ISOLATION AND SEQUENCE ANALYSIS OF A cDNA ENCODING A NOVEL PUTATIVE ESTERASE FROM THE MARINE MICROALGA ISOCHRYSIS GALBANA (PRYMNESIOPHYCEAE, HAPTOPHYTA)1
Received 3 April 2009. Accepted 28 January 2010.
Abstract
Microalgae constitute an interesting novel study area for characterizing new esterases, and so we decided to isolate a complete cDNA encoding a new putative microalgal esterase from the haptophyte Isochrysis galbana Parke. Rapid amplifications of both the 5′ and 3′ cDNA ends (RACE) were performed with specific primers, designed using an incomplete candidate gene from the I. galbana expressed sequence tag (EST) database. The full-length cDNA obtained was designated IgEst1. The coding sequence was 828 bp long, and the deduced amino acid sequence revealed a polypeptide of 275 amino acids with a predicted signal peptide of 23 residues in the N-terminal region. The following 252 amino acids formed, after in silico analysis, a mature protein with a molecular mass of ∼26.92 kDa and had a theoretical pI of 5.87. Alignment analyses revealed slight but significant identity and similarity with carboxylesterases, phospholipases, and lysophospholipases from various organisms including fungi, plants, and animals. The new sequence IgEst1 enclosed the catalytic triad Ser/Asp/His and the consensus pentapeptide Gly-X-Ser-X-Gly, two highly conserved patterns found in serine hydrolases. Phylogenetic analyses established a close relationship with putative esterases identified in microalgae genomes.
Abbreviations:
-
- DHA
-
- docosahexaenoic acid
-
- EPA
-
- eicosapentaenoic acid
-
- IgEst1
-
- Isochrysis galbana esterase number 1
-
- LC-PUFA
-
- long-chain polyunsaturated fatty acids
-
- ORF
-
- open reading frame
-
- RACE
-
- rapid amplification of cDNA ends
Lipolytic enzymes, such as lipases, carboxylesterases, and phospholipases, are carboxylic ester hydrolases (E.C. 3.1.1) involved in lipid metabolism that catalyze lipid hydrolysis and synthesis. They constitute biotechnological tools that are widely used in research and in various industrial applications due to various advantageous features, such as stability, broad substrate specificity, high stereoselectivity, regioselectivity, and the fact that they do not require cofactors (Bornscheuer 2002, Jaeger and Eggert 2002, Hasan et al. 2006). Therefore, many efforts are being made to screen, isolate, and fully characterize new lipolytic enzymes from animals, plants, fungi, and bacteria.
Very few studies have reported lipolytic activity in microalgae that would suggest the presence of lipolytic enzymes in these organisms (Bilinski et al. 1968, Antia et al. 1970, Pohnert 2002). Moreover, as far as we are aware, no ester hydrolase from either a microalga or a macroalga has yet been isolated and described. However, some observations suggest that microalgae could constitute an interesting source of new lipolytic enzymes with attractive specificities.
Eukaryotic microalgae are photosynthetic microorganisms living in the ocean or in freshwater. The group covers several phylogenetic lineages with different evolutionary histories. Some divisions would seem to date back to primary, secondary, or tertiary endosymbiosis events associated with gene loss and transfer between hosts and symbionts (Delwiche 1999, Keeling et al. 2004, Li et al. 2006). Microalgal evolution may also be closely related to the horizontal movement of genetic material from other unrelated species (Andersson 2005, Nosenko and Bhattacharya 2007). Consequently, microalgae might well be a source of innovative genomes, genes, and proteins.
Microalgal lipid and fatty acid composition are also unconventional. Many studies have demonstrated unexpectedly high intracellular level of lipids in microalgae. Some species can store significant amounts of lipids, corresponding to up to 86% of their total cell dry weight, as neutral lipids, particularly in response to environmental changes (Murphy 2001). Other species also contain large amounts of long-chain polyunsaturated fatty acids (LC-PUFAs) belonging to the omega-3 family, such as docosahexaenoic acid (DHA, C22:6) and eicosapentaenoic acid (EPA, C20:5) (Poisson et al. 2002, Pencreac’h et al. 2004, Guschina and Harwood 2006).
