulation with cytokines (Fig. 3B). It may be pertinent to note that total CRP synthesized in control and experimental groups is fully secreted because we found no CRP band on immunoblot analysis of cell extracts (data not shown).
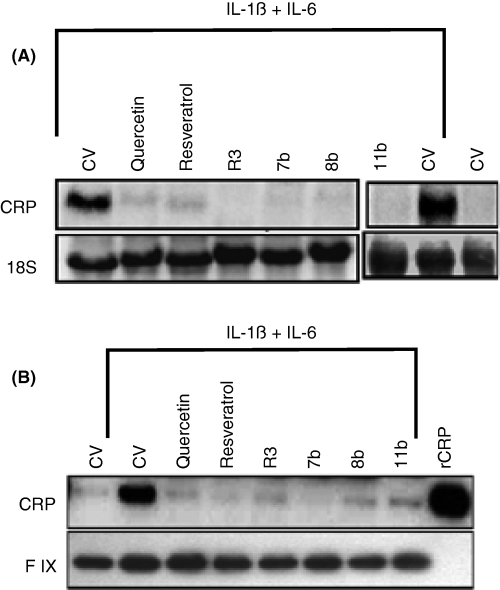
Quercetin, resveratrol and its derivatives suppress C-reactive protein (CRP) mRNA and protein synthesis. Hep3B cells were subjected to 1 h pretreatment with a control vehicle, resveratrol (50 μm), quercetin (50 μm), R3 (1 μm), 7b (1 μm), 8b (25 μm) or 11b (25 μm) in the presence of dexamethasone (1 μm). Then, the cells were stimulated with IL-1ß (10 ng mL−1) and IL-6 (20 ng mL−1) for 24 h. The conditioned media were removed, concentrated and subjected to immunoblot analysis with anti-CRP antibodies (top panel B) and antifactor IX to serve as a loading control (bottom panel B). Ten ng of recombinant CRP (rCRP) protein was loaded in the last lane. The cells were extracted with Trizol reagent to obtain total RNA, and 30 μg total RNA was subjected to Northern blot analysis and probed with 32P-labeled CRP cDNA (top panel A). The 18S RNA band of the same gel was shown for loading control (bottom panel A). CV denotes control vehicle.
Next, we examined whether these compounds could inhibit the cytokine-induced transcriptional activation of the CRP promoter. For these studies, Hep3B cells were transfected with a CRP promoter construct linked to a luciferase reporter (-157 CRP-LUC) that was shown to respond to IL-6 + IL-1ß treatment [40], and the transfected cells were pretreated with quercetin, resveratrol and resveratrol derivatives for 1 h before they were stimulated with IL-6 and IL-1ß. As expected, stimulation of the transfected cells with cytokines increased CRP promoter activity by 4- to 5-fold (Fig. 4). Pretreatment of the cells with quercetin, resveratrol and resveratrol derivatives inhibited the promoter activity by about 30 to 60%, indicating that the inhibitory actions of these compounds were mediated through the 157 bp of the CRP promoter.
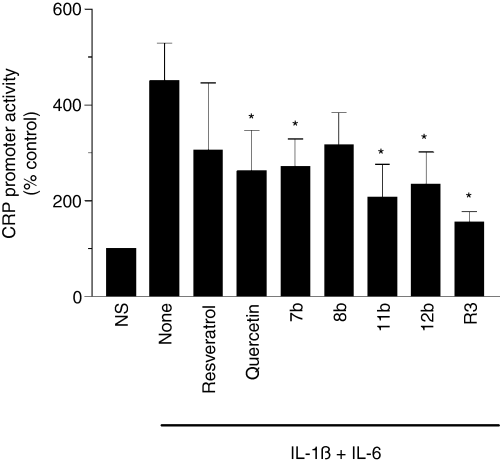
Wine phenolics inhibit transcriptional activation of C-reactive protein (CRP) promoter. Hep3B cells were transiently transfected with –157/+9 CRP promoter region linked to luciferase reporter gene. After 6 h, the transfected cells were treated with various phenolic compounds for 1 h, and then stimulated with IL-1ß and IL-6 for 24 h. The cells were extracted with reporter lysis buffer and the cell extracts were used for measurement of luciferase activity. The concentration of wine phenolics used in this study was same as in Fig. 3. NS denotes non-stimulated cells. *Denotes that the CRP promoter activity is significantly lower compared with stimulated cells that were exposed to a control vehicle.