This study reports, for the first time, the isolation and comparative and phylogenetic analyses of a complete cDNA encoding a putative esterase from I. galbana, a marine microalga belonging to the haptophyta. I. galbana is known to produce large amounts of DHA and is currently used in aquaculture to feed juvenile fish and crustaceans, as well as bivalve larvae in mollusk hatcheries. Previous work in our laboratory has shown that DHA constitutes 50% of the phospholipid fatty acids in this species, which makes membrane lipids from I. galbana an interesting, natural high-omega-3-PUFA lipid fraction with potential nutritive and therapeutic value (Poisson and Ergan 2001, Devos et al. 2006).
Experiments were carried out with I. galbana (CCAP 927/1) obtained from the Culture Collection of Algae and Protozoa, Centre for Coastal and Marine Sciences (Dunstaffnage Marine Laboratory, Oban, Scotland, UK). This strain was grown on Provasoli 1/3 medium as previously described by Robert et al. (2004). Batch cultures were carried out in flasks and in bioreactors under the pH, temperature, and illumination conditions defined by Poisson and Ergan (2001).
The strategy employed to isolate the gene of interest from I. galbana was to use RACE with purified mRNA. To design the specific internal primers required for RACE experiments, we consulted the I. galbana EST database (http://tbestdb.bcm.umontreal.ca/searches/organism.php?orgID=IS) and selected the EST numbered ISE00013401 because it exhibited significant similarities with lipolytic enzymes from various organisms. The selected EST was found to be incomplete and to correspond in fact to a segment of 577 bp without initiation and termination translation signals. To obtain full-length cDNA, 5′- and 3′-RACE were performed with the specific primers 5′-TTC ACA GCC TCC GGC GCC AGT ATG AAA-3′ and 5′-ATC CAC GGG CTG GGC GAC AGC AAC ATG-3′, respectively, using the SMARTTM RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA). Two major amplicons were obtained among unspecific amplifications and used as templates for a second nested amplification step. Specific primers 5′-AGG GTG TGT GAA TAC TGC AGC CCC TGT GAA GA-3′ and 5′-AAC GGC GGA ATG TCG ATG CCC AGT TGG TAT GA-3′ were used for the 5′ and 3′ nested amplifications, respectively. This second-round PCR generated single products of 696 and 964 bp, corresponding to the 5′ and 3′ cDNA ends, respectively. Both PCR products were sequenced (MillGen, Labège, France), and the complete cDNA sequence was reconstituted by overlapping the 5′ and 3′ sequences confirming the presence of a single gene. Finally, new primers designed on the basis of the coding sequence extremities were used to isolate the full-length coding sequence in a single amplification step. Restriction sites EcoRI and XbaI (underlined) were added to forward 5′-GGAATTCC ATG GCG CGA GGG CTT GCC GCG TGC A-3′ and reverse 5′-GCTCTAGAGC TCA GGC GTC CTC TGG CAG TCT GGC CT-3′ primers, respectively, for future cloning. The PCR product was purified and sequenced in forward and reverse directions.
Concerning the sequence analysis, signal peptides were predicted using the SignalP 3.0 server (Bendtsen et al. 2004). The complete coding sequence was compared using BLASTX (Altschul et al. 1990) with sequences of GenBank, Swiss-Prot, and Protein Data Bank (PDB) databases. Only hits with an E-value <10−10 and >25% identities were considered. Best BLASTX hits were aligned with I. galbana deduced protein using the MUSCLE 3.5 program (Edgar 2004). Multiple alignments were edited with GeneDoc 2.7 (Nicholas et al. 1997). Automatic molecular modeling of the three-dimensional (3-D) structure was performed with the Geno3D server (Combet et al. 2002) based on available structures of hydrolases referenced in the PDB database. For phylogenetic analysis, the program GBlocks 0.91b (Castresana 2000) was used to extract poorly matched regions and generate informative blocks from a multiple alignment generated from 54 various species sequences. The phylogenetic analysis was performed using the neighbor-joining (NJ) method and 1,000 random replicates (bootstraps) using PHYLO_WIN (Galtier et al. 1996). The consensus tree diagram was edited with TreeView (Page 1996).
Complete gene isolation and sequence analysis. The full-length cDNA consisted of 1,124 nucleotides including an 828 bp open reading frame (ORF) flanked by untranslated 5′ and 3′ regions. The deduced amino acid sequence consisted of 275 amino acid residues, taking the first ATG codon as the initiation codon. The first 23 amino acid residues constituted a hydrophobic N-terminal region and corresponded to a signal peptide according to the prediction server SignalP 3.0. After cleavage of the signal peptide, the following 252 residues formed a putative protein with a molecular mass of ∼26.92 kDa and a calculated isoelectric point of 5.87. The cDNA sequence was deposited in the GenBank database under accession number AM748093. The new sequence was designated IgEst1 for I. galbana Esterase number 1.
Homology search. The sequence IgEst1 was used to look for equivalent sequences in public microalgae data banks. The accessibility of some microalgae genomes made it possible to compare the IgEst1 sequence with predicted protein sequences of six different microalgae species. Alignment of IgEst1 with algal sequences generated identity values in the range of 26%–29% with proteins similar to lysophospholipases from green and red microalgae (Fig. 1, Table 1). Higher scores (33%–35%) were observed with putative esterases from diatoms. The best identity score was obtained with a putative esterase from the haptophyte Emiliania huxleyi, which displayed 58% identical residues. The coccolithophorid E. huxleyi is a dominant marine calcifying phytoplankton species. Although I. galbana has unmineralized body scales rather than calcium carbonate coccoliths, these two species are in fact closely related. They share many similar ultrastructural features and have been assigned to the same taxonomic order, Isochrysidales (Fujiwara et al. 2001).
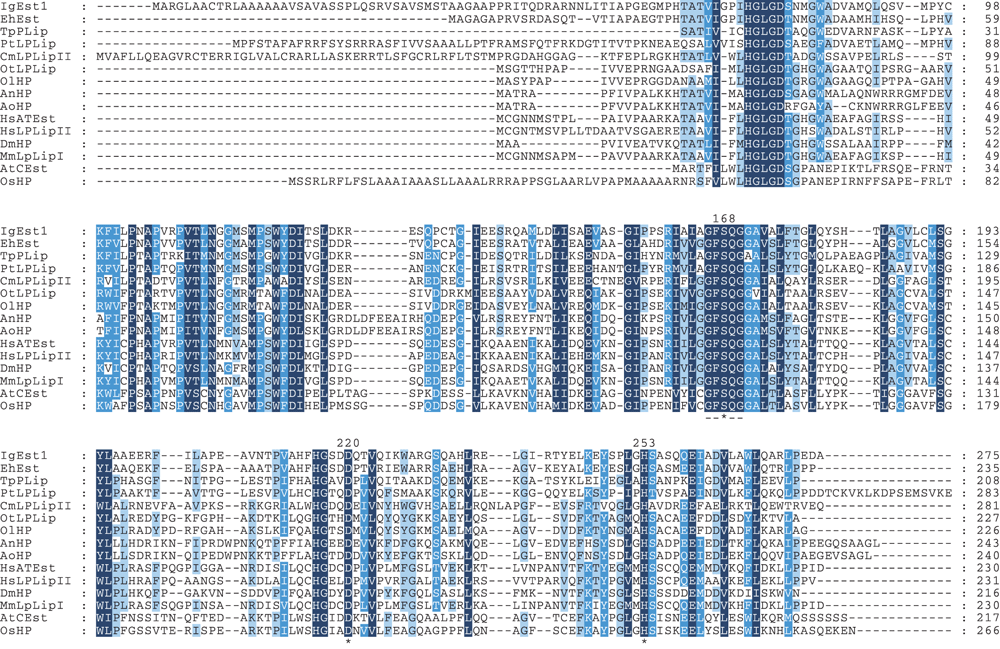
Amino acid sequence alignment of IgEst1 with other known or putative esterases from various organisms including insects, mammals, fungi, plants, and microalgae. Columns of identical residues are shaded. Three different shading levels indicative of the degree of conservation are differentiated. From dark to light shading, these levels correspond to 100% of conserved residues, 80% or more conserved, and 60% or more conserved. Unshaded columns have <60% conserved residues. The dashed line indicates the conserved region of the serine active site. Asterisks indicate the positions of the conserved residues Ser, Asp, and His forming the catalytic triad. Organism abbreviations: At, Arabidopsis thaliana; An, Aspergillus niger; Ao, Aspergillus oryzae; Cm, Cyanidioschyzon merolae; Dm, Drosophila melanogaster; Eh, Emiliania huxleyi; Hs, Homo sapiens; Mm, Mus musculus; Os, Oryza sativa; Ol, Ostreococcus lucimarinus; Ot, Ostreococcus tauri; Tp, Thalassiosira pseudonana; and Pt, Phaeodactylum tricornutum. Protein abbreviations: HP, hypothetical protein; LPLip (I, II), lysophospholipase (I, II); ATEst, acyl thioesterase; CEst, carboxylesterase; PLip, phospholipase; and Est, esterase.