Effect of wine phenolics on activation of signaling pathways that contribute to cytokine-induced CRP expression
To investigate potential mechanisms by which the wine phenolics may suppress CRP expression, we first evaluated the effect of various signaling pathway inhibitors on IL-6 + IL-1ß-induced CRP expression to identify pathways responsible for CRP expression in our model system. As shown in Fig. 5, the specific inhibitors of p38 and p44/42 MAPK pathways abolished cytokine-induced CRP expression. MG132, the proteosome inhibitor that often used to inhibit the NF-κB activation pathway, and curcumin, which inhibits NF-κB and PKC pathways, failed to suppress cytokine-induced CRP expression. Similarly, EGFR inhibitor also had no effect on the cytokine-induced CRP expression. Because earlier studies showed the involvement of STAT3 in CRP induction [44] and STAT3 is known to be regulated by IL-6 and other cytokines [45], we next evaluated the activation of STAT3. As shown in Fig. 6A, the cytokine treatment induced the phosphorylation of STAT3 in a time-dependent manner in Hep3B cells. Next, we investigated the effect of quercetin, resveratrol and its derivatives, in concentrations that were shown to inhibit CRP expression, on cytokine-induced activation of STAT3. None of the compounds inhibited the phosphorylation of STAT3 (Fig. 6B), indicating that the inhibition of STAT3 activation is an unlikely mechanism by which wine phenolics exert their suppressive effect on cytokine-induced CRP expression.
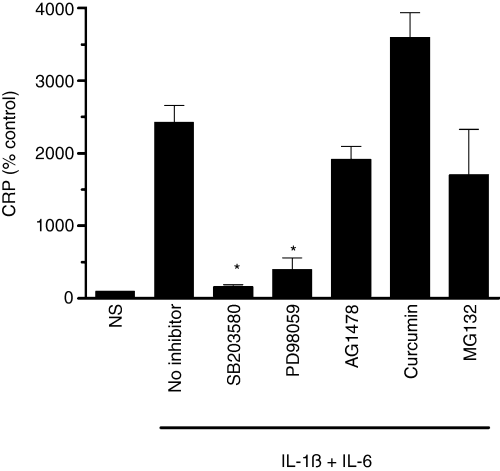
Effect of various signaling pathway inhibitors on cytokine-induced C-reactive protein (CRP) expression. Hep3B cells were pretreated with a control vehicle; p38 MAPK inhibitor, SB203580 (10 μm); p44/42 MAPK inhibitor, PD98059 (10 μm); EGFR inhibitor, AG1478 (10 μm); NF-κB inhibitors, curcumin (30 μm) and MG132 (1 μm); for 1 h and then stimulated with IL-1ß and IL-6 for 24 h. The conditioned media was used to measure CRP levels. NS denotes non-stimulated cells. *Denotes that the CRP levels were significantly lower compared with stimulated cells not exposed to inhibitors.
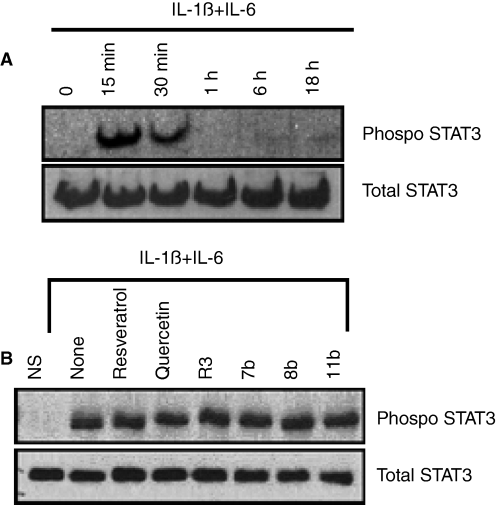
Wine phenolics do not suppress cytokine-induced STAT3 phosphorylation. Panel A: Hep3B cells were treated with IL-1ß and IL-6 for varying time periods, and cell extracts harvested at the end of the treatment period were subjected to immunoblot analysis to evaluate STAT3 phosphorylation. Panel B: Hep3B cells were pretreated with quercetin, resveratrol or resveratrol derivatives for 1 h, and then treated with IL-1ß and IL-6 for 15 min to activate STAT3. STAT3 phosphorylation was evaluated as in panel A. The concentration of wine phenolics used in this study was the same as in Fig. 3. NS denotes non-stimulated cells.
Next, we investigated the possible contribution of p38 and p44/42 MAPK pathways to cytokine-induced CRP expression. Treatment of Hep3B cells with IL-6 and IL-1ß induced the activation of p38 and p44/42 MAPK pathways (Fig. 7A,B) and the inhibition of these pathways with specific inhibitors abolished IL-6 + IL-1ß-induced CRP expression (Fig. 5). The involvement of p38 MAPK in CRP expression was reported recently [46]; however, the involvement of p44/42 MAPK in cytokine-induced CRP expression was not known. Next, we examined the effect of wine phenolics on cytokine-induced p38 and p44/22 MAPK activation. As shown in Fig. 7C,D, none of the compounds inhibited cytokine-induced activation of p38 and p44/42 MAPK.
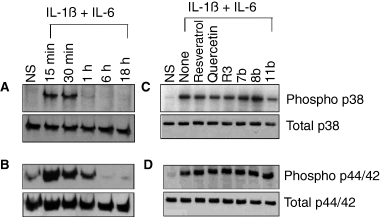
Effect of resveratrol and its derivatives on cytokine-induced activation of p38 and p44/42 MAPK. Hep3B cells were treated with IL-1ß and IL-6 for varying time periods, and cell extracts harvested at the end of the treatment period were subjected to immunoblot analysis to evaluate p38 (Panel A) or p44/42 MAPK (panel B) activation. Panels C and D: Hep3B cells were pretreated with quercetin, resveratrol or resveratrol derivatives for 1 h, and then treated with IL-1ß and IL-6 for 15 min, and the cell extracts were analyzed for p38 (panel C) and p44/42 (panel D) MAPK activation. The concentration of wine phenolics used in this study was the same as in Fig. 3.
Discussion
Resveratrol, a constituent of red wine, which has long been suspected to have cardioprotective effects [20,22,47], has received much attention in recent years for its therapeutic potential for many diseases and potential for prolonging the lifespan [48]. Although the mechanism by which resveratrol exerts a wide range of beneficial effects across disease models is unclear, multiple direct targets for this compound have been identified [48–53]. The protective effects of resveratrol against heart diseases are thought to stem from its ability to decrease platelet aggregation [29,54], promote vasorelaxation [55,56], reduce lipid peroxidation [22,24], improve serum cholesterol and triglyceride concentrations [57], and reduce procoagulant tissue factor [31,58]. The beneficial effects of red wine are possibly not only mediated through the phenolics, but also ethanol, as it was shown to modulate various biological activities including inhibition of platelet aggregation [59] and tissue factor expression in monocytes [60]. In the present study, we identified an additional potential mechanism by which resveratrol and other wine phenolics could provide protection against atherosclerosis. The data presented herein show that wine phenolics (resveratrol and quercetin) suppress the expression of CRP, a protein that is believed to play a key role in the pathogenesis of atherosclerosis [8].
A half maximal concentration of resveratrol and quercetin required to inhibit cytokine-induced expression of CRP in the present study is similar to the reported concentration of these compounds required to modulate various biological activities in in vitro studies [29–31,61]. Based on in vivo pharmacokinetics of these compounds [48,53], it is unlikely that such high concentrations of resveratrol and quercetin are ever achieved in vivo either after daily intake of two glasses of wine or even after administration of pharmacological doses. However, despite skepticism concerning the bioavailability, a growing body of recent in vivo studies provides evidence that resveratrol has protective effects in rodent models of stress and disease [48]. It has been shown that resveratrol or its metabolites may accumulate at much higher concentrations in tissues than in blood [62,63]. This, coupled with potential synergistic interaction of resveratrol and quercetin with other constituents of the diet, may explain how a relatively low dose of resveratrol obtained from red wine could produce measurable health benefits [48]. In this context, it may be pertinent to note that quercetin, whose concentration in red wine is 10 or more times higher than resveratrol, also suppressed CRP expression as effectively as resveratrol. This suggests that quercetin may be the principal active ingredient in red wine that is capable of suppressing CRP induction.