| Organism rank | Organism species | Enzymea | GenBank access | % identityb | % similarityb |
|---|---|---|---|---|---|
| Fungi | Aspergillus niger | Hypothetical protein | XP_001395827 | 31 | 44 |
| Fungi | Aspergillus oryzae | Hypothetical protein | XP_001823920 | 31 | 44 |
| Mammalia | Mus musculus | Lysophospholipase Ic | P97823 | 30 | 44 |
| Mammalia | Homo sapiens | Acylthioesterase, lysophospholipase Ic | O75608, 1fj2 | 29 | 44 |
| Mammalia | Homo sapiens | Lysophospholipase II | AAC72844 | 28 | 44 |
| Insecta | Drosophila melanogaster | Hypothetical protein | AAG22322 | 28 | 46 |
| Plantae | Oryza sativa | Hypothetical protein | CAE02816 | 28 | 43 |
| Plantae | Arabidopsis thaliana | Carboxylesterase | NP_193961 | 24 | 38 |
| Green alga | Ostreococcus tauri | Lysophospholipase | CAL54716 | 29 | 45 |
| Green alga | Ostreococcus lucimarinus | Predicted protein | XP_001418794 | 26 | 44 |
| Red alga | Cyanidioschyzon merolae | Similar to lysophospholipase II | CMF176C d | 27 | 41 |
| Diatom | Phaeodactylum tricornutum | Lysophospholipase | 21816 d | 33 | 46 |
| Diatom | Thalassiosira pseudonana | Phospholipase | EED92715 | 35 | 49 |
| Haptophyta | Emiliania huxleyi | Esterase | 74149 d | 58 | 68 |
- aThe corresponding proteins were chosen among the closest homologs to IgEst1 identified by a BLAST search. The protein names are given as originally designated by the authors.
- bValues correspond to percentage of amino acids that are identical (or similar) when sequences are aligned with IgEst1.
- cEnzymes that have been purified and biochemically characterized.
- dIdentification numbers were obtained from algae genome browsers rather than from GenBank.
Database searches also revealed sequence homology with lipolytic enzymes from diverse origins. Slight but significant estimated similarity and identity were detected between IgEst1 and sequences chosen for the multiple alignment (Fig. 1, Table 1). The percentage of identical residues with proteins from insects, fungi, mammals, and plants ranged from 24% to 31%. The highest identity scores were found with fungal hypothetical proteins with typical motifs from carboxylesterases from Aspergillus niger and Aspergillus oryzae. Low conservation rates are frequently observed for lipolytic enzymes, which may have evolved from a common ancestral protein by divergent evolutionary mechanisms (Holmquist 2000).
Conserved motifs. Most of the sequences cited above and used for the multiple alignment correspond to uncharacterized proteins predicted by sequence homology. Two of them, human acyl thioesterase and murine lysophospholipase, have been studied in detail (Wang et al. 1997a). This fact is why we used them to identify the IgEst1 residues most likely to belong to conserved motifs. Overall, the multiple alignment appeared imperfect in the N-terminal region, especially for the first 60 residues, whereas the other 200 residues of the IgEst1 sequence matched correctly (Fig. 1). Two highly conserved motifs in the serine hydrolases were detected in IgEst1. Sequence comparisons with the human and murine esterases allowed us to identify the Ser168 as the probable serine residue of the active site of IgEst1. Asp220 and His253 were also identified as the most likely candidates for participating in the catalytic triad of IgEst1 on the basis of sequence conservation. In the murine lysophospholipase, these three residues comprised the catalytic triad and have been shown to be essential for catalysis in site-directed mutagenesis experiments (Wang et al. 1997b). From positions 166 to 170, we also observed the consensus active site pentapeptide, Gly-X-Ser-X-Gly, which is found in serine hydrolases, including the serine catalytic residue, which is highly conserved here as Gly-Phe-Ser-Gln-Gly.
Structurally conserved elements. A putative 3-D structure of IgEst1 was constructed by comparison to the human acyl-protein thioesterase (PDB entry code: 1fj2). Interestingly, this small human enzyme was first described as a lysophospholipase hydrolyzing acyl chain in lysophospholipids (Wang et al. 1999) and more recently has also been shown to display significant acyl-protein thioesterase activity (Wang et al. 2000). Among the reported crystal structure of esterases, the human thioesterase shows evident structural similarity with the carboxylesterase (PDB entry code: 1auo) of the proteobacterium Pseudomonas fluorescens. Although both sequences are only 34% identical, the overall folding of both enzymes is very similar (Devedjiev et al. 2000). Human acyl-protein thioesterase has been shown to be the protein with the structural and functional features that are most similar to IgEst1 and was therefore chosen as the template for modeling. The resulting model remains hypothetical and has to be interpreted carefully because of the low identity between the two sequences (29%). The putative IgEst1 model displays a globular structure with central parallel β sheets surrounded by α helices (Fig. 2). This conformation corresponds to the emblematic structure of the typical α/β-hydrolase-fold superfamily. The structural organization required for catalytic activity also appears to be conserved in IgEst1. Presumed IgEst1 catalytic residues Ser168, Asp220, and His253 perfectly match the Ser119, Asp174, and His208 triad, which have been identified as residues involved in the catalytic mechanism of human thioesterase. As in the human template, despite having very different primary sequences, these three residues are oriented in such a way that they could form the charge-relay network necessary for catalysis. This specific conformation is characteristic of serine esterases and allows exposure and accessibility to the substrate.
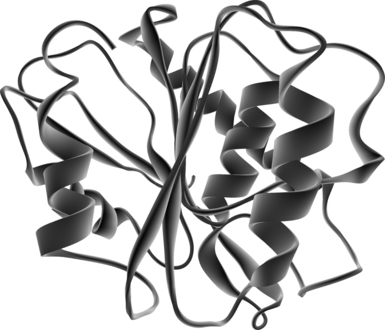
Structure modeling of IgESt1. The three-dimensional model of IgEst1 was constructed with the Geno3D program using the structure of the human acyl-protein thioesterase (code PDB 1fj2) as template.
Phylogenetic analysis. Based on our data and those available in the literature, a phylogenic tree was constructed to complete the comparative analyses and to position the newly identified IgEst1 sequence from I. galbana among other lineages (Fig. S1 in the supplementary material). Many studies have previously reported comparative and phylogenic analyses of lipolytic enzymes from bacterial, fungal, animal, and vegetal species (Fischer and Pleiss 2003). As far as we are aware, the work reported here is first to extend such analyses to algal lipolytic enzymes. The results demonstrate that several clades (plants, animals, fungi, microalgae) have strong bootstrap values consistent with generally accepted evolutionary concepts. The NJ analysis positioned both the IgEst1 sequence and the putative esterase of the coccolithophorid E. huxleyi in a group supported by a strong bootstrap value equal to 100. Both these haptophyte sequences are closely related to those of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. In contrast, the haptophyte sequences were more distantly related to green and red algae. Along with the esterases of the diatoms, they therefore constitute a new group that clusters with the esterases of animals and fungi rather than with those of photosynthetic land plants.
Conclusion. For the first time, a cDNA encoding a putative small esterase (27 kDa) has been isolated from the microalga I. galbana and has therefore been designated IgEst1. Primary sequence comparisons with other esterases revealed that the serine in position 168 occupies a central position in the conserved Gly-Phe-Ser-Gln-Gly sequence, which constitutes the conserved signature Gly-X-Ser-X-Gly of serine hydrolases. The other two structural amino acids in the catalytic triad are Asp in position 220 and His in position 253. Homology modeling of IgEst1 fits the α/β-hydrolase-fold, and places the triad residues in a favorable conformation for catalytic activity. These observations suggest that IgEst1 is likely to be a serine hydrolase belonging to the α/β-hydrolase-fold superfamily. This preliminary study shows that microalgae could therefore constitute a novel study area for characterizing new esterases. This finding is interesting not only from the fundamental point of view, but also because we are justified in believing that microalgae may have unusual lipolytic enzymes with potential new specificities useful for biotechnological applications.
Acknowledgments
We thank Isabelle Martin and Rose-Marie Leroux for their technical assistance. This research was financially supported by “Laval Agglomération” and the “Conseil Général de la Mayenne.”