Despite initial doubts regarding the physiological relevance of in vitro studies in which typically high concentrations of resveratrol were used, recent studies suggest the therapeutic potential for resveratrol [34,48]. This stirred interest in developing more potent resveratrol mimetics. Chemical structural alterations of the stilbene motif of resveratrol were shown to yield extremely effective analogs [38,39,64]. A cis- form of methyl derivative of resveratrol (R3) was shown to be hundredfold more potent than resveratrol in inhibiting cell proliferation by arresting the cell cycle at the G2/M phase transition in Caco-2 cells [38]. Similarly, resveratrol analogs bearing the 3,5-dimethoxy motif at the A phenyl ring with amino, methoxy and hydroxyl moieties at the 3′- and 4′-positions (7b, 8b, 11b, 12b and 12c) were found to be more effective than resveratrol as apoptosis-inducing agents [39]. In our recent studies, these derivatives were shown to be 2- to twenty-fivefold more effective, compared with resveratrol, in inhibiting TF expression in monocytes [65]. In the present study we found that a number of these derivatives, particularly R3 and 7b, suppressed CRP induction in Hep3B cells very effectively when compared with resveratrol. For example, 50 μm of resveratrol inhibited CRP induction by about 50%, whereas a similar inhibition was attained with 1 μm of R3 and 7b compounds. Thus, resveratrol derivatives may have potential in becoming viable therapeutic agents in the future. It would be interesting to examine whether quercetin derivatives would have much higher inhibitory activity than the derivatives of resveratrol, but unavailability of chemically synthesized quercetin derivatives prevented us from investigating this possibility.
Quercetin, resveratrol or its derivatives suppressed the induction of CRP expression primarily by inhibiting transcriptional activation of the CRP gene. Suppression of CRP antigen levels in cells treated with these compounds is associated with the lower accumulation of CRP mRNA. Consistent with these data, these phenolic compounds were shown to inhibit cytokine-induced CRP promoter activity.
Our data provide additional information on the mechanism of transcriptional induction of IL-1ß and IL-6-induced CRP expression in Hep3B cells. Previously, it has been shown that the transcription factors C/EBPß [66–68], STAT3 [69,70] and NF-κB [40,44,71] participate in IL-1ß and IL-6-induced CRP expression. It was shown that overexpressed NF-κB induced CRP expression [44,71]; however, it was not clear whether IL-1β worked through the activation of the NF-κB pathway in synergizing the effects of IL-6 on CRP expression [71]. Mutation of the κB site on the CRP proximal promoter did not abolish the synergistic effect of IL-1ß on IL-6 [71]. Our results showing that the inhibitors of the NF-κB pathway failed to suppress IL-6 and IL-1ß-induced-CRP expression support the view that the sole mechanism of IL-1ß action on the CRP promoter is not just NF-κB. However, it is possible that polyphenols may be inhibiting CRP transcription partially through the short promoter by perturbing the interaction of NF-κB and C/EBPß on the short promoter as NF-κB was shown to act synergistically with C/EBPß on the 157 bp CRP proximal promoter region to regulate CRP gene expression [71]. Here it may pertinent to note that wine phenolics inhibited only partly the cytokine-induced 157 bp promoter activity whereas they completely attenuated the cytokine-induced CRP mRNA and protein expression. These data suggest that that polyphenols may additionally regulate CRP expression through the promoter regions located outside the 157 bp promoter. In addition to the known transcription factors acting on the CRP proximal promoter, it has been recently reported that the p38 MAPK pathway also contributes to cytokine-induced CRP expression [46]. Our studies reported herein reveal that IL-6 and 1 L-1ß-induced expression of CRP in Hep3B cells also involves the participation of p44/42 MAPK activation. IL-6 and IL-1ß treatment induced the activation of p38 and p44/42 MAPK, and the specific inhibitors of p38 and p44/42 MAPK fully attenuated the cytokine-induced expression of CRP.
Surprisingly, we found that none of these mechanisms known to participate in inducing CRP expression was involved in the inhibition of CRP expression by resveratrol and its derivatives. Two other substances, nitric oxide donors and cholesterol-lowering agents statins, have also been reported to inhibit cytokine-induced CRP expression [70]. Probably, the mechanism of inhibition of CRP expression by these substances also did not involve the known pathways required for the induction of CRP expression. Combined data indicate that there is a common pathway, as yet unidentified, that mediates the inhibition of cytokine-induced CRP expression by nitric oxide, statins, resveratrol and its derivatives.
In summary, the data presented in the manuscript provide convincing evidence that resveratrol and quercetin suppress cytokine-induced CRP expression and illustrate the feasibility of developing effective drugs to suppress aberrant expression of CRP. As these drugs not only suppress CRP but many other mediators that contribute to atherosclerosis, they will have a good potential to become an effective treatment for atherosclerosis.
Acknowledgements
This work was supported primarily by a grant from the American Heart Association (Texas Affiliate), grant number 0355121Y, and partially by grants from the National Institute of Health, HL65500 and HL58869. The authors would like to thank M. Kaleemuddin for her technical help.
Disclosure of Conflict of Interests
The authors state that they have no conflict of interest.




